Scanning Probe Microscopies: Imaging and Biomechanics in Reproductive Medicine Research
Abstract
1. Multifunctional and High-Resolution Imaging Techniques for Application in Reproductive Medicine
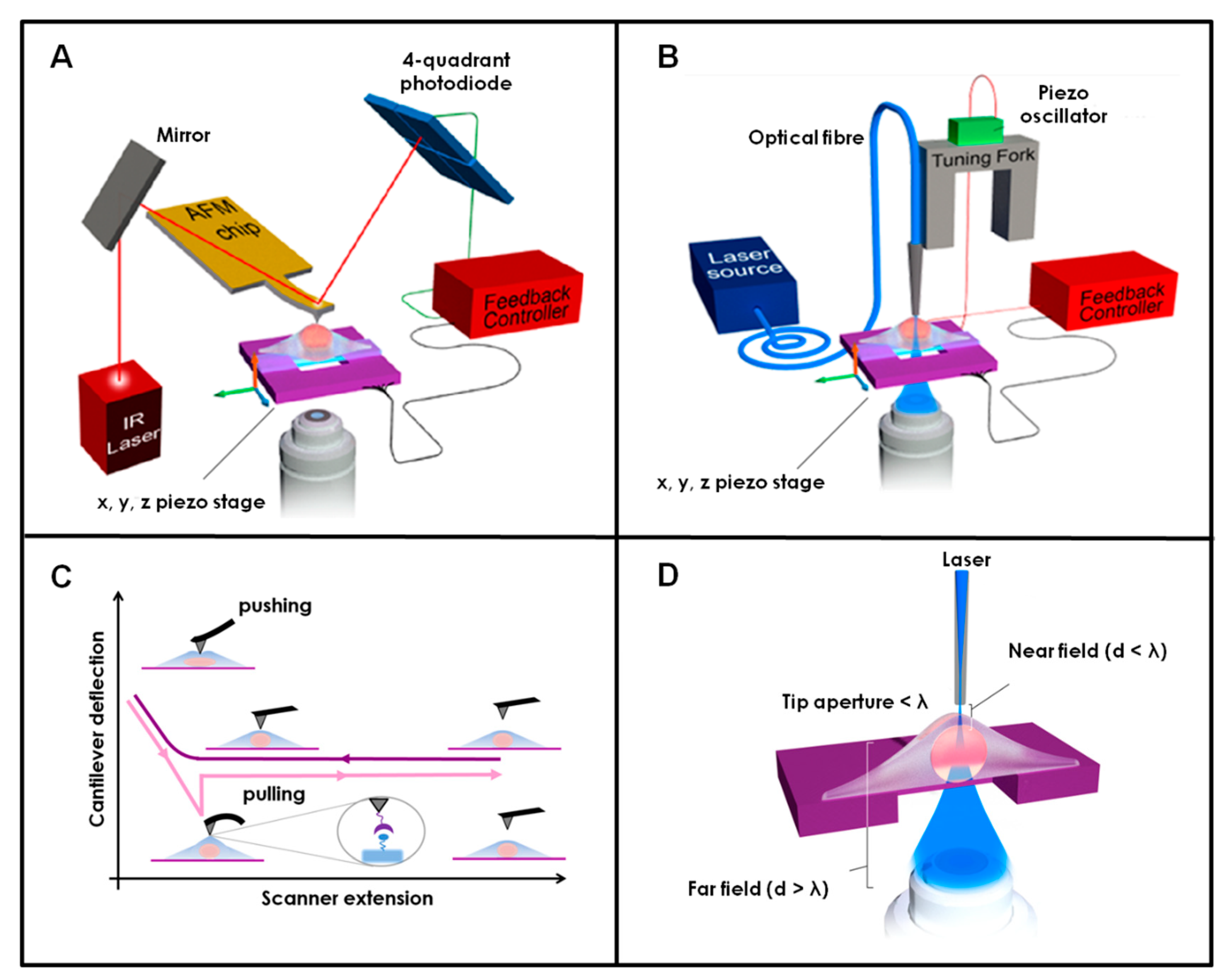
2. AFM and SNOM: The Use of Force and Light
3. Morphological Analysis of Gametes by AFM Imaging
4. The Importance of Mechanics in Gametes and Embryos
4.1. The Mechanical Properties of the Oocyte
4.2. Embryo: Mechanotransduction and Mechanical Properties
5. The Mechanical Properties of the Cells and Tissues in Female Reproductive Disorders
6. New Insights in Reproductive Medicine from the Study of Single-Molecule Interactions
7. From Ultrastructure to Single-Molecule Visualization in the Reproductive System
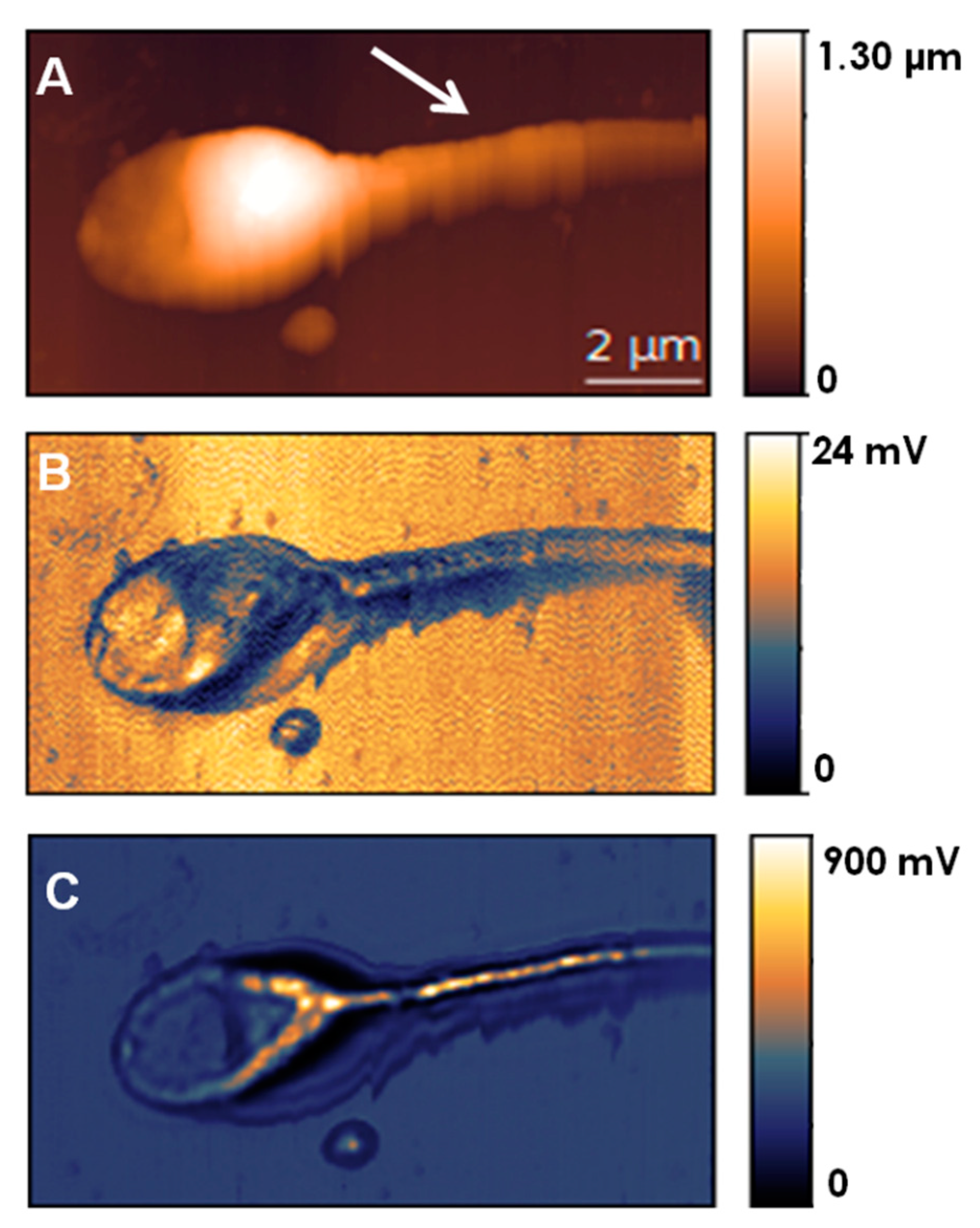
7.1. Inner Cellular Organization of Spermatozoa
7.2. Diseases of the Female Reproductive System
7.3. Diseases of the Male Reproductive System and Fertility
8. The Impact of AFM and SNOM Findings on Reproductive Medicine
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, G.D.; Takayama, S. Application of microfluidic technologies to human assisted reproduction. Mol. Hum. Reprod. 2017, 23, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Pascolo, L.; Bedolla, D.E.; Vaccari, L.; Venturin, I.; Cammisuli, F.; Gianoncelli, A.; Mitri, E.; Giolo, E.; Luppi, S.; Martinelli, M.; et al. Pitfalls and promises in FTIR spectromicroscopy analyses to monitor iron-mediated DNA damage in sperm. Reprod. Toxicol. 2016, 61, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Pascolo, L.; Zupin, L.; Gianoncelli, A.; Giolo, E.; Luppi, S.; Martinelli, M.; De Rocco, D.; Sala, S.; Crovella, S.; Ricci, G. XRF analyses reveal that capacitation procedures produce changes in magnesium and copper levels in human sperm. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 2019, 459, 120–124. [Google Scholar] [CrossRef]
- Zupin, L.; Pascolo, L.; Gianoncelli, A.; Gariani, G.; Luppi, S.; Giolo, E.; Ottaviani, G.; Crovella, S. Ricci Synchrotron radiation soft X-ray microscopy and low energy X-ray fluorescence to reveal elemental changes in spermatozoa treated with photobiomod-ulation therapy. Anal. Methods 2020, 12, 3691–3696. [Google Scholar] [CrossRef]
- Pachetti, M.; Zupin, L.; Venturin, I.; Mitri, E.; Boscolo, R.; D’Amico, F.; Vaccari, L.; Crovella, S.; Ricci, G.; Pascolo, L. FTIR Spectroscopy to Reveal Lipid and Protein Changes Induced on Sperm by Capacitation: Bases for an Improvement of Sample Selection in ART. Int. J. Mol. Sci. 2020, 21, 8659. [Google Scholar] [CrossRef]
- Jasensky, J.; Swain, J.E. Peering Beneath the Surface: Novel Imaging Techniques to Noninvasively Select Gametes and Em-bryos for ART. Biol. Reprod. 2013, 89, 1–12. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ajduk, A.; Szkulmowski, M. Light microscopy of mammalian gametes and embryos: Methods and applications. Int. J. Dev. Biol. 2019, 63, 235–244. [Google Scholar] [CrossRef]
- D’Amico, F.; Zucchiatti, P.; Latella, K.; Pachetti, M.; Gessini, A.; Masciovecchio, C.; Vaccari, L.; Pascolo, L. Investigation of genomic DNA methylation by ultraviolet resonant Raman spectroscopy. J. Biophotonics 2020, 13, 202000150. [Google Scholar] [CrossRef]
- Mallidis, C.; Sanchez, V.; Wistuba, J.; Wuebbeling, F.; Burger, M.; Fallnich, C.; Schlatt, S. Raman microspectroscopy: Shining a new light on reproductive medicine. Hum. Reprod. 2014, 20, 403–414. [Google Scholar] [CrossRef]
