Utilizing an Animal Model to Identify Brain Neurodegeneration-Related Biomarkers in Aging
Abstract
1. Introduction
2. Materials and Methods
2.1. Mouse GNMT Isolation and Generation
2.2. Protein Confirmation-Western Blotting
2.3. Tissue Preparation for Histology and Immunohistochemistry
2.4. Brain Proteome Research
2.5. Statistics Analysis
3. Results and Discussions
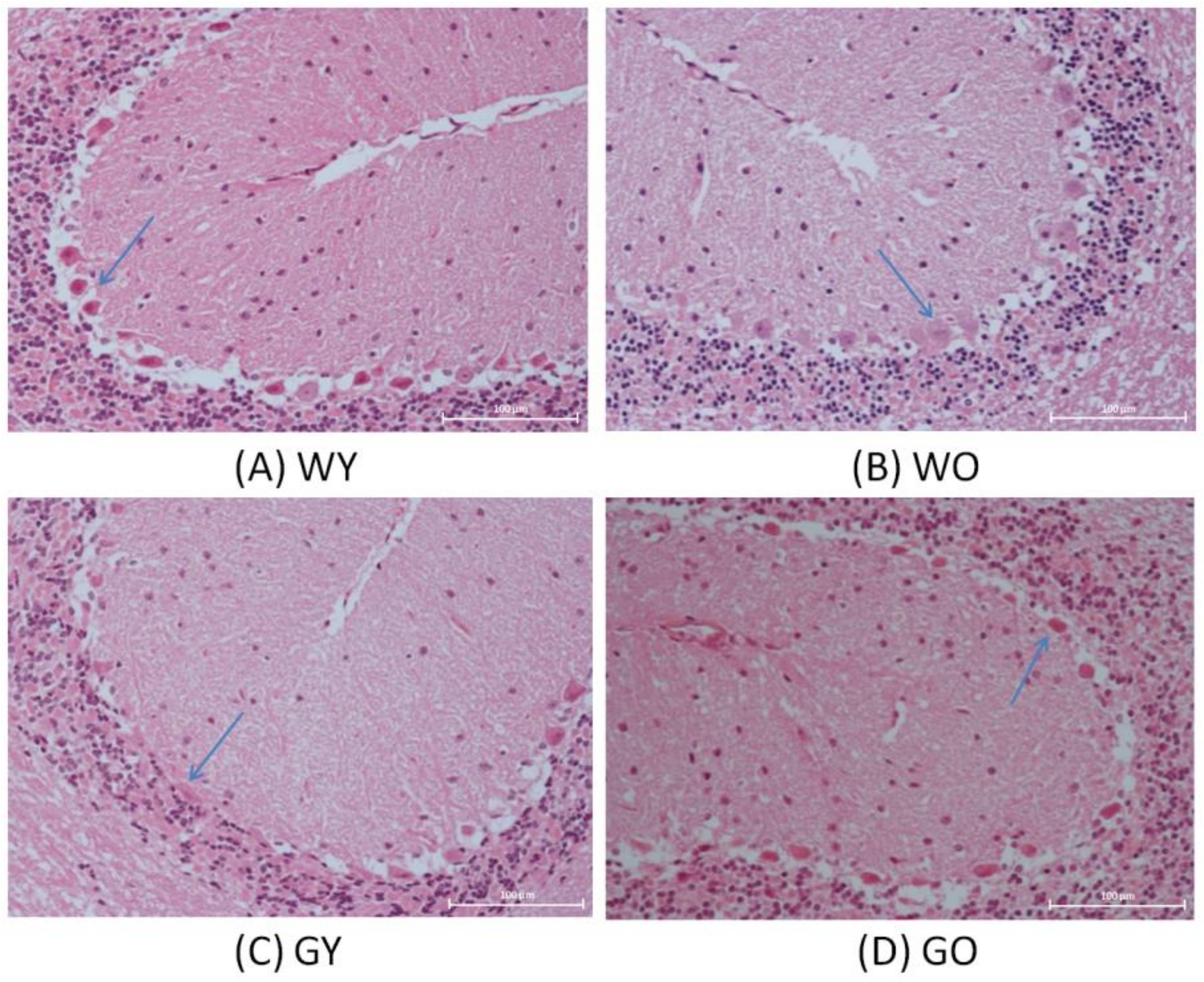
3.1. H&E Stain
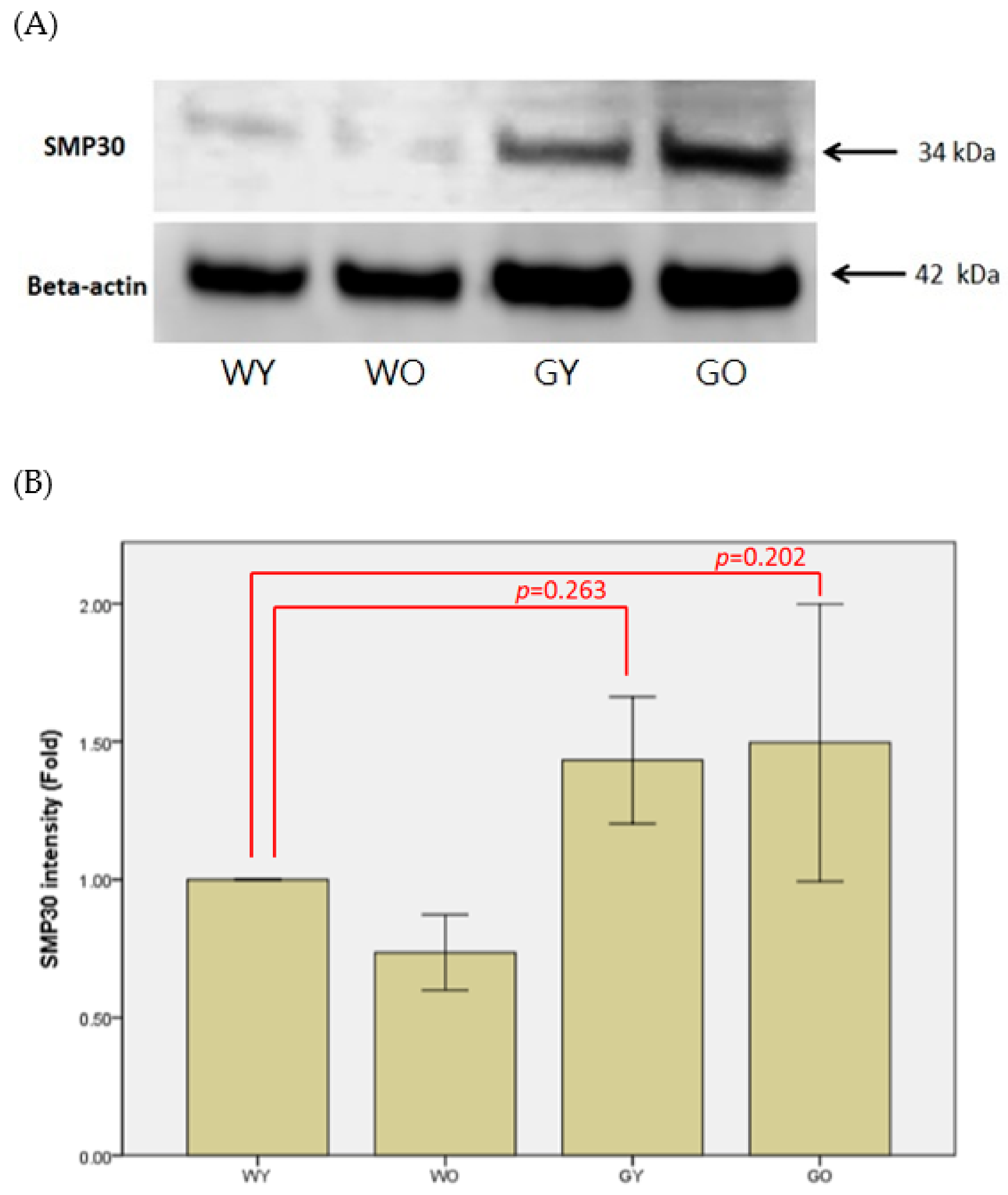
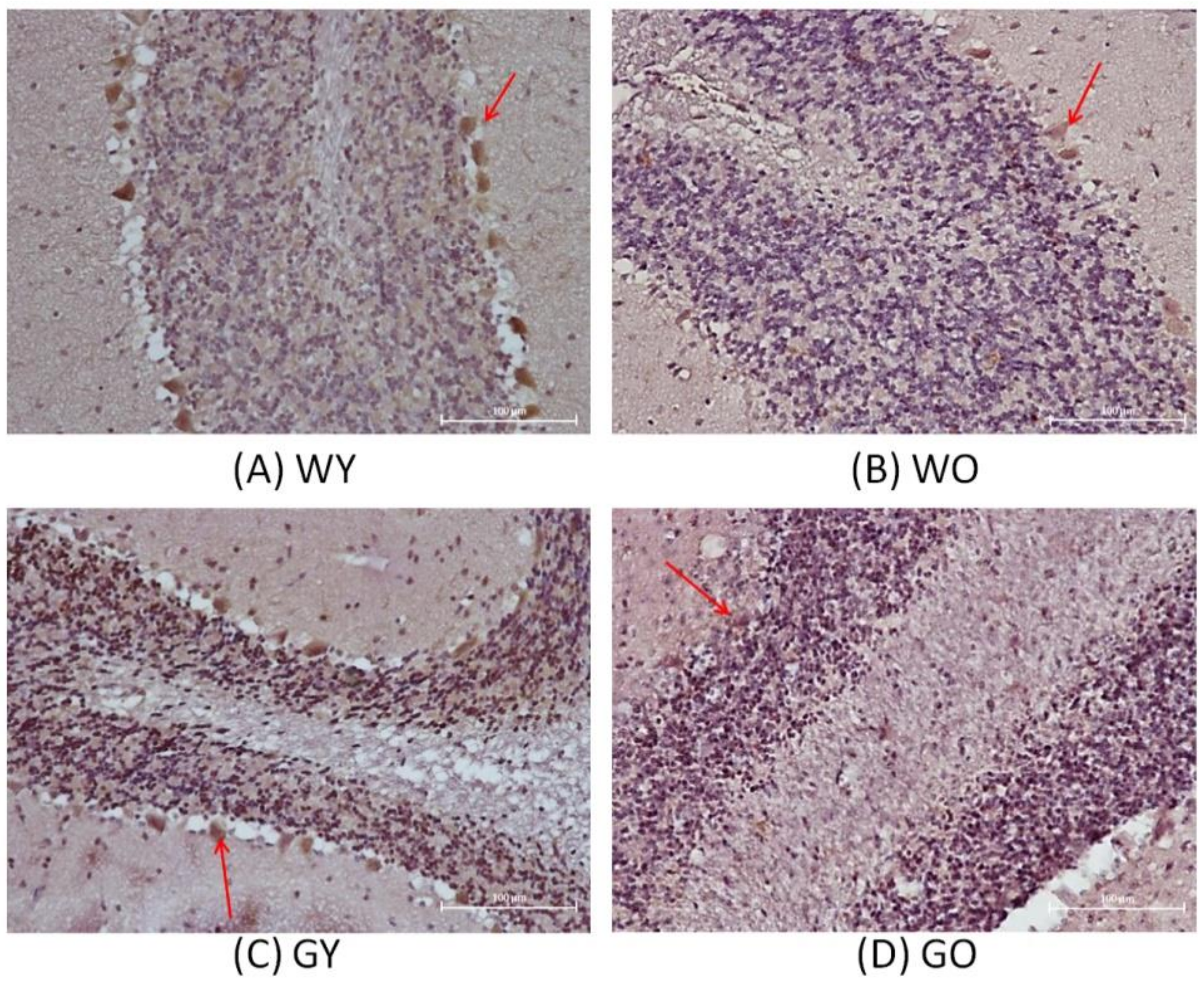
