MGMT-Methylation in Non-Neoplastic Diseases of the Central Nervous System
Abstract
1. Introduction
2. Results
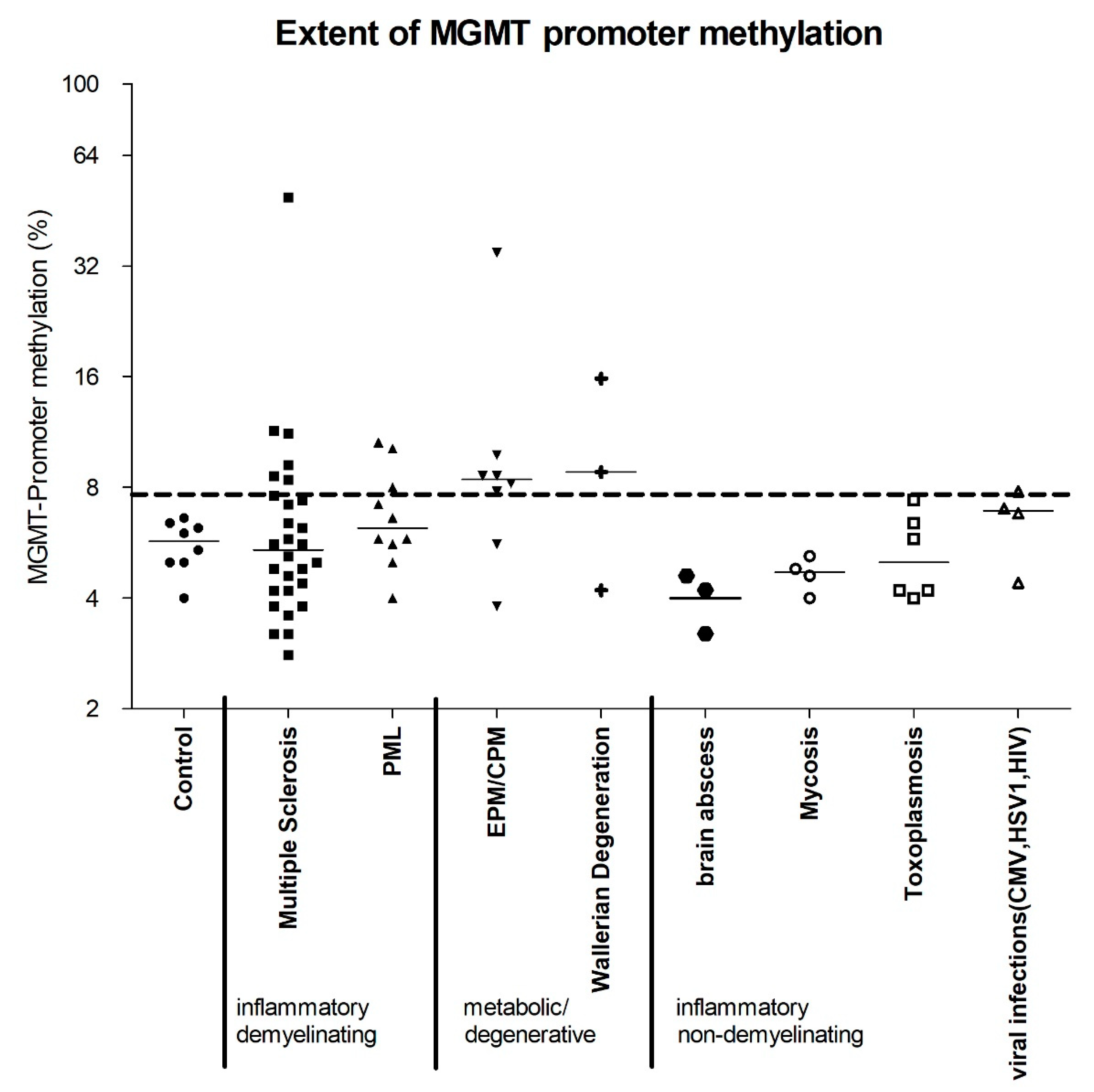
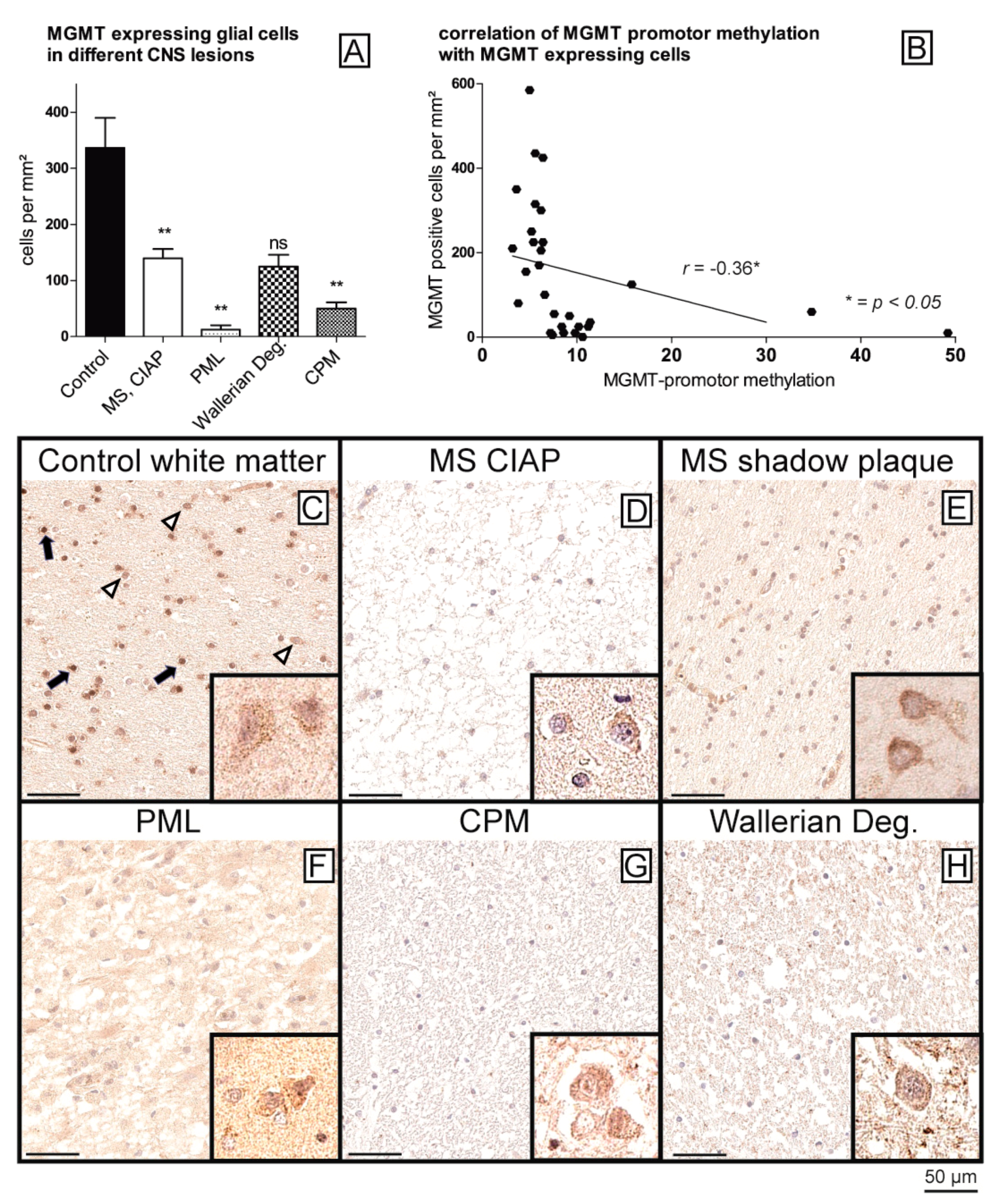
2.1. The MGMT Promoter Is Variably Methylated in Various Non-Neoplastic CNS Diseases
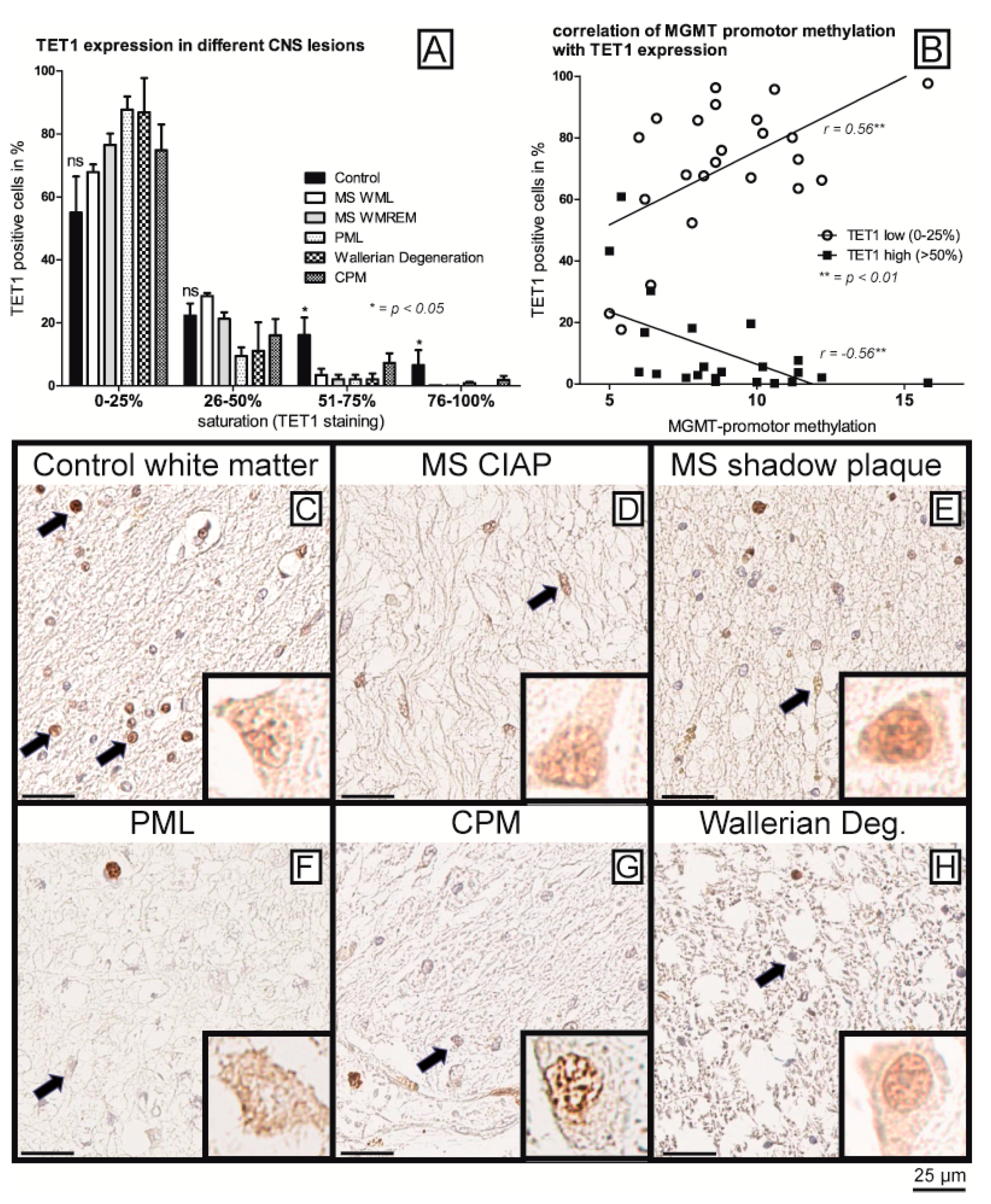
2.2. The Protein Expression of Demethylase TET1 Is Associated with MGMT Promoter Methylation
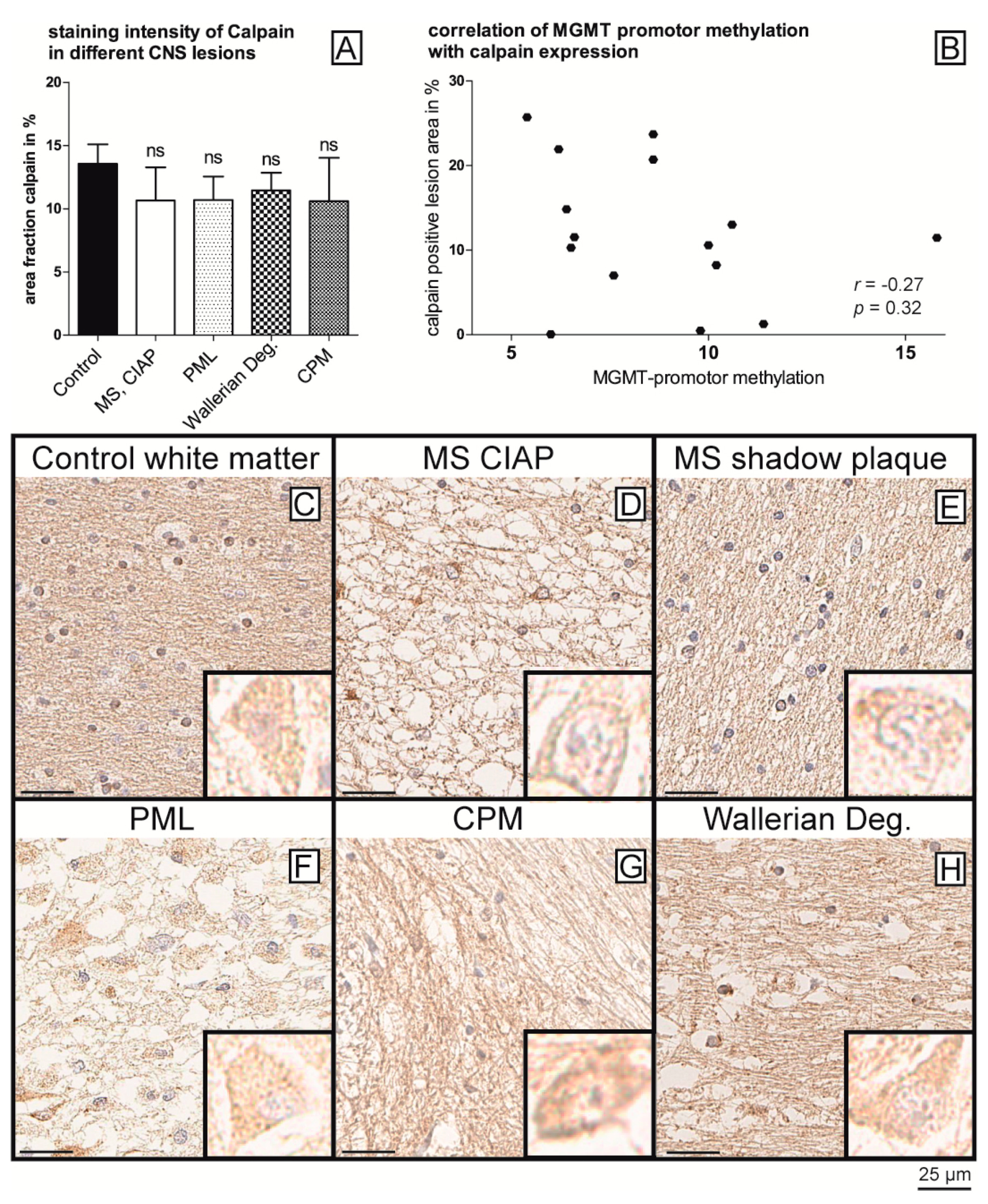
2.3. Calpain-1 Does Not Regulate TET1 Expression in Hypermethylated Non-Neoplastic CNS Diseases
3. Discussion
4. Materials and Methods
4.1. Human Brain Tissue
4.2. Histology and Immunohistochemistry
4.3. cDNA Synthesis and Quantitative PCR
4.4. MGMT Promoter Methylation Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christmann, M.; Verbeek, B.; Roos, W.P.; Kaina, B. O(6)-Methylguanine-DNA methyltransferase (MGMT) in normal tissues and tumors: Enzyme activity, promoter methylation and immunohistochemistry. Biochim. Biophys. Acta 2011, 1816, 179–190. [Google Scholar] [CrossRef]
- Chen, Y.P.; Hou, X.Y.; Yang, C.S.; Jiang, X.X.; Yang, M.; Xu, X.F.; Feng, S.X.; Liu, Y.Q.; Jiang, G. DNA methylation and histone acetylation regulate the expression of MGMT and chemosensitivity to temozolomide in malignant melanoma cell lines. Tumour Biol. 2016, 37, 11209–11218. [Google Scholar] [CrossRef]
- Do, H.; Wong, N.C.; Murone, C.; John, T.; Solomon, B.; Mitchell, P.L.; Dobrovic, A. A critical re-assessment of DNA repair gene promoter methylation in non-small cell lung carcinoma. Sci. Rep. 2014, 4, 4186. [Google Scholar] [CrossRef]
- Gu, C.; Lu, J.; Cui, T.; Lu, C.; Shi, H.; Xu, W.; Yuan, X.; Yang, X.; Huang, Y.; Lu, M. Association between MGMT promoter methylation and non-small cell lung cancer: A meta-analysis. PLoS ONE 2013, 8, e72633. [Google Scholar] [CrossRef]
- Paska, A.V.; Hudler, P. Aberrant methylation patterns in cancer: A clinical view. Biochem. Med. 2015, 25, 161–176. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Mollemann, M.; Wolter, M.; Felsberg, J.; Collins, V.P.; Reifenberger, G. Frequent promoter hypermethylation and low expression of the MGMT gene in oligodendroglial tumors. Int. J. Cancer 2005, 113, 379–385. [Google Scholar] [CrossRef]
