Targeting Protein Kinases in Blood Cancer: Focusing on CK1α and CK2
Abstract
1. Introduction
2. General Features of CK1α and CK2: Structure, Regulation, and Function
2.1. CK1α
2.2. CK2
3. Protein Kinases CK1α and CK2 in Hematological Malignancies
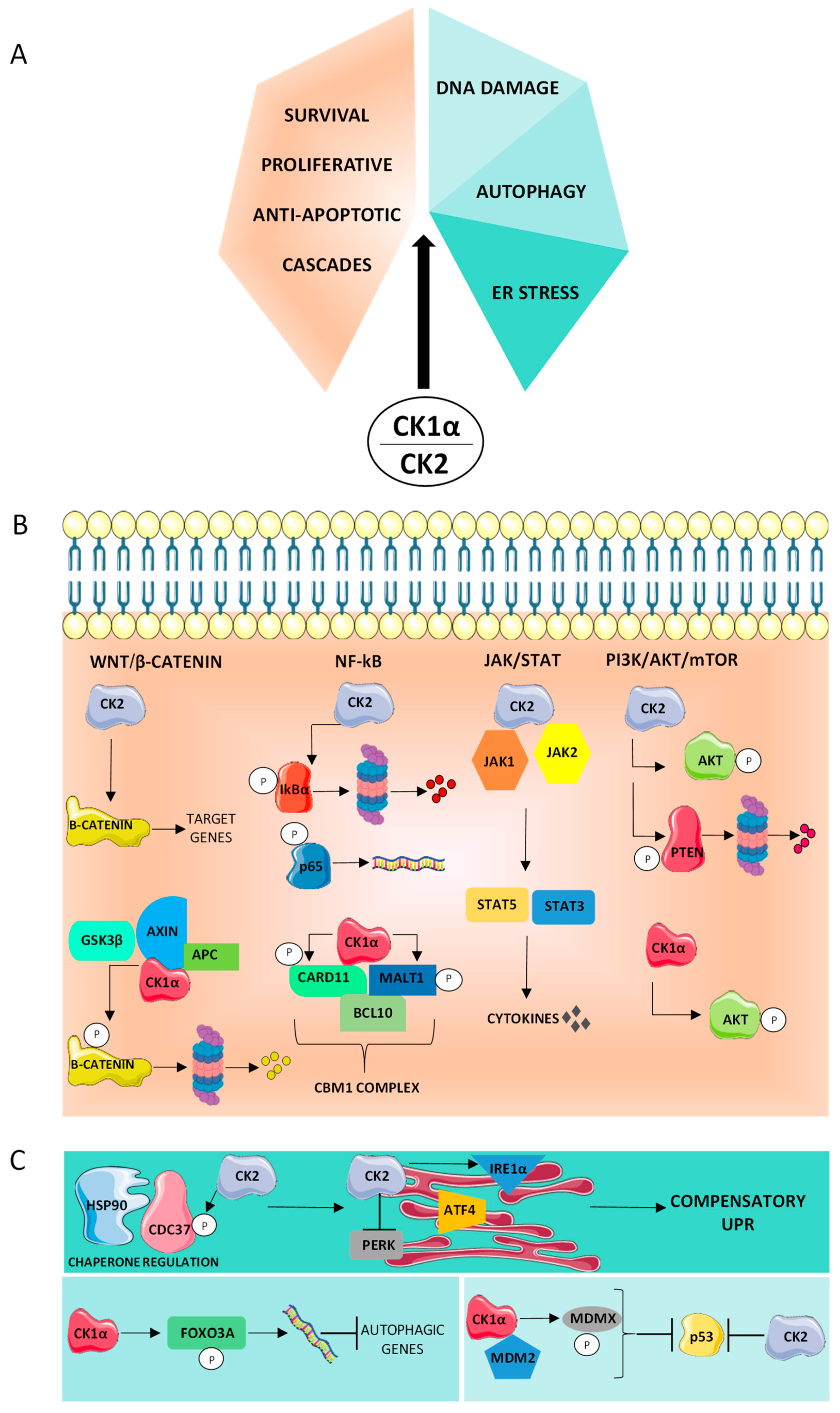
The Importance of Being “Non-Oncogene”
4. CK1α and CK2 Sustain Proliferation and Survival, and Counteract Apoptosis
4.1. PI3K/AKT
4.2. NF-κB
4.3. Wnt/β-Catenin Pathway
4.4. JAK/STAT Signaling
5. Supportive Role of CK1α and CK2 in the Cancer Stress Phenotype
5.1. Proteotoxic Stress
5.2. DNA Damage Stress
5.3. Autophagy
6. Therapeutic Strategies
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALL | Acute Lymphoblastic Leukemia |
| AML | Acute myeloid leukemia |
| ATF4 | transcription factor 4 |
| ATF6 | Activating Transcription Factor-6 |
| BCR | B-cell receptor |
| CHOP | C/EBP-homologous protein |
| CK1 | Casein kinase 1 |
| CK2 | Casein kinase 2 |
| CLL | Chronic Lymphocytic Leukemia |
| CMD | Chronic Myeloproliferative disorders |
| CML | Chronic Myeloid Leukemia |
| DEPTOR | DEP domain-containing mTOR-interacting protein |
| DLBCL | Diffuse large B-cell Lymphoma |
| Dvl | Dishevelled |
| eIF2α | eukaryotic translation initiation factor 2α |
| ER | Endoplasmic reticulum |
| FL | Follicular Lymphoma |
| FOXO3A | Forkhead Box O3 |
| Grp78/Bip | glucose regulated protein 78/Binding immunoglobulin protein |
| GSK3-β | Glycogen synthase kinase 3-β |
| Hsp90 | Heat Shock Protein 90 |
| IL-6 | Interleukin-6 |
| IRE1α | Inositol Requiring Enzyme 1 |
| JAK | Janus associated kinase-signal |
| LSCs | Leukemic-stem cells |
| MCL | Mantle Cell Lymphoma |
| MDM2 | Mouse Double Minute 2 |
| MDMX | Murine Double minute X |
| MDS | Myelodysplastic syndrome |
| MM | Multiple Myeloma |
| mTOR | Mammalian target of rapamycine |
| NF-kB | Nuclear Factor kappa-light-chain-enhancer of activated B cells |
| NHL | Non-Hodgkin Lymphoma |
| PDGFR | Platelet-derived growth factor receptors |
| PERK | PKR-like ER kinase |
| PI3K | Phosphoinositide 3-kinase |
| PIP3 | Phosphatidylinositol (3,4,5)-trisphosphate |
| PTEN | Phosphate and tensin homolog |
| PTM | Post translational modification |
| STAT3 | Signal transducer and activator of transcription 3 |
| UPR | Unfolded Protein response |
References
- Manning, G. The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Burnett, G.; Kennedy, E.P. The Enzymatic Phosphorylation of Proteins. J. Biol. Chem. 1954, 211, 969–980. [Google Scholar] [CrossRef]
- Venerando, A.; Ruzzene, M.; Pinna, L.A. Casein Kinase: The Triple Meaning of a Misnomer. Biochem. J. 2014, 460, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Knippschild, U.; Krüger, M.; Richter, J.; Xu, P.; García-Reyes, B.; Peifer, C.; Halekotte, J.; Bakulev, V.; Bischof, J. The CK1 Family: Contribution to Cellular Stress Response and Its Role in Carcinogenesis. Front. Oncol. 2014, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, M.; Sun, J.; Yang, X. Casein Kinase 1α: Biological Mechanisms and Theranostic Potential. Cell Commun. Signal. 2018, 16, 23. [Google Scholar] [CrossRef] [PubMed]
- Chua, M.M.J.; Ortega, C.E.; Sheikh, A.; Lee, M.; Abdul-Rassoul, H.; Hartshorn, K.L.; Dominguez, I. CK2 in Cancer: Cellular and Biochemical Mechanisms and Potential Therapeutic Target. Pharmaceuticals 2017, 10, 18. [Google Scholar] [CrossRef]
- Piazza, F.; Manni, S.; Colpo, A.; Quotti Tubi, L.; Gurrieri, C.; Semenzato, G. Serine-Threonine Protein Kinases CK1, CK2 and GSK3 in Normal and Malignant Haematopoiesis. Curr. Signal Transduct. Ther. 2011, 6, 88–98. [Google Scholar] [CrossRef]
- Martins, L.R.; Lúcio, P.; Silva, M.C.; Anderes, K.L.; Gameiro, P.; Silva, M.G.; Barata, J.T. Targeting CK2 Overexpression and Hyperactivation as a Novel Therapeutic Tool in Chronic Lymphocytic Leukemia. Blood 2010, 116, 2724–2731. [Google Scholar] [CrossRef]
- Piazza, F.; Manni, S.; Ruzzene, M.; Pinna, L.A.; Gurrieri, C.; Semenzato, G. Protein Kinase CK2 in Hematologic Malignancies: Reliance on a Pivotal Cell Survival Regulator by Oncogenic Signaling Pathways. Leukemia 2012, 26, 1174–1179. [Google Scholar] [CrossRef]
- Martins, L.R.; Perera, Y.; Lúcio, P.; Silva, M.G.; Perea, S.E.; Barata, J.T. Targeting Chronic Lymphocytic Leukemia Using CIGB-300, a Clinical-Stage CK2-Specific Cell-Permeable Peptide Inhibitor. Oncotarget 2014, 5, 258–263. [Google Scholar] [CrossRef]