- Schermelleh, L.; Ferrand, A.; Huser, T.; Eggeling, C.; Sauer, M.; Biehlmaier, O.; Drummen, G.P.C. Super-resolution micros-copy demystified. Nat. Cell Biol. 2019, 21, 72–84. [Google Scholar] [CrossRef]
- Sherlekar, A.; Mundhe, G.; Richa, P.; Dey, B.; Sharma, S.; Rikhy, R. F-BAR domain protein Syndapin regulates actomyosin dynamics during apical cap remodeling in syncytial Drosophila embryos. J. Cell Sci. 2020, 133, jcs235846. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.T.; Bershadsky, A.; Sheetz, M.P. Mechanobiology. J. R. Soc. Interface 2010, 7, 291–293. [Google Scholar] [CrossRef]
- Moeendarbary, E.; Harris, A.R. Cell mechanics: Principles, practices, and prospects. Wiley Interdiscip. Rev. Syst. Biol. Med. 2014, 6, 371–388. [Google Scholar] [CrossRef]
- Schwarz, U.S. Mechanobiology by the numbers: A close relationship between biology and physics. Nat. Rev. Mol. Cell Biol. 2017, 18, 711–712. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-H.; Aroush, D.R.-B.; Asnacios, A.; Chen, W.-C.; Dokukin, M.E.; Doss, B.L.; Durand-Smet, P.; Ekpenyong, A.; Guck, J.; Guz, N.V.; et al. A comparison of methods to assess cell mechanical properties. Nat. Methods 2018, 15, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Krieg, M.; Fläschner, G.; Alsteens, D.; Gaub, B.M.; Roos, W.H.; Wuite, G.J.L.; Gaub, H.E.; Gerber, C.; Dufrêne, Y.F.; Müller, D.J. Atomic force microscopy-based mechanobiology. Nat. Rev. Phys. 2019, 1, 41–57. [Google Scholar] [CrossRef]
- Raigoza, A.F.; Dugger, J.W.; Webb, L.J. Recent Advances and Current Challenges in Scanning Probe Microscopy of Bio-molecular Surfaces and Interfaces. Appl. Mater. Interfaces 2013, 5, 9249–9261. [Google Scholar] [CrossRef]
- Roca-Cusachs, P.; Conte, V.; Trepat, X. Quantifying forces in cell biology. Nat. Cell Biol. 2017, 19, 742–751. [Google Scholar] [CrossRef]
- Dufrêne, Y.F.; Ando, T.; Garcia, R.; Alsteens, Y.F.D.D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Müller, D.M.-M.D.J. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 2017, 12, 295–307. [Google Scholar] [CrossRef]
- Ando, T. Directly watching biomolecules in action by high-speed atomic force microscopy. Biophys. Rev. 2017, 9, 1–9. [Google Scholar] [CrossRef]
- Alsteens, D.; Gaub, H.E.; Newton, R.; Pfreundschuh, M.; Gerber, C.; Müller, D.J. Atomic force microscopy-based characteri-zation and design of biointerfaces. Nat. Rev. Mat. 2017, 2, 17008. [Google Scholar] [CrossRef]
- Müller, D.J.; Dufrêne, Y.F. Atomic force microscopy: A nanoscopic window on the cell surface. Trends Cell Biol. 2011, 21, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, Y.F.; Pelling, A.E. Force nanoscopy of cell mechanics and cell adhesion. Nanoscale 2013, 5, 4094–4104. [Google Scholar] [CrossRef] [PubMed]
- Rheinlaender, J.; Dimitracopoulos, A.; Wallmeyer, B.; Kronenberg, N.M.; Chalut, K.J.; Gather, M.C.; Betz, T.; Charras, G.; Franze, K. Cortical cell stiffness is independent of substrate mechanics. Nat. Mater. 2020, 19, 1019–1025. [Google Scholar] [CrossRef]
- Bazylewski, P.; Ezugwu, S.; Fanchini, G. A Review of Three-Dimensional Scanning Near-Field Optical Microscopy (3D-SNOM) and Its Applications in Nanoscale Light Management. Appl. Sci. 2017, 7, 973. [Google Scholar] [CrossRef]