- Nutt, C.L.; Noble, M.; Chambers, A.F.; Cairncross, J.G. Differential expression of drug resistance genes and chemosensitivity in glial cell lineages correlate with differential response of oligodendrogliomas and astrocytomas to chemotherapy. Cancer Res. 2000, 60, 4812–4818. [Google Scholar] [PubMed]
- Lu, C.; Ward, P.S.; Kapoor, G.S.; Rohle, D.; Turcan, S.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.H.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.T.; et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Noushmehr, H.; Weisenberger, D.J.; Diefes, K.; Phillips, H.S.; Pujara, K.; Berman, B.P.; Pan, F.; Pelloski, C.E.; Sulman, E.P.; Bhat, K.P.; et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010, 17, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.Y.; Ho, H.L.; Chang-Chien, Y.C.; Chang, Y.W.; Ho, D.M. MGMT promoter methylation in non-neoplastic brain. J. Neurooncol. 2015, 121, 459–467. [Google Scholar] [CrossRef]
- Kang, G.H.; Lee, H.J.; Hwang, K.S.; Lee, S.; Kim, J.H.; Kim, J.S. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am. J. Pathol. 2003, 163, 1551–1556. [Google Scholar] [CrossRef]
- Matsumura, S.; Oue, N.; Ito, R.; Nakayama, H.; Kitadai, Y.; Yokozaki, H.; Chayama, K.; Yasui, W. The promoter methylation status of the DNA repair gene O6-methylguanine-DNA methyltransferase in ulcerative colitis. Virchows Arch. 2003, 443, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Zekri, A.N.; Raafat, A.M.; Elmasry, S.; Bahnassy, A.A.; Saad, Y.; Dabaon, H.A.; El-Kassas, M.; Shousha, H.I.; Nassar, A.A.; El-Dosouky, M.A.; et al. Promotor methylation: Does it affect response to therapy in chronic hepatitis C (G4) or fibrosis? Ann. Hepatol. 2014, 13, 518–524. [Google Scholar]
- Chang, M.S.; Uozaki, H.; Chong, J.M.; Ushiku, T.; Sakuma, K.; Ishikawa, S.; Hino, R.; Barua, R.R.; Iwasaki, Y.; Arai, K.; et al. CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clin. Cancer Res. 2006, 12, 2995–3002. [Google Scholar] [CrossRef] [PubMed]
- Dhiab, M.B.; Ziadi, S.; Mestiri, S.; Gacem, R.B.; Ksiaa, F.; Trimeche, M. DNA methylation patterns in EBV-positive and EBV-negative Hodgkin lymphomas. Cell. Oncol. 2015, 38, 453–462. [Google Scholar] [CrossRef]
- Su, P.F.; Lee, T.C.; Lin, P.J.; Lee, P.H.; Jeng, Y.M.; Chen, C.H.; Liang, J.D.; Chiou, L.L.; Huang, G.T.; Lee, H.S. Differential DNA methylation associated with hepatitis B virus infection in hepatocellular carcinoma. Int. J. Cancer 2007, 121, 1257–1264. [Google Scholar] [CrossRef]