- Manni, S.; Toscani, D.; Mandato, E.; Brancalion, A.; Quotti Tubi, L.; Macaccaro, P.; Cabrelle, A.; Adami, F.; Zambello, R.; Gurrieri, C.; et al. Bone Marrow Stromal Cell-Fueled Multiple Myeloma Growth and Osteoclastogenesis Are Sustained by Protein Kinase CK2. Leukemia 2014, 28, 2094–2097. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Song, W.; Cirstea, D.; Lu, D.; Munshi, N.C.; Anderson, K.C. CSNK1α1 Mediates Malignant Plasma Cell Survival. Leukemia 2015, 29, 474–482. [Google Scholar] [CrossRef][Green Version]
- Manni, S.; Carrino, M.; Piazza, F. Role of Protein Kinases CK1α and CK2 in Multiple Myeloma: Regulation of Pivotal Survival and Stress-Managing Pathways. J. Hematol. Oncol. 2017, 10, 157. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Song, C.; Ding, Y.; Tan, B.-H.; Desai, D.; Sharma, A.; Gowda, R.; Yue, F.; Huang, S.; Spiegelman, V.; et al. Dual Targeting of MTOR as a Novel Therapeutic Approach for High-Risk B-Cell Acute Lymphoblastic Leukemia. Leukemia 2021. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Solimini, N.L.; Elledge, S.J. Principles of Cancer Therapy: Oncogene and Non-Oncogene Addiction. Cell 2009, 136, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Ruzzene, M.; Pinna, L.A. Addiction to Protein Kinase CK2: A Common Denominator of Diverse Cancer Cells? Biochim. Biophys. Acta BBA Proteins Proteom. 2010, 1804, 499–504. [Google Scholar] [CrossRef]
- Mandato, E.; Manni, S.; Zaffino, F.; Semenzato, G.; Piazza, F. Targeting CK2-Driven Non-Oncogene Addiction in B-Cell Tumors. Oncogene 2016, 35, 6045–6052. [Google Scholar] [CrossRef]
- Fulcher, L.J.; Sapkota, G.P. Functions and Regulation of the Serine/Threonine Protein Kinase CK1 Family: Moving beyond Promiscuity. Biochem. J. 2020, 477, 4603–4621. [Google Scholar] [CrossRef]
- Bustos, V.H.; Marin, O.; Meggio, F.; Cesaro, L.; Allende, C.C.; Allende, J.E.; Pinna, L.A. Generation of Protein Kinase Ck1α Mutants Which Discriminate between Canonical and Non-Canonical Substrates. Biochem. J. 2005, 391, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Knippschild, U.; Gocht, A.; Wolff, S.; Huber, N.; Löhler, J.; Stöter, M. The Casein Kinase 1 Family: Participation in Multiple Cellular Processes in Eukaryotes. Cell. Signal. 2005, 17, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Järås, M.; Miller, P.G.; Chu, L.P.; Puram, R.V.; Fink, E.C.; Schneider, R.K.; Al-Shahrour, F.; Peña, P.; Breyfogle, L.J.; Hartwell, K.A.; et al. Csnk1a1 Inhibition Has P53-Dependent Therapeutic Efficacy in Acute Myeloid Leukemia. J. Exp. Med. 2014, 211, 605–612. [Google Scholar] [CrossRef]
- Bidère, N.; Ngo, V.N.; Lee, J.; Collins, C.; Zheng, L.; Wan, F.; Davis, R.E.; Lenz, G.; Anderson, D.E.; Arnoult, D.; et al. Casein Kinase 1α Governs Antigen-Receptor-Induced NF-ΚB Activation and Human Lymphoma Cell Survival. Nature 2009, 458, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Janovská, P.; Normant, E.; Miskin, H.; Bryja, V. Targeting Casein Kinase 1 (CK1) in Hematological Cancers. Int. J. Mol. Sci. 2020, 21, 9026. [Google Scholar] [CrossRef] [PubMed]
- Manni, S.; Carrino, M.; Manzoni, M.; Gianesin, K.; Nunes, S.C.; Costacurta, M.; Tubi, L.Q.; Macaccaro, P.; Taiana, E.; Cabrelle, A.; et al. Inactivation of CK1α in Multiple Myeloma Empowers Drug Cytotoxicity by Affecting AKT and β-Catenin Survival Signaling Pathways. Oncotarget 2017, 8, 14604–14619. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.K.; Ademà, V.; Heckl, D.; Järås, M.; Mallo, M.; Lord, A.M.; Chu, L.P.; McConkey, M.E.; Kramann, R.; Mullally, A.; et al. Role of Casein Kinase 1A1 in the Biology and Targeted Therapy of Del(5q) MDS. Cancer Cell 2014, 26, 509–520. [Google Scholar] [CrossRef]
- Litchfield, D.W. Protein Kinase CK2: Structure, Regulation and Role in Cellular Decisions of Life and Death. Biochem. J. 2003, 369, 1–15. [Google Scholar] [CrossRef]
- Bibby, A.C.; Litchfield, D.W. The Multiple Personalities of the Regulatory Subunit of Protein Kinase CK2: CK2 Dependent and CK2 Independent Roles Reveal a Secret Identity for CK2β. Int. J. Biol. Sci. 2005, 67–79. [Google Scholar] [CrossRef]
- Nuñez de Villavicencio-Diaz, T.; Rabalski, A.; Litchfield, D. Protein Kinase CK2: Intricate Relationships within Regulatory Cellular Networks. Pharmaceuticals 2017, 10, 27. [Google Scholar] [CrossRef]
- Salvi, M.; Sarno, S.; Cesaro, L.; Nakamura, H.; Pinna, L.A. Extraordinary Pleiotropy of Protein Kinase CK2 Revealed by Weblogo Phosphoproteome Analysis. Biochim. Biophys. Acta BBA Mol. Cell Res. 2009, 1793, 847–859. [Google Scholar] [CrossRef]