- Biagioni, P.; Celebrano, M.; Polli, D.; Labardi, M.; Zavelani-Rossi, M.; Cerullo, G.; Finazzi, M.; Duò, L. Nonlinear optics and spectroscopy at the nanoscale with a hollow-pyramid aperture SNOM. J. Phys. Conf. Ser. 2007, 61, 125–129. [Google Scholar] [CrossRef]
- Richards, D. Near-field microscopy: Throwing light on the nanoworld. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2003, 361, 2843–2857. [Google Scholar] [CrossRef]
- Nagy, P.; Jenei, A.; Kirsch, A.K.; Szöllosi, J.; Damjanovich, S.; Jovin, T.M. Activation-dependent clustering of the erbB2 recep-tor tyrosine kinase detected by scanning near-field optical microscopy. J. Cell Sci. 1999, 112, 1733–1741. [Google Scholar]
- Hinterdorfer, P.; Garcia-Parajo, M.F.; Dufrêne, Y.F. Single-Molecule Imaging of Cell Surfaces Using Near-Field Nanoscopy. Accounts Chem. Res. 2012, 45, 327–336. [Google Scholar] [CrossRef]
- De Lange, F.; Cambi, A.; Huijbens, R.; De Bakker, B.; Rensen, W.; Garcia-Parajo, M.; Van Hulst, N.; Figdor, C.G. Cell biology beyond the diffraction limit: Near-field scanning optical. J. Cell Sci. 2001, 114, 4153. [Google Scholar]
- Bulat, K.; Rygula, A.; Szafraniec, E.; Ozaki, Y.; Baranska, M. Live endothelial cells imaged by Scanning Near-field Optical Microscopy (SNOM): Capabilities and challenges. J. Biophotonics 2016, 10, 928–938. [Google Scholar] [CrossRef]
- Zhang, Y.; Kyle, J.R.; Penchev, M.; Yazdanpanah, V.; Yu, J.; Li, Y.; Yang, M.; Budak, G.; Ozbay, E.; Ozkan, M.; et al. Transmission Near-Field Scanning Optical Microscopy Investigation on Cellular Uptake Behavior of Iron Oxide Nanoparticles. BioNanoScience 2012, 2, 135–143. [Google Scholar] [CrossRef]
- Naumenko, D.; Cassese, D.; Lazzarino, M.; Bek, A.; Benfenati, F. (Eds.) Novel Approaches for Single Molecule Activation and Detection. Advances in Atom and Single Molecules Machine; Springer: Berlin/Heidelberg, Germany, 2014; pp. 61–83. [Google Scholar]
- Mortimer, D. The functional anatomy of the human spermatozoon: Relating ultrastructure and function. Mol. Hum. Reprod. 2018, 24, 567–592. [Google Scholar] [CrossRef]
- Ricci, G.; Andolfi, L.; Zabucchi, G.; Luppi, S.; Boscolo, R.; Martinelli, M.; Zweyer, M.; Trevisan, E. Ultrastructural Morphology of Sperm from Human Globozoospermia. BioMed Res. Int. 2015, 2015, 798754. [Google Scholar] [CrossRef] [PubMed]
- Nottola, S.A.; Macchiarelli, G.; Familiari, G. Fine Structural Markers of Human Oocyte Quality in Assisted Reproduction. Austin J. Reprod. Med. Infertil. 2014, 1, 5. [Google Scholar]
- Kumar, S.; Chaudhury, K.; Sen, P.; Guha, S.K. Atomic force microscopy: A powerful tool for high-resolution imaging of spermatozoa. J. Nanobiotechnol. 2005, 3, 9. [Google Scholar] [CrossRef]
- Mai, A.; Weerachatyanuku, W.; Tomietto, M.; Wayner, D.D.M.; Wells, G.; Balhorn, R.; Leader, A.; Cyr, J.L.; Tanphaichitr, N. Use of Atomic Force Microscopy for Morphological and Morphometric Analyses of Acrosome Intact and Acrosome-Reacted Human Sperm. Mol. Reprod. Dev. 2002, 63, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Sunanda, P.; Panda, B.; Dash, C.; Padhy, R.N.; Routray, P. An illustration of human sperm morphology and their functional ability among different group of subfertile males. Andrology 2018, 6, 680–689. [Google Scholar] [CrossRef]
- Danis, R.B.; Samplaski, M.K. Sperm Morphology: History, Challenges, and Impact on Natural and Assisted Fertility. Curr. Urol. Rep. 2019, 20, 43. [Google Scholar] [CrossRef]