- Fritsche, L.; Teuber-Hanselmann, S.; Soub, D.; Harnisch, K.; Mairinger, F.; Junker, A. MicroRNA profiles of MS gray matter lesions identify modulators of the synaptic protein synaptotagmin-7. Brain Pathol. 2020, 30, 524–540. [Google Scholar] [CrossRef]
- Arthur-Farraj, P.; Moyon, S. DNA methylation in Schwann cells and in oligodendrocytes. Glia 2020, 68, 1568–1583. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y. Regulation of TET protein stability by calpains. Cell Rep. 2014, 6, 278–284. [Google Scholar] [CrossRef]
- Diaz-Sanchez, M.; Williams, K.; DeLuca, G.C.; Esiri, M.M. Protein co-expression with axonal injury in multiple sclerosis plaques. Acta Neuropathol. 2006, 111, 289–299. [Google Scholar] [CrossRef]
- Hoffmann, D.B.; Williams, S.K.; Bojcevski, J.; Muller, A.; Stadelmann, C.; Naidoo, V.; Bahr, B.A.; Diem, R.; Fairless, R. Calcium influx and calpain activation mediate preclinical retinal neurodegeneration in autoimmune optic neuritis. J. Neuropathol. Exp. Neurol. 2013, 72, 745–757. [Google Scholar] [CrossRef]
- Shields, D.C.; Tyor, W.R.; Deibler, G.E.; Banik, N.L. Increased calpain expression in experimental demyelinating optic neuritis: An immunocytochemical study. Brain Res. 1998, 784, 299–304. [Google Scholar] [CrossRef]
- Shields, D.C.; Tyor, W.R.; Deibler, G.E.; Hogan, E.L.; Banik, N.L. Increased calpain expression in activated glial and inflammatory cells in experimental allergic encephalomyelitis. Proc. Natl. Acad. Sci. USA 1998, 95, 5768–5772. [Google Scholar] [CrossRef]
- Mokarram, P.; Kavousipour, S.; Sarabi, M.M.; Mehrabani, G.; Fahmidehkar, M.A.; Shamsdin, S.A.; Alipour, A.; Naini, M.A. MGMT-B gene promoter hypermethylation in patients with inflammatory bowel disease—A novel finding. Asian Pac. J. Cancer Prev. 2015, 16, 1945–1952. [Google Scholar] [CrossRef] [PubMed]
- Locke, W.J.; Guanzon, D.; Ma, C.; Liew, Y.J.; Duesing, K.R.; Fung, K.Y.C.; Ross, J.P. DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front. Genet. 2019, 10, 1150. [Google Scholar] [CrossRef] [PubMed]
- Coppede, F.; Tannorella, P.; Stoccoro, A.; Chico, L.; Siciliano, G.; Bonuccelli, U.; Migliore, L. Methylation analysis of DNA repair genes in Alzheimer’s disease. Mech. Ageing Dev. 2017, 161 Pt A, 105–111. [Google Scholar] [CrossRef]