- Buchou, T.; Vernet, M.; Blond, O.; Jensen, H.H.; Pointu, H.; Olsen, B.B.; Cochet, C.; Issinger, O.-G.; Boldyreff, B. Disruption of the Regulatory β Subunit of Protein Kinase CK2 in Mice Leads to a Cell-Autonomous Defect and Early Embryonic Lethality. Mol. Cell. Biol. 2003, 23, 908–915. [Google Scholar] [CrossRef]
- Ortega, C.E.; Seidner, Y.; Dominguez, I. Mining CK2 in Cancer. PLoS ONE 2014, 9, e115609. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Reichert, A.; Cunnick, J.; Senadheera, D.; Hemmeryckx, B.; Heisterkamp, N.; Groffen, J. Protein Kinase CKIIα Interacts with the Bcr Moiety of Bcr/Abl and Mediates Proliferation of Bcr/Abl-Expressing Cells. Oncogene 2003, 22, 8255–8262. [Google Scholar] [CrossRef][Green Version]
- Quotti Tubi, L.; Canovas Nunes, S.; Brancalion, A.; Doriguzzi Breatta, E.; Manni, S.; Mandato, E.; Zaffino, F.; Macaccaro, P.; Carrino, M.; Gianesin, K.; et al. Protein Kinase CK2 Regulates AKT, NF-ΚB and STAT3 Activation, Stem Cell Viability and Proliferation in Acute Myeloid Leukemia. Leukemia 2017, 31, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Piazza, F.A.; Ruzzene, M.; Gurrieri, C.; Montini, B.; Bonanni, L.; Chioetto, G.; Di Maira, G.; Barbon, F.; Cabrelle, A.; Zambello, R.; et al. Multiple Myeloma Cell Survival Relies on High Activity of Protein Kinase CK2. Blood 2006, 108, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Manni, S.; Brancalion, A.; Mandato, E.; Tubi, L.Q.; Colpo, A.; Pizzi, M.; Cappellesso, R.; Zaffino, F.; Di Maggio, S.A.; Cabrelle, A.; et al. Protein Kinase CK2 Inhibition Down Modulates the NF-ΚB and STAT3 Survival Pathways, Enhances the Cellular Proteotoxic Stress and Synergistically Boosts the Cytotoxic Effect of Bortezomib on Multiple Myeloma and Mantle Cell Lymphoma Cells. PLoS ONE 2013, 8, e75280. [Google Scholar] [CrossRef]
- Pizzi, M.; Piazza, F.; Agostinelli, C.; Fuligni, F.; Benvenuti, P.; Mandato, E.; Casellato, A.; Rugge, M.; Semenzato, G.; Pileri, S.A. Protein Kinase CK2 Is Widely Expressed in Follicular, Burkitt and Diffuse Large B-Cell Lymphomas and Propels Malignant B-Cell Growth. Oncotarget 2015, 6, 6544–6552. [Google Scholar] [CrossRef]
- Gross, S.; Rahal, R.; Stransky, N.; Lengauer, C.; Hoeflich, K.P. Targeting Cancer with Kinase Inhibitors. J. Clin. Investig. 2015, 125, 1780–1789. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Bhanumathy, K.K.; Balagopal, A.; Vizeacoumar, F.S.; Vizeacoumar, F.J.; Freywald, A.; Giambra, V. Protein Tyrosine Kinases: Their Roles and Their Targeting in Leukemia. Cancers 2021, 13, 184. [Google Scholar] [CrossRef]
- Siveen, K.S.; Prabhu, K.S.; Achkar, I.W.; Kuttikrishnan, S.; Shyam, S.; Khan, A.Q.; Merhi, M.; Dermime, S.; Uddin, S. Role of Non Receptor Tyrosine Kinases in Hematological Malignances and Its Targeting by Natural Products. Mol. Cancer 2018, 17, 31. [Google Scholar] [CrossRef]
- Joshi, S.K.; Davare, M.A.; Druker, B.J.; Tognon, C.E. Revisiting NTRKs as an Emerging Oncogene in Hematological Malignancies. Leukemia 2019, 33, 2563–2574. [Google Scholar] [CrossRef]
- An, X.; Tiwari, A.K.; Sun, Y.; Ding, P.-R.; Ashby, C.R.; Chen, Z.-S. BCR-ABL Tyrosine Kinase Inhibitors in the Treatment of Philadelphia Chromosome Positive Chronic Myeloid Leukemia: A Review. Leuk. Res. 2010, 34, 1255–1268. [Google Scholar] [CrossRef]
- Burger, J.A.; Wiestner, A. Targeting B Cell Receptor Signalling in Cancer: Preclinical and Clinical Advances. Nat. Rev. Cancer 2018, 18, 148–167. [Google Scholar] [CrossRef]
- Visentin, A.; Frezzato, F.; Severin, F.; Imbergamo, S.; Pravato, S.; Romano Gargarella, L.; Manni, S.; Pizzo, S.; Ruggieri, E.; Facco, M.; et al. Lights and Shade of Next-Generation Pi3k Inhibitors in Chronic Lymphocytic Leukemia. OncoTargets Ther. 2020, 13, 9679–9688. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.; Wang, J.; Shi, Y.; Qian, H.; Liu, P. Inhibitors Targeting Bruton’s Tyrosine Kinase in Cancers: Drug Development Advances. Leukemia 2021, 35, 312–332. [Google Scholar] [CrossRef]
- Bello, E.; Pellagatti, A.; Shaw, J.; Mecucci, C.; Kušec, R.; Killick, S.; Giagounidis, A.; Raynaud, S.; Calasanz, M.J.; Fenaux, P.; et al. CSNK1A1 Mutations and Gene Expression Analysis in Myelodysplastic Syndromes with Del(5q). Br. J. Haematol. 2015, 171, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, Q.; Chen, L.; Zhang, H.; Schonbrunn, E.; Chen, J. Tumor-Derived CK1α Mutations Enhance MDMX Inhibition of P53. Oncogene 2020, 39, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Broséus, J.; Chen, G.; Hergalant, S.; Ramstein, G.; Mounier, N.; Guéant, J.-L.; Feugier, P.; Gisselbrecht, C.; Thieblemont, C.; Houlgatte, R. Relapsed Diffuse Large B-Cell Lymphoma Present Different Genomic Profiles between Early and Late Relapses. Oncotarget 2016, 7, 83987–84002. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Nagata, Y.; Kitanaka, A.; Shiraishi, Y.; Shimamura, T.; Yasunaga, J.; Totoki, Y.; Chiba, K.; Sato-Otsubo, A.; Nagae, G.; et al. Integrated Molecular Analysis of Adult T Cell Leukemia/Lymphoma. Nat. Genet. 2015, 47, 1304–1315. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lu, Q.; Wang, Z.; Cai, P.; Zeng, Z.; Zhang, L.; Wang, M.; Ma, L.; Ruan, C.; Chen, S. Identification of a Novel CSNK2A1-PDGFRB Fusion Gene in a Patient with Myeloid Neoplasm with Eosinophilia. Cancer Res. Treat. 2020. [Google Scholar] [CrossRef]