- Tang, S.; Wang, X.; Li, L.; Yang, X.; Li, Z.; Liu, W.; Li, C.; Zhu, Z.; Wang, L.; Wang, J.; et al. Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility with Multiple Morphological Abnormalities of the Sperm Flagella. Am. J. Hum. Genet. 2017, 100, 854–864. [Google Scholar] [CrossRef]
- Kumar, S.; Chaudhury, K.; Sen, P.; Guha, S.K. Quantitative analysis of surface micro-roughness alterations in human sper-matozoa using atomic force microscopy. J. Microsc. 2007, 227, 118–123. [Google Scholar] [CrossRef]
- Jensen, C.F.S.; Østergren, P.; Dupree, J.M.; Ohl, D.A.; Sønksen, J.; Fode, M. Varicocele and male infertility. Nat. Rev. 2017, 14, 523. [Google Scholar] [CrossRef] [PubMed]
- Gabrielsen, A.; Lindenberg, S.; Petersen, K. The impact of zona pellucida thickness variation of human embryos on preg-nancy outcome in relation to suboptimal embryo development. A prospective randomized controlled study. Hum. Reprod. 2001, 16, 2166–2170. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, V.; Macchiarelli, G.; Borini, A.; Lappi, M.; Cecconi, S.; Miglietta, S.; Familiari, G.; Nottola, S.A. Fine morphological assessment of quality of human mature oocytes after slow freezing or vitrification with a closed device: A comparative analysis. Reprod. Biol. Endocrinol. 2014, 12, 110. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sauerbrun-Cutler, M.-T.; Vega, M.; Bręborowicz, A.; Gonzales, E.; Stein, D.; Lederman, M.; Keltz, M. Oocyte zona pellucida dysmorphology is associated with diminished in-vitro fertilization success. J. Ovarian Res. 2015, 8, 5. [Google Scholar] [CrossRef]
- Papi, M.; Brunelli, R.; Sylla, L.; Parasassi, T.; Monaci, M.; Maulucci, G.; Missori, M.; Arcovito, G.; Ursini, F.; De Spirito, M. Mechanical properties of zona pellucida hardening. Eur. Biophys. J. 2009, 39, 987–992. [Google Scholar] [CrossRef]
- Cross, S.E.; Jin, Y.-S.; Rao, J.; Gimzewski, J.K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2007, 2, 780–783. [Google Scholar] [CrossRef]
- Andolfi, L.; Bourkoula, E.; Migliorini, E.; Palma, A.; Pucer, A.; Skrap, M.; Scoles, G.; Beltrami, A.P.; Cesselli, D.; Lazzarino, M. Investigation of Adhesion and Mechanical Properties of Human Glioma Cells by Single Cell Force Spectroscopy and Atomic Force Microscopy. PLoS ONE 2014, 9, e112582. [Google Scholar] [CrossRef] [PubMed]
- Lekka, M. Discrimination Between Normal and Cancerous Cells Using AFM. BioNanoScience 2016, 6, 65–80. [Google Scholar] [CrossRef]
- Rosendahl, P.; Plak, K.; Jacobi, A.; Kraeter, M.; Toepfner, N.; Otto, O.; Herold, C.; Winzi, M.; Herbig, M.; Ge, Y.; et al. Real-time fluorescence and deformability cytometry. Nat. Methods 2018, 15, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Faramarzi, A.; Khalili, M.A.; Omidi, M. Morphometric analysis of human oocytes using time lapse: Does it predict embryo developmental outcomes? Hum. Fertil. 2017, 22, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Barberet, J.; Bruno, C.; Valot, E.; Antunes-Nunes, C.; Jonval, L.; Chammas, J.; Choux, C.; Ginod, P.; Sagot, P.; Soudry-Faure, A.; et al. Can novel early non-invasive biomarkers of embryo quality be identified with time-lapse imaging to predict live birth? Hum. Reprod. 2019, 34, 1439–1449. [Google Scholar] [CrossRef]