- Alvarez, M.C.; Santos, J.C.; Maniezzo, N.; Ladeira, M.S.; da Silva, A.L.; Scaletsky, I.C.; Pedrazzoli, J., Jr.; Ribeiro, M.L. MGMT and MLH1 methylation in Helicobacter pylori-infected children and adults. World J. Gastroenterol. 2013, 19, 3043–3051. [Google Scholar] [CrossRef]
- Zekri, A.R.; Bahnasy, A.A.; Shoeab, F.E.; Mohamed, W.S.; El-Dahshan, D.H.; Ali, F.T.; Sabry, G.M.; Dasgupta, N.; Daoud, S.S. Methylation of multiple genes in hepatitis C virus associated hepatocellular carcinoma. J. Adv. Res. 2014, 5, 27–40. [Google Scholar] [CrossRef]
- Guzman, L.; Depix, M.S.; Salinas, A.M.; Roldan, R.; Aguayo, F.; Silva, A.; Vinet, R. Analysis of aberrant methylation on promoter sequences of tumor suppressor genes and total DNA in sputum samples: A promising tool for early detection of COPD and lung cancer in smokers. Diagn. Pathol. 2012, 7, 87. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Gorlia, T.; Erridge, S.C.; Perry, J.; Hong, Y.K.; Aldape, K.D.; Lhermitte, B.; Pietsch, T.; Grujicic, D.; et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2014, 15, 1100–1108. [Google Scholar] [CrossRef]
- Bakulski, K.M.; Dolinoy, D.C.; Sartor, M.A.; Paulson, H.L.; Konen, J.R.; Lieberman, A.P.; Albin, R.L.; Hu, H.; Rozek, L.S. Genome-wide DNA methylation differences between late-onset Alzheimer’s disease and cognitively normal controls in human frontal cortex. J. Alzheimers Dis. 2012, 29, 571–588. [Google Scholar] [CrossRef]
- Liu, Y.; Aryee, M.J.; Padyukov, L.; Fallin, M.D.; Hesselberg, E.; Runarsson, A.; Reinius, L.; Acevedo, N.; Taub, M.; Ronninger, M.; et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 2013, 31, 142–147. [Google Scholar] [CrossRef]
- Zhang, B.G.; Hu, L.; Zang, M.D.; Wang, H.X.; Zhao, W.; Li, J.F.; Su, L.P.; Shao, Z.; Zhao, X.; Zhu, Z.G.; et al. Helicobacter pylori CagA induces tumor suppressor gene hypermethylation by upregulating DNMT1 via AKT-NFkappaB pathway in gastric cancer development. Oncotarget 2016, 7, 9788–9800. [Google Scholar] [CrossRef]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef] [PubMed]
- Mc Guire, C.; Prinz, M.; Beyaert, R.; van Loo, G. Nuclear factor kappa B (NF-kappaB) in multiple sclerosis pathology. Trends Mol. Med. 2013, 19, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Haenold, R.; Weih, F.; Herrmann, K.H.; Schmidt, K.F.; Krempler, K.; Engelmann, C.; Nave, K.A.; Reichenbach, J.R.; Lowel, S.; Witte, O.W.; et al. NF-kappaB controls axonal regeneration and degeneration through cell-specific balance of RelA and p50 in the adult CNS. J. Cell Sci. 2014, 127 Pt 14, 3052–3065. [Google Scholar] [CrossRef]