- Nagel, R.; Semenova, E.A.; Berns, A. Drugging the Addict: Non-oncogene Addiction as a Target for Cancer Therapy. EMBO Rep. 2016, 17, 1516–1531. [Google Scholar] [CrossRef]
- Zheng, M.; Jiang, J.; Tang, Y.; Liang, X. Oncogene and Non-Oncogene Addiction in Inflammation-Associated Cancers. Future Oncol. 2013, 9, 561–573. [Google Scholar] [CrossRef]
- Weinstein, I.B. CANCER: Enhanced: Addiction to Oncogenes--the Achilles Heal of Cancer. Science 2002, 297, 63–64. [Google Scholar] [CrossRef]
- Solimini, N.L.; Luo, J.; Elledge, S.J. Non-Oncogene Addiction and the Stress Phenotype of Cancer Cells. Cell 2007, 130, 986–988. [Google Scholar] [CrossRef]
- Piazza, F.; Manni, S.; Arjomand, A.; Visentin, A.; Trentin, L.; Semenzato, G. New Responsibilities for Aged Kinases in B-lymphomas. Hematol. Oncol. 2020, 38, 3–11. [Google Scholar] [CrossRef]
- Jiang, N.; Dai, Q.; Su, X.; Fu, J.; Feng, X.; Peng, J. Role of PI3K/AKT Pathway in Cancer: The Framework of Malignant Behavior. Mol. Biol. Rep. 2020, 47, 4587–4629. [Google Scholar] [CrossRef] [PubMed]
- Ruzzene, M.; Bertacchini, J.; Toker, A.; Marmiroli, S. Cross-Talk between the CK2 and AKT Signaling Pathways in Cancer. Adv. Biol. Regul. 2017, 64, 1–8. [Google Scholar] [CrossRef]
- Di Maira, G.; Salvi, M.; Arrigoni, G.; Marin, O.; Sarno, S.; Brustolon, F.; Pinna, L.A.; Ruzzene, M. Protein Kinase CK2 Phosphorylates and Upregulates Akt/PKB. Cell Death Differ. 2005, 12, 668–677. [Google Scholar] [CrossRef]
- Vazquez, F.; Grossman, S.R.; Takahashi, Y.; Rokas, M.V.; Nakamura, N.; Sellers, W.R. Phosphorylation of the PTEN Tail Acts as an Inhibitory Switch by Preventing Its Recruitment into a Protein Complex. J. Biol. Chem. 2001, 276, 48627–48630. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.M.; Soares, M.V.D.; Ribeiro, P.; Caldas, J.; Povoa, V.; Martins, L.R.; Melao, A.; Serra-Caetano, A.; de Sousa, A.B.; Lacerda, J.F.; et al. Adult B-Cell Acute Lymphoblastic Leukemia Cells Display Decreased PTEN Activity and Constitutive Hyperactivation of PI3K/Akt Pathway despite High PTEN Protein Levels. Haematologica 2014, 99, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.; Schnabl, S.; Demirtas, D.; Hilgarth, M.; Hubmann, R.; Ponath, E.; Badrnya, S.; Lehner, C.; Hoelbl, A.; Duechler, M.; et al. Reconstitution of PTEN Activity by CK2 Inhibitors and Interference with the PI3-K/Akt Cascade Counteract the Antiapoptotic Effect of Human Stromal Cells in Chronic Lymphocytic Leukemia. Blood 2010, 116, 2513–2521. [Google Scholar] [CrossRef]
- Duan, S.; Skaar, J.R.; Kuchay, S.; Toschi, A.; Kanarek, N.; Ben-Neriah, Y.; Pagano, M. MTOR Generates an Auto-Amplification Loop by Triggering the ΒTrCP- and CK1α-Dependent Degradation of DEPTOR. Mol. Cell 2011, 44, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Imbert, V.; Peyron, J.-F. NF-ΚB in Hematological Malignancies. Biomedicines 2017, 5, 27. [Google Scholar] [CrossRef] [PubMed]
- Grondona, P.; Bucher, P.; Schulze-Osthoff, K.; Hailfinger, S.; Schmitt, A. NF-ΚB Activation in Lymphoid Malignancies: Genetics, Signaling, and Targeted Therapy. Biomedicines 2018, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Gehring, T.; Erdmann, T.; Rahm, M.; Graß, C.; Flatley, A.; O’Neill, T.J.; Woods, S.; Meininger, I.; Karayel, O.; Kutzner, K.; et al. MALT1 Phosphorylation Controls Activation of T Lymphocytes and Survival of ABC-DLBCL Tumor Cells. Cell Rep. 2019, 29, 873–888. [Google Scholar] [CrossRef]
- McElhinny, J.A.; Trushin, S.A.; Bren, G.D.; Chester, N.; Paya, C.V. Casein Kinase II Phosphorylates I Kappa B Alpha at S-283, S-289, S-293, and T-291 and Is Required for Its Degradation. Mol. Cell. Biol. 1996, 16, 899–906. [Google Scholar] [CrossRef]
- Wang, D.; Westerheide, S.D.; Hanson, J.L.; Baldwin, A.S. Tumor Necrosis Factor Alpha-Induced Phosphorylation of RelA/P65 on Ser529 Is Controlled by Casein Kinase II. J. Biol. Chem. 2000, 275, 32592–32597. [Google Scholar] [CrossRef]