- Caamano, J.N.; Munoz, M.; Diez, C.; Gomez, E. Polarized Light Microscopy in Mammalian Oocytes. Reprod. Dom. Anim. 2010, 45, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Zaninovic, N.; Rosenwaks, Z. Artificial intelligence in human in vitro fertilization and embryology. Fertil. Steril. 2020, 114, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Murayama, Y.; Mizuno, J.; Kamakura, H.; Fueta, Y.; Nakamura, H.; Akaishi, K.; Anzai, K.; Watanabe, A.; Inui, H.; Omata, S. Mouse zona pellucida dynamically changes its elasticity during oocyte maturation, fertilization and early embryo development. Hum. Cell 2006, 19, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Boccaccio, A.; Frassanito, M.C.; Lamberti, L.; Brunelli, R.; Maulucci, G.; Monaci, M.; Papi, M.; Pappalettere, C.; Parasassi, T.; Sylla, L.; et al. Nanoscale characterization of the biomechanical hardening of Bovine zona pellucida. J. R. Soc. Interface 2012, 9, 2871–2882. [Google Scholar] [CrossRef]
- Miao, Y.-L.; Kikuchi, K.; Su, Q.-Y.; Schatten, H. Oocyte aging: Cellular and molecular changes, developmental potential and reversal possibility. Hum. Reprod. 2009, 15, 573–585. [Google Scholar] [CrossRef]
- Lord, T.; Aitken, R.J. Oxidative stress and ageing of the post-ovulatory oocyte in Reproduction. Reproduction 2013, 146, R217–R227. [Google Scholar] [CrossRef]
- Yanez, L.Z.; Camarillo, D.B. Microfluidic analysis of oocyte and embryo biomechanical properties to improve outcomes in assisted reproductive technologies. Mol. Hum. Reprod. 2017, 23, 235–247. [Google Scholar] [CrossRef]
- Sun, Y.; Wan, K.-T.; Roberts, K.P.; Bischof, J.C.; Nelson, B.J. Mechanical property characterization of mouse zona pellucida. IEEE Trans. NanoBiosci. 2003, 2, 279–286. [Google Scholar] [CrossRef]
- Liu, X.; Shi, J.; Zong, Z.; Wan, K.-T.; Sun, Y. Elastic and Viscoelastic Characterization of Mouse Oocytes Using Micropipette Indentation. Ann. Biomed. Eng. 2012, 40, 2122–2130. [Google Scholar] [CrossRef] [PubMed]
- Khalilian, M.; Navidbakhsh, M.; Valojerdi, M.R.; Chizari, M.; Yazdi, P.E. Estimating Young’s modulus of zona pellucida by micropipette aspiration in combination with theoretical models of ovum. J. R. Soc. Interface 2009, 7, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Yanez, L.Z.; Han, J.; Behr, B.B.; Reijo Pera, R.A.; Camarillo, D.B. Human oocyte developmental potential is predicted by mechanical properties within hours after fertilization. Nat. Commun. 2016, 7, 10809. [Google Scholar] [CrossRef]
- Krause, M.; Te Riet, J.; Wolf, K. Probing the compressibility of tumor cell nuclei by combined atomic force–confocal microscopy. Phys. Biol. 2013, 10, 065002. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Benet, E.; Sridhar, S.L.; Abadie, J.; Piat, E.; Vernerey, F.J. Separating the contributions of zona pellucida and cyto-plasm in the viscoelastic response of human oocytes. Acta Biomater. 2019, 85, 253–262. [Google Scholar] [CrossRef]
- Andolfi, L.; Masiero, E.; Giolo, E.; Martinelli, M.; Luppi, S.; Zilio, S.D.; Delfino, I.; Bortul, R.; Zweyer, M.; Ricci, G.; et al. Investigating the mechanical properties of zona pellucida of whole human oocytes by atomic force spectroscopy. Integr. Biol. 2016, 8, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Giolo, E.; Martinelli, M.; Luppi, S.; Romano, F.; Ricci, G.; Lazzarino, M.; Andolfi, L. Study of the mechanical properties of fresh and cryopreserved individual human oocytes. Eur. Biophys. J. 2019, 48, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Andolfi, L.; Greco, S.L.M.; Tierno, D.; Chignola, R.; Martinelli, M.; Giolo, E.; Luppi, S.; Delfino, I.; Zanetti, M.; Battistella, A.; et al. A novel AFM macro-probe to study the biomechanical properties of large cells and 3D cell spheroids. Acta Biomater. 2019, 94, 505–513. [Google Scholar] [CrossRef]