- White, M.K.; Bellizzi, A.; Ibba, G.; Pietropaolo, V.; Palamara, A.T.; Wollebo, H.S. The DNA damage response promotes polyomavirus JC infection by nucleus to cytoplasm NF- kappaB activation. Virol. J. 2017, 14, 31. [Google Scholar] [CrossRef] [PubMed]
- Chomyk, A.M.; Volsko, C.; Tripathi, A.; Deckard, S.A.; Trapp, B.D.; Fox, R.J.; Dutta, R. DNA methylation in demyelinated multiple sclerosis hippocampus. Sci. Rep. 2017, 7, 8696. [Google Scholar] [CrossRef]
- Huynh, J.L.; Garg, P.; Thin, T.H.; Yoo, S.; Dutta, R.; Trapp, B.D.; Haroutunian, V.; Zhu, J.; Donovan, M.J.; Sharp, A.J.; et al. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat. Neurosci. 2014, 17, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Voigt, D.; Scheidt, U.; Derfuss, T.; Bruck, W.; Junker, A. Expression of the Antioxidative Enzyme Peroxiredoxin 2 in Multiple Sclerosis Lesions in Relation to Inflammation. Int. J. Mol. Sci. 2017, 18, 760. [Google Scholar] [CrossRef]
- Junker, A.; Krumbholz, M.; Eisele, S.; Mohan, H.; Augstein, F.; Bittner, R.; Lassmann, H.; Wekerle, H.; Hohlfeld, R.; Meinl, E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 2009, 132 Pt 12, 3342–3352. [Google Scholar] [CrossRef]
- Bienkowski, M.; Berghoff, A.S.; Marosi, C.; Wohrer, A.; Heinzl, H.; Hainfellner, J.A.; Preusser, M. Clinical Neuropathology practice guide 5-2015: MGMT methylation pyrosequencing in glioblastoma: Unresolved issues and open questions. Clin. Neuropathol. 2015, 34, 250–257. [Google Scholar] [CrossRef] [PubMed]
| Case | MGMT Promoter Methylation (%) at Different Positions | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | Sex | Cause of Death | Disease Duration | Biopsy/Autopsy | Pos. 1 | Pos. 2 | Pos. 3 | Pos. 4 | Pos. 5 | Mean (1–5) | |
| Control | |||||||||||
| control 1 | 75 | f | multiorgan failure | na | autopsy | 2 | 6 | 5 | 7 | 5 | 5 |
| control 2 | 77 | f | sepsis | na | autopsy | 2 | 5 | 8 | 6 | 10 | 6.2 |
| control 3 | 61 | m | heart failure | na | autopsy | 4 | 5 | 6 | 7 | 10 | 6.4 |
| control 4 | 54 | f | respiratory failure | na | autopsy | 2 | 4 | 8 | 6 | 7 | 5.4 |
| control 5 | 57 | f | heart failure | na | autopsy | 3 | 4 | 9 | 7 | 10 | 6.6 |