- Buontempo, F.; Orsini, E.; Lonetti, A.; Cappellini, A.; Chiarini, F.; Evangelisti, C.; Evangelisti, C.; Melchionda, F.; Pession, A.; Bertaina, A.; et al. Synergistic Cytotoxic Effects of Bortezomib and CK2 Inhibitor CX-4945 in Acute Lymphoblastic Leukemia: Turning off the Prosurvival ER Chaperone BIP/Grp78 and Turning on the pro-Apoptotic NF-ΚB. Oncotarget 2016, 7, 1323–1340. [Google Scholar] [CrossRef]
- Liu, C.; Li, Y.; Semenov, M.; Han, C.; Baeg, G.-H.; Tan, Y.; Zhang, Z.; Lin, X.; He, X. Control of β-Catenin Phosphorylation/Degradation by a Dual-Kinase Mechanism. Cell 2002, 108, 837–847. [Google Scholar] [CrossRef]
- Cheong, J.K.; Virshup, D.M. Casein Kinase 1: Complexity in the Family. Int. J. Biochem. Cell Biol. 2011, 43, 465–469. [Google Scholar] [CrossRef]
- Li, B.; Orton, D.; Neitzel, L.R.; Astudillo, L.; Shen, C.; Long, J.; Chen, X.; Kirkbride, K.C.; Doundoulakis, T.; Guerra, M.L.; et al. Differential Abundance of CK1α Provides Selectivity for Pharmacological CK1α Activators to Target WNT-Dependent Tumors. Sci. Signal. 2017, 10, eaak9916. [Google Scholar] [CrossRef]
- van Andel, H.; Kocemba, K.A.; Spaargaren, M.; Pals, S.T. Aberrant Wnt Signaling in Multiple Myeloma: Molecular Mechanisms and Targeting Options. Leukemia 2019, 33, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- de Groot, R.E.A.; Rappel, S.B.; Lorenowicz, M.J.; Korswagen, H.C. Protein Kinase CK2 Is Required for Wntless Internalization and Wnt Secretion. Cell. Signal. 2014, 26, 2601–2605. [Google Scholar] [CrossRef]
- Seldin, D.C.; Landesman-Bollag, E.; Farago, M.; Currier, N.; Lou, D.; Dominguez, I. CK2 as a Positive Regulator of Wnt Signalling and Tumourigenesis. Mol. Cell. Biochem. 2005, 274, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Willert, K.; Brink, M.; Wodarz, A.; Varmus, H.; Nusse, R. Casein Kinase 2 Associates with and Phosphorylates Dishevelled. EMBO J. 1997, 16, 3089–3096. [Google Scholar] [CrossRef]
- Song, D.H.; Sussman, D.J.; Seldin, D.C. Endogenous Protein Kinase CK2 Participates in Wnt Signaling in Mammary Epithelial Cells. J. Biol. Chem. 2000, 275, 23790–23797. [Google Scholar] [CrossRef] [PubMed]
- Song, D.H.; Dominguez, I.; Mizuno, J.; Kaut, M.; Mohr, S.C.; Seldin, D.C. CK2 Phosphorylation of the Armadillo Repeat Region of Beta-Catenin Potentiates Wnt Signaling. J. Biol. Chem. 2003, 278, 24018–24025. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jones, K.A. CK2 Controls the Recruitment of Wnt Regulators to Target Genes In Vivo. Curr. Biol. 2006, 16, 2239–2244. [Google Scholar] [CrossRef]
- Wu, C.; Lao, F.S.; Nan, E.; Li, H.; Choi, M.Y.; Messer, K.; Carson, D.A. Inhibition of Casein Kinase 2 Impairs Wnt Signaling and Cell Survival in Chronic Lymphocytic Leukemia. Blood 2016, 128, 2050. [Google Scholar] [CrossRef]
- Vainchenker, W.; Constantinescu, S.N. JAK/STAT Signaling in Hematological Malignancies. Oncogene 2013, 32, 2601–2613. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Qin, H.; Frank, S.J.; Deng, L.; Litchfield, D.W.; Tefferi, A.; Pardanani, A.; Lin, F.-T.; Li, J.; Sha, B.; et al. A CK2-Dependent Mechanism for Activation of the JAK-STAT Signaling Pathway. Blood 2011, 118, 156–166. [Google Scholar] [CrossRef]
- Hazan-Halevy, I.; Harris, D.; Liu, Z.; Liu, J.; Li, P.; Chen, X.; Shanker, S.; Ferrajoli, A.; Keating, M.J.; Estrov, Z. STAT3 Is Constitutively Phosphorylated on Serine 727 Residues, Binds DNA, and Activates Transcription in CLL Cells. Blood 2010, 115, 2852–2863. [Google Scholar] [CrossRef] [PubMed]
- Rozovski, U.; Harris, D.M.; Li, P.; Liu, Z.; Jain, P.; Veletic, I.; Ferrajoli, A.; Burger, J.; O’Brien, S.; Bose, P.; et al. Constitutive Phosphorylation of STAT3 by the CK2-BLNK-CD5 Complex. Mol. Cancer Res. 2017, 15, 610–618. [Google Scholar] [CrossRef] [PubMed]
- White-Gilbertson, S.; Hua, Y.; Liu, B. The Role of Endoplasmic Reticulum Stress in Maintaining and Targeting Multiple Myeloma: A Double-Edged Sword of Adaptation and Apoptosis. Front. Genet. 2013, 4, 109. [Google Scholar] [CrossRef] [PubMed]
- Martelli, A.M.; Paganelli, F.; Chiarini, F.; Evangelisti, C.; McCubrey, J.A. The Unfolded Protein Response: A Novel Therapeutic Target in Acute Leukemias. Cancers 2020, 12, 333. [Google Scholar] [CrossRef]
- Ampofo, E.; Sokolowsky, T.; Götz, C.; Montenarh, M. Functional Interaction of Protein Kinase CK2 and Activating Transcription Factor 4 (ATF4), a Key Player in the Cellular Stress Response. Biochim. Biophys. Acta BBA Mol. Cell Res. 2013, 1833, 439–451. [Google Scholar] [CrossRef]