- Chen, X.; Bonfiglio, R.; Banerji, S.; Jackson, D.G.; Salustri, A.; Richter, R.P. Micromechanical Analysis of the Hyaluronan-Rich Matrix Surrounding the Oocyte Reveals a Uniquely Soft and Elastic Composition. Biophys. J. 2016, 110, 2779–2789. [Google Scholar] [CrossRef]
- Mammoto, T.; Ingber, D.E. Mechanical control of tissue and organ development. Development 2010, 137, 1407–1420. [Google Scholar] [CrossRef]
- Piccolo, S. Mechanics in the embryo. Nat. Cell Biol. 2013, 504, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Maître, J.-L. Mechanics of blastocyst morphogenesis. Biol. Cell 2017, 109, 323–338. [Google Scholar] [CrossRef]
- Chan, C.J.; Costanzo, M.; Ruiz-Herrero, T.; Mönke, G.; Petrie, R.J.; Bergert, M.; Diz-Muñoz, A.; Mahadevan, L.; Hiiragi, T. Hydraulic control of mammalian embryo size and cell fate. Nat. Cell Biol. 2019, 571, 112–116. [Google Scholar] [CrossRef]
- D’Angelo, M.; Benedetti, E.; Tupone, M.G.; Catanesi, M.; Castelli, V.; Antonosante, A.; Cimini, A. The Role of Stiness in Cell Reprogramming: A Potential Role for Biomaterials in Inducing Tissue Regeneration. Cells 2019, 8, 1036. [Google Scholar] [CrossRef]
- Heisenberg, C.-P.; Bellaïche, Y. Forces in Tissue Morphogenesis and Patterning. Cell 2013, 153, 948–962. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Z.; Tao, H.; Liu, J.; Hopyan, S.; Sun, Y. Characterizing Inner Pressure and Stiffness of Trophoblast and Inner Cell Mass of Blastocysts. Biophys. J. 2018, 115, 2443–2450. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Pillai, E.K.; Dimov, I.B.; Foster, S.K.; Holt, C.E.; Franze, K. Rapid changes in tissue mechanics regulate cell behaviour in the developing embryonic brain. eLife 2019, 8, e39356. [Google Scholar] [CrossRef]
- Kort, J.; Behr, B. Biomechanics and developmental potential of oocytes and embryos. Fertil. Steril. 2017, 108, 738–741. [Google Scholar] [CrossRef] [PubMed]
- Baah-Dwomoh, A.; McGuire, J.; Tan, T.; De Vita, R. Mechanical Properties of Female Reproductive Organs and Supporting Connective Tissues: A Review of the Current State of Knowledge. Appl. Mech. Rev. 2016, 68, 060801. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Lee, J.R. Fertility preservation option in young women with ovarian cancer. Future Oncol. 2016, 12, 1695–1698. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, H.; Hao, J.; Bai, X.; Li, H.; Zhang, Z.; Wang, H.; Tang, J. Single molecular recognition force spectroscopy study of a DNA aptamer with the target epithelial cell adhesion molecule. Analyst 2015, 140, 6226–6229. [Google Scholar] [CrossRef]
- Xu, W.; Mezencev, R.; Kim, B.; Wang, L.; McDonald, J.; Sulchek, T. Cell Stiffness Is a Biomarker of the Metastatic Potential of Ovarian Cancer Cells. PLoS ONE 2012, 7, e46609. [Google Scholar] [CrossRef]
- McKenzie, A.J.; Hicks, S.R.; Svec, K.V.; Naughton, H.; Edmunds, Z.L.; Howe, A.K. The mechanical microenvironment regulates ovarian cancer cell morphology, migration, and spheroid disaggregation. Sci. Rep. 2018, 8, 7228. [Google Scholar] [CrossRef]