| control 6 | 56 | m | multiorgan failure | na | autopsy | 2 | 5 | 7 | 4 | 12 | 6 |
| control 7 | 58 | m | heart failure | na | autopsy | 3 | 4 | 5 | 4 | 4 | 4 |
| control 8 | 78 | m | heart failure | na | autopsy | 4 | 5 | 6 | 5 | 5 | 5 |
| Multiple Sclerosis | |||||||||||
| MS autopsy 1 | 75 | f | pneumonia | autopsy | 5 | 5 | 9 | 7 | 16 | 8.4 | |
| MS autopsy 2 | 49 | m | pneumonia | autopsy | 2 | 4 | 4 | 2 | 4 | 3.2 | |
| MS autopsy 3 | 57 | f | respiratory failure | autopsy | 2 | 6 | 11 | 6 | 18 | 8.6 | |
| MS autopsy 4 | 52 | m | multiorgan failure | autopsy | 2 | 56 | 100 | 44 | 44 | 49.2 | |
| MS autopsy 5 | 68 | f | pneumonia | autopsy | 3 | 6 | 9 | 3 | 8 | 5.8 | |
| MS autopsy 6 | 62 | f | cachexia and pulmonary insufficiency | autopsy | 1 | 6 | 9 | 5 | 7 | 5.6 | |
| MS autopsy 7 | 44 | m | multiorgan failure | autopsy | 1 | 6 | 4 | 4 | 3 | 3.6 | |
| MS autopsy 8 | 57 | f | respiratory failure | autopsy | 9 | 15 | 10 | 8 | 14 | 11.2 | |
| MS autopsy 9 | 78 | f | stroke | autopsy | 9 | 10 | 12 | 12 | 14 | 11.4 | |
| MS autopsy 10 | 55 | m | respiratory insufficiency complicating pneumonia and urosepsis | autopsy | 4 | 6 | 7 | 5 | 10 | 6.4 | |
| MS autopsy 11 | 56 | f | respiratory insufficiency in pneumonia | autopsy | 3 | 6 | 4 | 6 | 7 | 5.2 | |
| MS autopsy 12 | 44 | m | aspiration pneumonia | autopsy | 2 | 4 | 9 | 7 | 9 | 6.2 | |
| MS autopsy 13 | 63 | m | pneumonia | autopsy | 6 | 9 | 5 | 6 | 10 | 7.2 | |
| MS autopsy 14 | 53 | m | assisted suicide | autopsy | 4 | 3 | 4 | 3 | 5 | 3.8 | |
| MS autopsy 15 | 54 | f | heart failure | autopsy | 3 | 4 | 5 | 2 | 7 | 4.2 | |
| MS autopsy 16 | 48 | f | respiratory failure | autopsy | 4 | 6 | 9 | 8 | 11 | 7.6 | |
| MS autopsy 17 | 58 | m | terminal renal failure | autopsy | 4 | 7 | 3 | 3 | 11 | 5.6 | |
| MS autopsy 18 | 66 | f | cancer metastases in the liver resulting in severe failure of the liver functions | autopsy | 3 | 4 | 5 | 3 | 8 | 4.6 | |
| MS autopsy 19 | 56 | f | respiratory insufficiency in pneumonia | autopsy | 6 | 7 | 9 | 7 | 8 | 7.4 | |
| MS autopsy 20 | 26 | f | multiorgan failure | autopsy | 6 | 8 | 9 | 8 | 15 | 9.2 | |
| MS biopsy 1 | 43 | m | na | na | biopsy | 3 | 4 | 5 | 3 | 6 | 4.2 |
| MS biopsy 2 | 35 | f | na | 1 month | biopsy | 3 | 5 | 4 | 2 | 5 | 3.8 |
| MS biopsy 3 | 9 | m | na | <1 month | biopsy | 4 | 5 | 4 | 4 | 8 | 5 |