- Schneider, C.C.; Ampofo, E.; Montenarh, M. CK2 Regulates ATF4 and CHOP Transcription within the Cellular Stress Response Signalling Pathway. Cell. Signal. 2012, 24, 1797–1802. [Google Scholar] [CrossRef]
- Jin, S.; Wei, J.; You, L.; Liu, H.; Qian, W. Autophagy Regulation and Its Dual Role in Blood Cancers: A Novel Target for Therapeutic Development (Review). Oncol. Rep. 2018, 39, 2473–2481. [Google Scholar] [CrossRef]
- Manni, S.; Brancalion, A.; Tubi, L.Q.; Colpo, A.; Pavan, L.; Cabrelle, A.; Ave, E.; Zaffino, F.; Di Maira, G.; Ruzzene, M.; et al. Protein Kinase CK2 Protects Multiple Myeloma Cells from ER Stress–Induced Apoptosis and from the Cytotoxic Effect of HSP90 Inhibition through Regulation of the Unfolded Protein Response. Clin. Cancer Res. 2012, 18, 1888–1900. [Google Scholar] [CrossRef]
- Bandhakavi, S.; McCann, R.O.; Hanna, D.E.; Glover, C.V.C. A Positive Feedback Loop between Protein Kinase CKII and Cdc37 Promotes the Activity of Multiple Protein Kinases. J. Biol. Chem. 2003, 278, 2829–2836. [Google Scholar] [CrossRef]
- Miyata, Y.; Nishida, E. CK2 Controls Multiple Protein Kinases by Phosphorylating a Kinase-Targeting Molecular Chaperone, Cdc37. Mol. Cell. Biol. 2004, 24, 4065–4074. [Google Scholar] [CrossRef]
- Miyata, Y. Protein Kinase CK2 in Health and Disease: CK2: The Kinase Controlling the Hsp90 Chaperone Machinery. Cell. Mol. Life Sci. 2009, 66, 1840–1849. [Google Scholar] [CrossRef] [PubMed]
- Buontempo, F.; Orsini, E.; Martins, L.R.; Antunes, I.; Lonetti, A.; Chiarini, F.; Tabellini, G.; Evangelisti, C.; Evangelisti, C.; Melchionda, F.; et al. Cytotoxic Activity of the Casein Kinase 2 Inhibitor CX-4945 against T-Cell Acute Lymphoblastic Leukemia: Targeting the Unfolded Protein Response Signaling. Leukemia 2014, 28, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Rabalski, A.J.; Gyenis, L.; Litchfield, D.W. Molecular Pathways: Emergence of Protein Kinase CK2 (CSNK2) as a Potential Target to Inhibit Survival and DNA Damage Response and Repair Pathways in Cancer Cells. Clin. Cancer Res. 2016, 22, 2840–2847. [Google Scholar] [CrossRef]
- Venerando, A.; Marin, O.; Cozza, G.; Bustos, V.H.; Sarno, S.; Pinna, L.A. Isoform Specific Phosphorylation of P53 by Protein Kinase CK1. Cell. Mol. Life Sci. 2010, 67, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- MacLaine, N.J.; Øster, B.; Bundgaard, B.; Fraser, J.A.; Buckner, C.; Lazo, P.A.; Meek, D.W.; Höllsberg, P.; Hupp, T.R. A Central Role for CK1 in Catalyzing Phosphorylation of the P53 Transactivation Domain at Serine 20 after HHV-6B Viral Infection. J. Biol. Chem. 2008, 283, 28563–28573. [Google Scholar] [CrossRef]
- Huart, A.-S.; MacLaine, N.J.; Meek, D.W.; Hupp, T.R. CK1α Plays a Central Role in Mediating MDM2 Control of P53 and E2F-1 Protein Stability. J. Biol. Chem. 2009, 284, 32384–32394. [Google Scholar] [CrossRef]
- Huart, A.-S.; MacLaine, N.J.; Narayan, V.; Hupp, T.R. Exploiting the MDM2-CK1α Protein-Protein Interface to Develop Novel Biologics That Induce UBL-Kinase-Modification and Inhibit Cell Growth. PLoS ONE 2012, 7, e43391. [Google Scholar] [CrossRef]
- Chen, L.; Li, C.; Pan, Y.; Chen, J. Regulation of P53-MDMX Interaction by Casein Kinase 1 Alpha. Mol. Cell. Biol. 2005, 25, 6509–6520. [Google Scholar] [CrossRef]
- Wu, S.; Chen, L.; Becker, A.; Schonbrunn, E.; Chen, J. Casein Kinase 1 Regulates an MDMX Intramolecular Interaction To Stimulate P53 Binding. Mol. Cell. Biol. 2012, 32, 4821–4832. [Google Scholar] [CrossRef]
- Xu, W.; Huang, Z.; Gan, Y.; Chen, R.; Huang, Y.; Xue, B.; Jiang, S.; Yu, Z.; Yu, K.; Zhang, S. Casein Kinase 1α Inhibits P53 Downstream of MDM2-mediated Autophagy and Apoptosis in Acute Myeloid Leukemia. Oncol. Rep. 2020, 44, 1895–1904. [Google Scholar] [CrossRef] [PubMed]
- Ström, C.E.; Mortusewicz, O.; Finch, D.; Parsons, J.L.; Lagerqvist, A.; Johansson, F.; Schultz, N.; Erixon, K.; Dianov, G.L.; Helleday, T. CK2 Phosphorylation of XRCC1 Facilitates Dissociation from DNA and Single-Strand Break Formation during Base Excision Repair. DNA Repair 2011, 10, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Melander, F.; Bekker-Jensen, S.; Falck, J.; Bartek, J.; Mailand, N.; Lukas, J. Phosphorylation of SDT Repeats in the MDC1 N Terminus Triggers Retention of NBS1 at the DNA Damage–Modified Chromatin. J. Cell Biol. 2008, 181, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Olsen, B.B.; Wang, S.-Y.; Svenstrup, T.H.; Chen, B.P.; Guerra, B. Protein Kinase CK2 Localizes to Sites of DNA Double-Strand Break Regulating the Cellular Response to DNA Damage. BMC Mol. Biol. 2012, 13, 7. [Google Scholar] [CrossRef]