- Ansardamavandi, A.; Tafazzoli- Shadpour, M.; Omidvar, R.; Nili, F. An AFM-Based Nanomechanical Study of Ovarian Tis-sues with Pathological Conditions. Int. J. Nanomed. 2020, 15, 4333–4350. [Google Scholar] [CrossRef]
- Stylianou, A.; Lekka, M.; Stylianopoulos, T. AFM assessing of nanomechanical fingerprints for cancer early diagnosis and classification: From single cell to tissue level. Nanoscale 2018, 10, 20930–20945. [Google Scholar] [CrossRef]
- Wang, C.; Hu, R.; Morrissey, J.J.; Kharasch, E.D.; Singamaneni, S. Single Molecule Force Spectroscopy to Compare Natural versus Artificial Antibody-Antigen Interaction. Small 2017, 13, 1604255. [Google Scholar] [CrossRef] [PubMed]
- Koehler, M.; Fis, A.; Gruber, H.J.; Hinterdorfer, P. AFM-Based Force Spectroscopy Guided by Recognition Imaging: A New Mode for Mapping and Studying Interaction Sites at Low Lateral Density. Methods Protoc. 2019, 2, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, Z.; Liu, H.; Nash, M.A. Next Generation Methods for Single-Molecule Force Spectroscopy on Polyproteins and Receptor-Ligand Complexes. Front. Mol. Biosci. 2020, 7, 85. [Google Scholar] [CrossRef] [PubMed]
- Avella, M.A.; Baibakov, B.; Dean, J. A single domain of the ZP2 zona pellucida protein mediates gamete recognition in mice and humans. J. Cell Biol. 2014, 205, 801–809. [Google Scholar] [CrossRef]
- Tokuhiro, K.; Dean, J. Glycan-Independent Gamete Recognition Triggers Egg Zinc Sparks and ZP2 Cleavage to Prevent Polyspermy. Dev. Cell 2018, 46, 627–640.e5. [Google Scholar] [CrossRef]
- Baibakov, B.; Gauthier, L.; Talbot, P.; Rankin, T.L.; Dean, J. Sperm binding to the zona pellucida is not sufficient to induce acrosome exocytosis. Development 2007, 134, 933–943. [Google Scholar] [CrossRef]
- Chalbi, M.; Barraud-Lange, V.; Ravaux, B.; Howan, K.; Rodriguez, N.; Soule, P.; Ndzoudi, A.; Boucheix, C.; Rubinstein, E.; Wolf, J.P.; et al. Binding of sperm protein Izumo1 and its egg receptor Juno drives Cd9 accumulation in the intercellular contact area prior to fusion during mammalian fertilization. Development 2014, 141, 3732–3739. [Google Scholar] [CrossRef]
- Andolfi, L.; Trevisan, E.; Troian, B.; Prato, S.; Boscolo, R.; Giolo, E.; Luppi, S.; Martinelli, M.; Ricci, G.; Zweyer, M. The application of scanning near field optical imaging to the study of human sperm morphology. J. Nanobiotech. 2015, 13, 2. [Google Scholar] [CrossRef]
- Troian, B.; Boscolo, R.; Ricci, G.; Lazzarino, M.; Zito, G.; Prato, S.; Andolfi, L. Ultra-structural analysis of human spermatozoa by aperture scanning near-field optical microscopy. J. Biophotonics 2020, 13, e2418. [Google Scholar] [CrossRef]
- Fishman, E.L.; Jo, K.; Nguyen, Q.P.H.; Kong, D.; Royfman, R.; Cekic, A.R.; Khanal, S.; Miller, A.L.; Simerly, C.; Schatten, G.; et al. A novel atypical sperm centriole is functional during human fertilization. Nat. Commun. 2018, 9, 2210. [Google Scholar] [CrossRef]
- Yong, Y.-C.; Wang, Y.-Z.; Zhong, J.-J. Nano-spectroscopic imaging of proteins with near-field scanning optical microscopy (NSOM). Curr. Opin. Biotechnol. 2018, 54, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, D.E.; Morais, C.L.M.; Lima, K.M.G.; Trevisan, J.; Siggel-King, M.R.F.; Craig, T.; Ingham, J.; Martin, D.S.; Heys, K.A.; Kyrgiou, M.; et al. Imaging cervical cytology with scanning near-field optical microscopy (SNOM) coupled with an IR-FEL. Sci. Rep. 2016, 6, 29494. [Google Scholar] [CrossRef] [PubMed]