| MS biopsy 4 | 25 | f | na | na | biopsy | 3 | 4 | 6 | 4 | 7 | 4.8 |
| MS biopsy 5 | 31 | m | na | na | biopsy | 2 | 4 | 6 | 4 | 8 | 4.8 |
| MS biopsy 6 | 46 | f | na | <1 month | biopsy | 2 | 3 | 3 | 3 | 3 | 2.8 |
| MS biopsy 7 | 61 | m | na | na | biopsy | 4 | 5 | 5 | 4 | 4 | 4.4 |
| MS biopsy 8 | 35 | f | na | 15 years | biopsy | 3 | 3 | 4 | 3 | 3 | 3.2 |
| PML | |||||||||||
| PML 1 | 34 | m | na | na | biopsy | 4 | 6 | 8 | 4 | 6 | 5.6 |
| PML 2 | 31 | m | na | na | biopsy | 6 | 6 | 7 | 6 | 8 | 6.6 |
| PML 3 | 41 | m | renal failure | na | autopsy | 6 | 8 | 11 | 8 | 7 | 8 |
| PML 4 | 51 | m | na | na | autopsy | 8 | 13 | 7 | 12 | 13 | 10.6 |
| PML 5 | 65 | f | na | na | autopsy | 7 | 8 | 15 | 10 | 11 | 10.2 |
| PML 6 | 58 | f | na | na | autopsy | 3 | 6 | 6 | 3 | 7 | 5 |
| PML 7 | 77 | f | na | na | biopsy | 4 | 6 | 7 | 5 | 7 | 5.8 |
| PML 8 | 66 | f | na | na | biopsy | 3 | 5 | 7 | 5 | 9 | 5.8 |
| PML 9 | 41 | m | renal failure | na | autopsy | 4 | 6 | 8 | 6 | 12 | 7.2 |
| PML 10 | 59 | f | na | na | biopsy | 3 | 4 | 5 | 4 | 4 | 4 |
| CPM/EPM | |||||||||||
| CPM 1 | 54 | m | sepsis | na | autopsy | 6 | 6 | 10 | 6 | 15 | 8.6 |
| CPM 2 | 52 | f | na | na | autopsy | 5 | 8 | 6 | 7 | 17 | 8.6 |
| CPM 3 | 54 | m | sepsis | na | autopsy | 5 | 7 | 10 | 7 | 12 | 8.2 |
| CPM 4 | 52 | f | na | na | autopsy | 23 | 35 | 30 | 44 | 42 | 34.8 |
| CPM 5 | 53 | m | CPM, cerebral hemorrhage | na | autopsy | 3 | 5 | 3 | 4 | 4 | 3.8 |
| CPM 6 | 86 | m | CPM | na | autopsy | 7 | 6 | 7 | 5 | 3 | 5.6 |
| CPM 7 | 41 | m | CPM, cerebral hemorrhage | na | autopsy | 3 | 7 | 10 | 9 | 10 | 7.8 |
| CPM 8 | 55 | m | stroke | na | autopsy | 8 | 10 | 11 | 9 | 11 | 9.8 |
| Wallerian degeneration | |||||||||||
| WAL 1 | 59 | m | multiorgan failure | na | autopsy | 12 | 19 | 13 | 14 | 21 | 15.8 |
| WAL 2 | 50 | m | central regulatory failure | na | autopsy | 5 | 7 | 12 | 7 | 13 | 8.8 |
| WAL 3 | 65 | m | stroke | na | autopsy | 2 | 5 | 5 | 2 | 7 | 4.2 |
| brain abscess | |||||||||||
| ABS 1 | 42 | m | na | na | biopsy | 2 | 4 | 4 | 2 | 4 | 3.2 |
| ABS 2 | 45 | m | na | na | biopsy | 3 | 4 | 5 | 4 | 5 | 4.2 |
| ABS 3 | 3 | f | na | na | biopsy | 3 | 5 | 5 | 4 | 6 | 4.6 |
| mycosis | |||||||||||
| Myc 1 | 55 | m | na | na | autopsy | 2 | 4 | 5 | 4 | 5 | 4 |
| Myc 2 | 48 | f | na | na | biopsy | 3 | 5 | 5 | 4 | 6 | 4.6 |
| Myc 3 | 56 | f | stroke, sepsis | na | autopsy | 4 | 4 | 7 | 4 | 7 | 5.2 |