- Guerra, B.; Iwabuchi, K.; Issinger, O.-G. Protein Kinase CK2 Is Required for the Recruitment of 53BP1 to Sites of DNA Double-Strand Break Induced by Radiomimetic Drugs. Cancer Lett. 2014, 345, 115–123. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Bliesath, J.; Macalino, D.; Omori, M.; Huser, N.; Streiner, N.; Ho, C.B.; Anderes, K.; Proffitt, C.; O’Brien, S.E.; et al. CK2 Inhibitor CX-4945 Suppresses DNA Repair Response Triggered by DNA-Targeted Anticancer Drugs and Augments Efficacy: Mechanistic Rationale for Drug Combination Therapy. Mol. Cancer Ther. 2012, 11, 994–1005. [Google Scholar] [CrossRef]
- Keller, D.M.; Zeng, X.; Wang, Y.; Zhang, Q.H.; Kapoor, M.; Shu, H.; Goodman, R.; Lozano, G.; Zhao, Y.; Lu, H. A DNA Damage–Induced P53 Serine 392 Kinase Complex Contains CK2, HSpt16, and SSRP1. Mol. Cell 2001, 7, 283–292. [Google Scholar] [CrossRef]
- Hjerrild, M.; Milne, D.; Dumaz, N.; Hay, T.; Issinger, O.-G.; Meek, D. Phosphorylation of Murine Double Minute Clone 2 (MDM2) Protein at Serine-267 by Protein Kinase CK2 in Vitro and in Cultured Cells. Biochem. J. 2001, 355, 347. [Google Scholar] [CrossRef]
- Allende-Vega, N.; Dias, S.; Milne, D.; Meek, D. Phosphorylation of the Acidic Domain of Mdm2 by Protein Kinase CK2. Mol. Cell. Biochem. 2005, 274, 85–90. [Google Scholar] [CrossRef]
- Khoronenkova, S.V.; Dianova, I.I.; Ternette, N.; Kessler, B.M.; Parsons, J.L.; Dianov, G.L. ATM-Dependent Downregulation of USP7/HAUSP by PPM1G Activates P53 Response to DNA Damage. Mol. Cell 2012, 45, 801–813. [Google Scholar] [CrossRef]
- Landesman-Bollag, E.; Channavajhala, P.L.; Cardiff, R.D.; Seldin, D.C. P53 Deficiency and Misexpression of Protein Kinase CK2α Collaborate in the Development of Thymic Lymphomas in Mice. Oncogene 1998, 16, 2965–2974. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Eom, J.I.; Cheong, J.-W.; Choi, A.J.; Lee, J.K.; Yang, W.I.; Min, Y.H. Protein Kinase CK2α as an Unfavorable Prognostic Marker and Novel Therapeutic Target in Acute Myeloid Leukemia. Clin. Cancer Res. 2007, 13, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Quotti Tubi, L.; Gurrieri, C.; Brancalion, A.; Bonaldi, L.; Bertorelle, R.; Manni, S.; Pavan, L.; Lessi, F.; Zambello, R.; Trentin, L.; et al. Inhibition of Protein Kinase CK2 with the Clinical-Grade Small ATP-Competitive Compound CX-4945 or by RNA Interference Unveils Its Role in Acute Myeloid Leukemia Cell Survival, P53-Dependent Apoptosis and Daunorubicin-Induced Cytotoxicity. J. Hematol. Oncol. 2013, 6, 78. [Google Scholar] [CrossRef]
- Cheong, J.K.; Zhang, F.; Chua, P.J.; Bay, B.H.; Thorburn, A.; Virshup, D.M. Casein Kinase 1α–Dependent Feedback Loop Controls Autophagy in RAS-Driven Cancers. J. Clin. Investig. 2015, 125, 1401–1418. [Google Scholar] [CrossRef] [PubMed]
- Carrino, M.; Quotti Tubi, L.; Fregnani, A.; Canovas Nunes, S.; Barilà, G.; Trentin, L.; Zambello, R.; Semenzato, G.; Manni, S.; Piazza, F. Prosurvival Autophagy Is Regulated by Protein Kinase CK1 Alpha in Multiple Myeloma. Cell Death Discov. 2019, 5, 98. [Google Scholar] [CrossRef]
- Silva-Pavez, E.; Tapia, J.C. Protein Kinase CK2 in Cancer Energetics. Front. Oncol. 2020, 10, 893. [Google Scholar] [CrossRef]
- Minzel, W.; Venkatachalam, A.; Fink, A.; Hung, E.; Brachya, G.; Burstain, I.; Shaham, M.; Rivlin, A.; Omer, I.; Zinger, A.; et al. Small Molecules Co-Targeting CKIα and the Transcriptional Kinases CDK7/9 Control AML in Preclinical Models. Cell 2018, 175, 171–185.e25. [Google Scholar] [CrossRef]
- List, A.; Dewald, G.; Bennett, J.; Giagounidis, A.; Raza, A.; Feldman, E.; Powell, B.; Greenberg, P.; Thomas, D.; Stone, R.; et al. Lenalidomide in the Myelodysplastic Syndrome with Chromosome 5q Deletion. N. Engl. J. Med. 2006, 355, 1456–1465. [Google Scholar] [CrossRef]
- Krönke, J.; Fink, E.C.; Hollenbach, P.W.; MacBeth, K.J.; Hurst, S.N.; Udeshi, N.D.; Chamberlain, P.P.; Mani, D.R.; Man, H.W.; Gandhi, A.K.; et al. Lenalidomide Induces Ubiquitination and Degradation of CK1α in Del(5q) MDS. Nature 2015, 523, 183–188. [Google Scholar] [CrossRef]
- Cozza, G.; Pinna, L.; Moro, S. Kinase CK2 Inhibition: An Update. Curr. Med. Chem. 2013, 20, 671–693. [Google Scholar] [CrossRef]
- Qiao, Y.; Chen, T.; Yang, H.; Chen, Y.; Lin, H.; Qu, W.; Feng, F.; Liu, W.; Guo, Q.; Liu, Z.; et al. Small Molecule Modulators Targeting Protein Kinase CK1 and CK2. Eur. J. Med. Chem. 2019, 181, 111581. [Google Scholar] [CrossRef]