- Craig, T.; Smith, A.D.; Holder, G.M.; Ingham, J.; Smith, C.I.; Varro, A.; Pritchard, D.M.; Barrett, S.D.; Martin, D.S.; Harrison, P.; et al. SNOM Imaging of a Crypt-Like Feature in Adenocarcinoma Associated with Barrett’s Oesophagus. Phys. Status Solidi B 2018, 255, 1700518. [Google Scholar] [CrossRef]
- Walker, K.-A.D.; Morgan, C.; Doak, S.H.; Dunstan, P.R. Quantum Dots for Multiplexed Detection and Characterisation of Prostate Cancer Cells Using a Scanning Near-Field Optical Microscope. PLoS ONE 2012, 7, e31592. [Google Scholar] [CrossRef]
- Hausmann, M.; Liebe, B.; Perner, B.; Jerratsch, M.; Greulich, K.-O.; Scherthan, H. Imaging of human meiotic chromosomes by scanning near-field optical microscopy (SNOM). Micron 2003, 34, 441–447. [Google Scholar] [CrossRef]
- Rocca, M.S.; Foresta, C.; Ferlin, A. Telomere length: Lights and shadows on their role in human reproduction. Biol. Reprod. 2018, 100, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Umakoshi, T.; Fukuda, S.; Iino, R.; Uchihashi, T.; Ando, T. High-speed near-field fluorescence microscopy combined with high-speed atomic force microscopy for biological studies. Biochim. Biophys. Acta (BBA) Gen. Subj. 2020, 1864, 129325. [Google Scholar] [CrossRef] [PubMed]
- Coronado, S.P.; Severac, C.; Martinez-Rivas, A.; Dague, E. Beyond the paradigm of nanomechanical measurements on cells using AFM: An automated methodology to rapidly analyse thousands of cells. Nanoscale Horiz. 2019, 5, 131–138. [Google Scholar] [CrossRef]


| Method | Force Loading/Pressure Applied | Deformation/Aspiration Length/ Indentation | Young Modulus | Model |
|---|---|---|---|---|
| Micropipette indentation | ||||
| 7.5 µN | 44 µm | 17.9 kPa | Biomembrane Point Load model [61] | |
| 0.5 μN | 35 µm | 3.1 kPa | Maugis-Dugdale model [62] | |
| Micropipette aspiration | ||||
| 0.8 kPa | 40 µm | 2.41 kPa | Cortical shell-liquid core model [63] | |
| 2.20 kPa | 16 µm | 0.056 N/m | Zener model [64] | |
| Microtactile sensor (MTS) | ||||
| 10 µm | 8.26 kPa | Hertz model [56] | ||
| AFM-indentation | ||||
| 0.001–0.002 μN | 1–2 µm | 0.05–0.1 kPa | Double Hertz model [67,68] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andolfi, L.; Battistella, A.; Zanetti, M.; Lazzarino, M.; Pascolo, L.; Romano, F.; Ricci, G. Scanning Probe Microscopies: Imaging and Biomechanics in Reproductive Medicine Research. Int. J. Mol. Sci. 2021, 22, 3823. https://doi.org/10.3390/ijms22083823
Andolfi L, Battistella A, Zanetti M, Lazzarino M, Pascolo L, Romano F, Ricci G. Scanning Probe Microscopies: Imaging and Biomechanics in Reproductive Medicine Research. International Journal of Molecular Sciences. 2021; 22(8):3823. https://doi.org/10.3390/ijms22083823
Chicago/Turabian StyleAndolfi, Laura, Alice Battistella, Michele Zanetti, Marco Lazzarino, Lorella Pascolo, Federico Romano, and Giuseppe Ricci. 2021. "Scanning Probe Microscopies: Imaging and Biomechanics in Reproductive Medicine Research" International Journal of Molecular Sciences 22, no. 8: 3823. https://doi.org/10.3390/ijms22083823
APA StyleAndolfi, L., Battistella, A., Zanetti, M., Lazzarino, M., Pascolo, L., Romano, F., & Ricci, G. (2021). Scanning Probe Microscopies: Imaging and Biomechanics in Reproductive Medicine Research. International Journal of Molecular Sciences, 22(8), 3823. https://doi.org/10.3390/ijms22083823