| Myc 4 | 84 | f | na | na | biopsy | 4 | 5 | 5 | 4 | 6 | 4.8 |
| toxoplasmosis | 6 | 6 | 9 | 7 | 9 | 7.4 | |||||
| Toxo 1 | 59 | f | na | na | biopsy | 3 | 3 | 6 | 4 | 5 | 4.2 |
| Toxo 2 | 47 | m | na | na | biopsy | 6 | 5 | 8 | 7 | 6 | 6.4 |
| Toxo 3 | 64 | m | na | na | biopsy | 3 | 3 | 5 | 4 | 6 | 4.2 |
| Toxo 4 | 29 | f | na | na | biopsy | 3 | 4 | 4 | 4 | 5 | 4 |
| Toxo 5 | 40 | m | na | na | biopsy | 5 | 6 | 8 | 3 | 7 | 5.8 |
| Toxo 6 | 22 | m | na | na | biopsy | 6 | 6 | 9 | 7 | 9 | 7.4 |
| CMV | |||||||||||
| CMV | 38 | m | na | autopsy | 6 | 5 | 12 | 3 | 8 | 6.8 | |
| HSV | |||||||||||
| HSV | 37 | m | HSV-encephalitis | 4 weeks | autopsy | 1 | 6 | 4 | 6 | 5 | 4.4 |
| HIV | |||||||||||
| HIV 1 | 44 | m | respiratory failure | na | autopsy | 4 | 7 | 9 | 5 | 10 | 7 |
| HIV 2 | 51 | m | multiorgan failure | na | autopsy | 5 | 8 | 8 | 7 | 11 | 7.8 |
| Antigen | Company | Pre-Treatment | Dilution |
|---|---|---|---|
| MGMT, ab39253, mouse monoclonal | Abcam | EDTA | 1:50 |
| TET1, HPA019032, rabbit polyclonal | Sigma | citrate | 1:200 |
| TET2, ab94580, rabbit polyclonal | Abcam | citrate | 1:100 |
| Calpain1, ALS16293, goat polyclonal | BioMol | citrate | 1:50 |
| caspase3, 9661, rabbit polyclonal | Cell Signaling | citrate | 1:200 |
| SMI31, 801601, mouse monoclonal | Biolegend | EDTA | 1:1000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teuber-Hanselmann, S.; Worm, K.; Macha, N.; Junker, A. MGMT-Methylation in Non-Neoplastic Diseases of the Central Nervous System. Int. J. Mol. Sci. 2021, 22, 3845. https://doi.org/10.3390/ijms22083845
Teuber-Hanselmann S, Worm K, Macha N, Junker A. MGMT-Methylation in Non-Neoplastic Diseases of the Central Nervous System. International Journal of Molecular Sciences. 2021; 22(8):3845. https://doi.org/10.3390/ijms22083845
Chicago/Turabian StyleTeuber-Hanselmann, Sarah, Karl Worm, Nicole Macha, and Andreas Junker. 2021. "MGMT-Methylation in Non-Neoplastic Diseases of the Central Nervous System" International Journal of Molecular Sciences 22, no. 8: 3845. https://doi.org/10.3390/ijms22083845
APA StyleTeuber-Hanselmann, S., Worm, K., Macha, N., & Junker, A. (2021). MGMT-Methylation in Non-Neoplastic Diseases of the Central Nervous System. International Journal of Molecular Sciences, 22(8), 3845. https://doi.org/10.3390/ijms22083845