- Kim, H.; Choi, K.; Kang, H.; Lee, S.-Y.; Chi, S.-W.; Lee, M.-S.; Song, J.; Im, D.; Choi, Y.; Cho, S. Identification of a Novel Function of CX-4945 as a Splicing Regulator. PLoS ONE 2014, 9, e94978. [Google Scholar] [CrossRef]
- Kim, H.; Lee, K.-S.; Kim, A.-K.; Choi, M.; Choi, K.; Kang, M.; Chi, S.-W.; Lee, M.-S.; Lee, J.-S.; Lee, S.-Y.; et al. A Chemical with Proven Clinical Safety Rescues Down-Syndrome-Related Phenotypes in through DYRK1A Inhibition. Dis. Model. Mech. 2016, 9, 839–848. [Google Scholar] [CrossRef]
- Brear, P.; De Fusco, C.; Hadje Georgiou, K.; Francis-Newton, N.J.; Stubbs, C.J.; Sore, H.F.; Venkitaraman, A.R.; Abell, C.; Spring, D.R.; Hyvönen, M. Specific Inhibition of CK2α from an Anchor Outside the Active Site. Chem. Sci. 2016, 7, 6839–6845. [Google Scholar] [CrossRef] [PubMed]
- De Fusco, C.; Brear, P.; Iegre, J.; Georgiou, K.H.; Sore, H.F.; Hyvönen, M.; Spring, D.R. A Fragment-Based Approach Leading to the Discovery of a Novel Binding Site and the Selective CK2 Inhibitor CAM4066. Bioorg. Med. Chem. 2017, 25, 3471–3482. [Google Scholar] [CrossRef] [PubMed]
- Iegre, J.; Brear, P.; De Fusco, C.; Yoshida, M.; Mitchell, S.L.; Rossmann, M.; Carro, L.; Sore, H.F.; Hyvönen, M.; Spring, D.R. Second-Generation CK2α Inhibitors Targeting the ΑD Pocket. Chem. Sci. 2018, 9, 3041–3049. [Google Scholar] [CrossRef]
- Brear, P.; North, A.; Iegre, J.; Hadje Georgiou, K.; Lubin, A.; Carro, L.; Green, W.; Sore, H.F.; Hyvönen, M.; Spring, D.R. Novel Non-ATP Competitive Small Molecules Targeting the CK2 α/β Interface. Bioorg. Med. Chem. 2018, 26, 3016–3020. [Google Scholar] [CrossRef] [PubMed]
- Bestgen, B.; Krimm, I.; Kufareva, I.; Kamal, A.A.M.; Seetoh, W.-G.; Abell, C.; Hartmann, R.W.; Abagyan, R.; Cochet, C.; Le Borgne, M.; et al. 2-Aminothiazole Derivatives as Selective Allosteric Modulators of the Protein Kinase CK2. 1. Identification of an Allosteric Binding Site. J. Med. Chem. 2019, 62, 1803–1816. [Google Scholar] [CrossRef]
- D’Amore, C.; Borgo, C.; Sarno, S.; Salvi, M. Role of CK2 Inhibitor CX-4945 in Anti-Cancer Combination Therapy–Potential Clinical Relevance. Cell. Oncol. 2020, 43, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Prins, R.C.; Burke, R.T.; Tyner, J.W.; Druker, B.J.; Loriaux, M.M.; Spurgeon, S.E. CX-4945, a Selective Inhibitor of Casein Kinase-2 (CK2), Exhibits Anti-Tumor Activity in Hematologic Malignancies Including Enhanced Activity in Chronic Lymphocytic Leukemia When Combined with Fludarabine and Inhibitors of the B-Cell Receptor Pathway. Leukemia 2013, 27, 2094–2096. [Google Scholar] [CrossRef]
- Mandato, E.; Nunes, S.C.; Zaffino, F.; Casellato, A.; Macaccaro, P.; Tubi, L.Q.; Visentin, A.; Trentin, L.; Semenzato, G.; Piazza, F. CX-4945, a Selective Inhibitor of Casein Kinase 2, Synergizes with B Cell Receptor Signaling Inhibitors in Inducing Diffuse Large B Cell Lymphoma Cell Death. Curr. Cancer Drug Targets 2018, 18, 608–616. [Google Scholar] [CrossRef] [PubMed]
| SIGNALLING | CK1α TARGETS | BLOOD TUMOR | CK2 TARGETS | BLOOD TUMOR |
|---|---|---|---|---|
| PI3K/AKT | AKT | MM | AKT/PTEN | B-ALL, CLL, LSCs, DLBCL |
| NF-κB | CARD11/MALT | DLBCL | RelA/p65 | MM, MCL, DLBLC, BL; ALL |
| Wnt/β-catenin | β-catenin | MM | β-catenin, Dvl | CLL |
| JAK/STAT | JAK1/2, STAT3/5 | CMD, MM, CLL, AML, ALL |
| STRESS PHENOTYPE | CK1α TARGETS | BLOOD TUMOR | CK2 TARGETS | BLOOD TUMOR |
|---|---|---|---|---|
| ER STRESS/UPR | Cdc37/Bip/Grp78, IRE1α, PERK | MM, NHL, T/B-ALL | ||
| DNA DAMAGE | p53 | MM, AML | p53 | AML |
| AUTOPHAGY | FOXO3A | MM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spinello, Z.; Fregnani, A.; Quotti Tubi, L.; Trentin, L.; Piazza, F.; Manni, S. Targeting Protein Kinases in Blood Cancer: Focusing on CK1α and CK2. Int. J. Mol. Sci. 2021, 22, 3716. https://doi.org/10.3390/ijms22073716
Spinello Z, Fregnani A, Quotti Tubi L, Trentin L, Piazza F, Manni S. Targeting Protein Kinases in Blood Cancer: Focusing on CK1α and CK2. International Journal of Molecular Sciences. 2021; 22(7):3716. https://doi.org/10.3390/ijms22073716
Chicago/Turabian StyleSpinello, Zaira, Anna Fregnani, Laura Quotti Tubi, Livio Trentin, Francesco Piazza, and Sabrina Manni. 2021. "Targeting Protein Kinases in Blood Cancer: Focusing on CK1α and CK2" International Journal of Molecular Sciences 22, no. 7: 3716. https://doi.org/10.3390/ijms22073716
APA StyleSpinello, Z., Fregnani, A., Quotti Tubi, L., Trentin, L., Piazza, F., & Manni, S. (2021). Targeting Protein Kinases in Blood Cancer: Focusing on CK1α and CK2. International Journal of Molecular Sciences, 22(7), 3716. https://doi.org/10.3390/ijms22073716