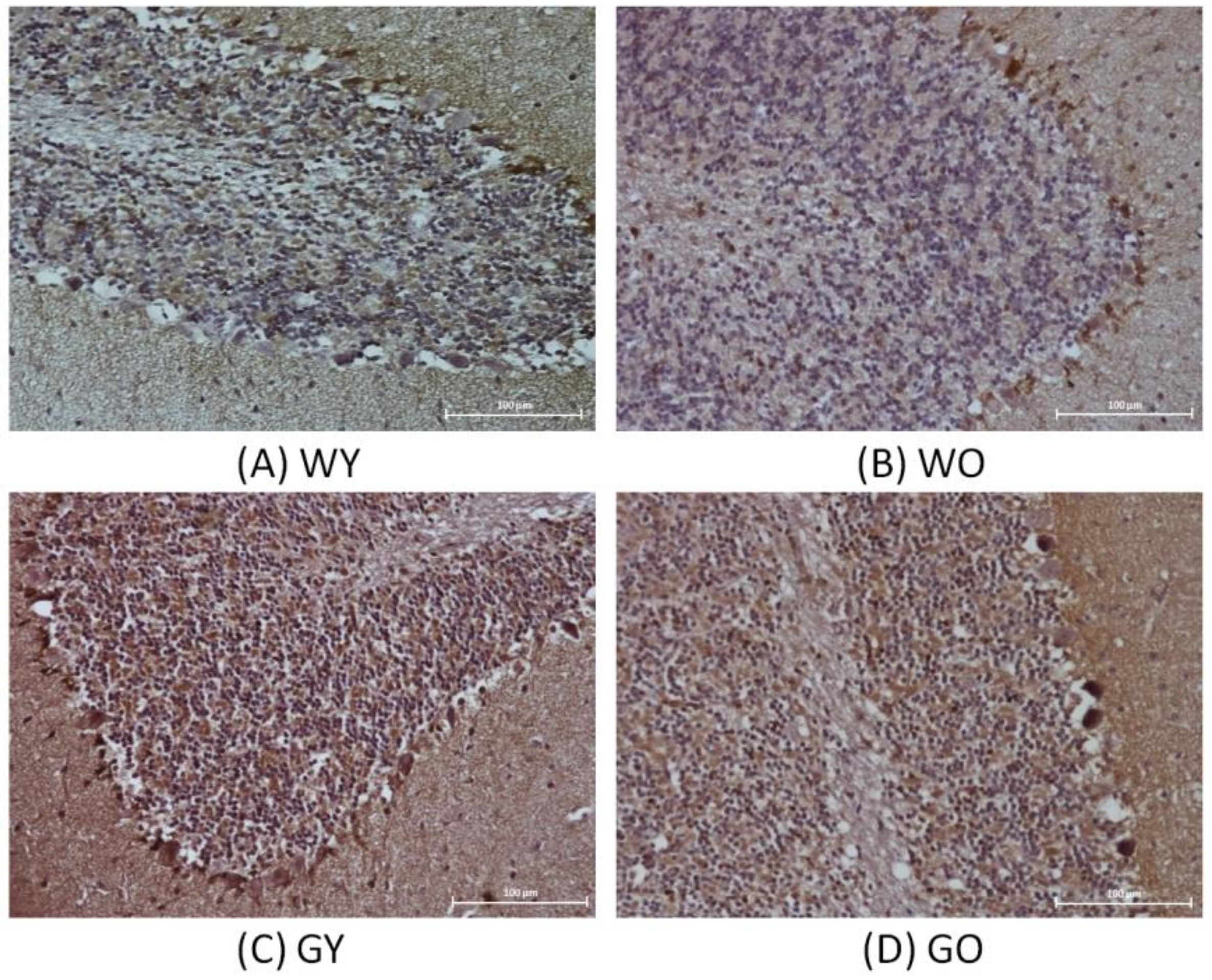
3.2. Senescence Marker Protein 30 (SMP30)
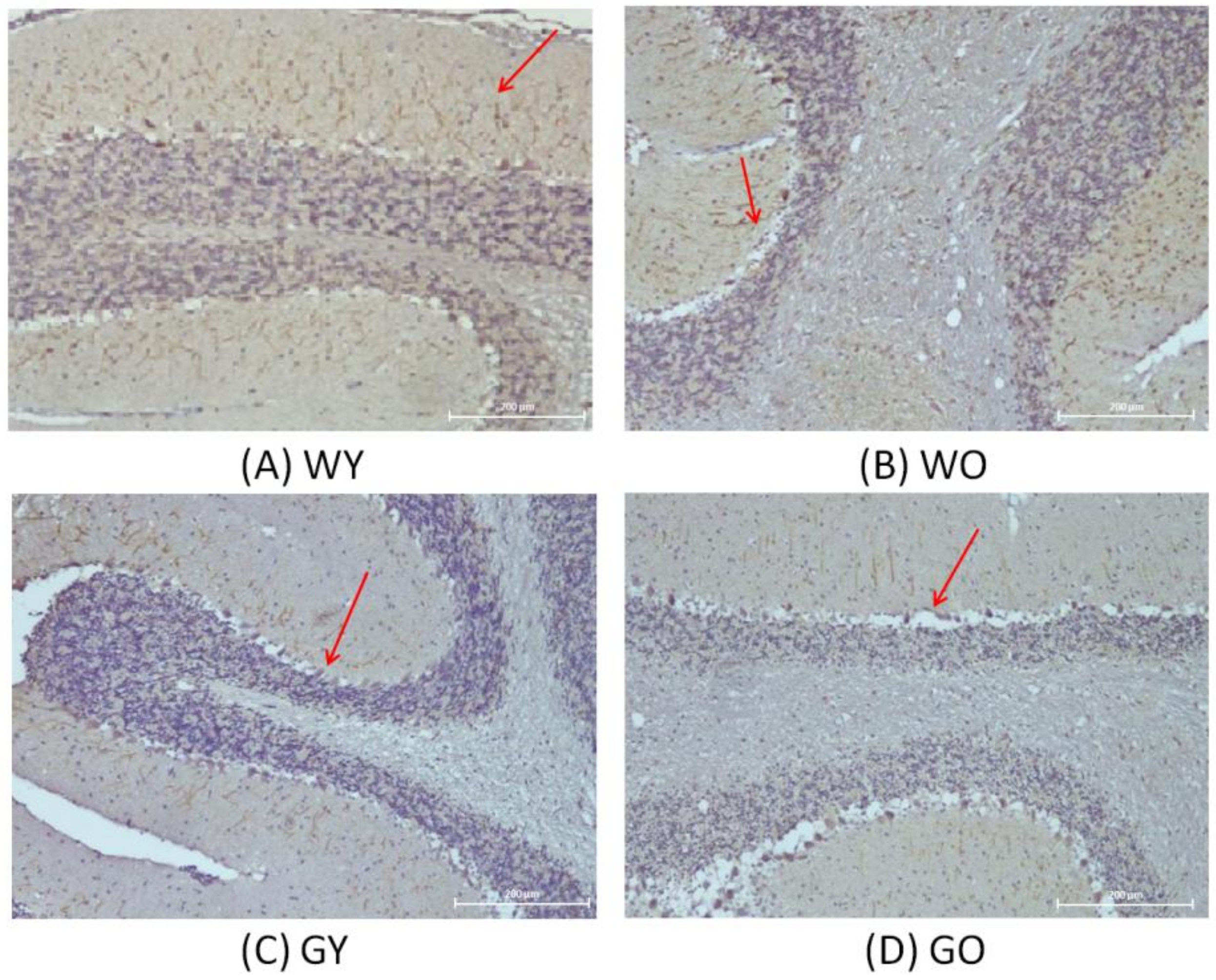
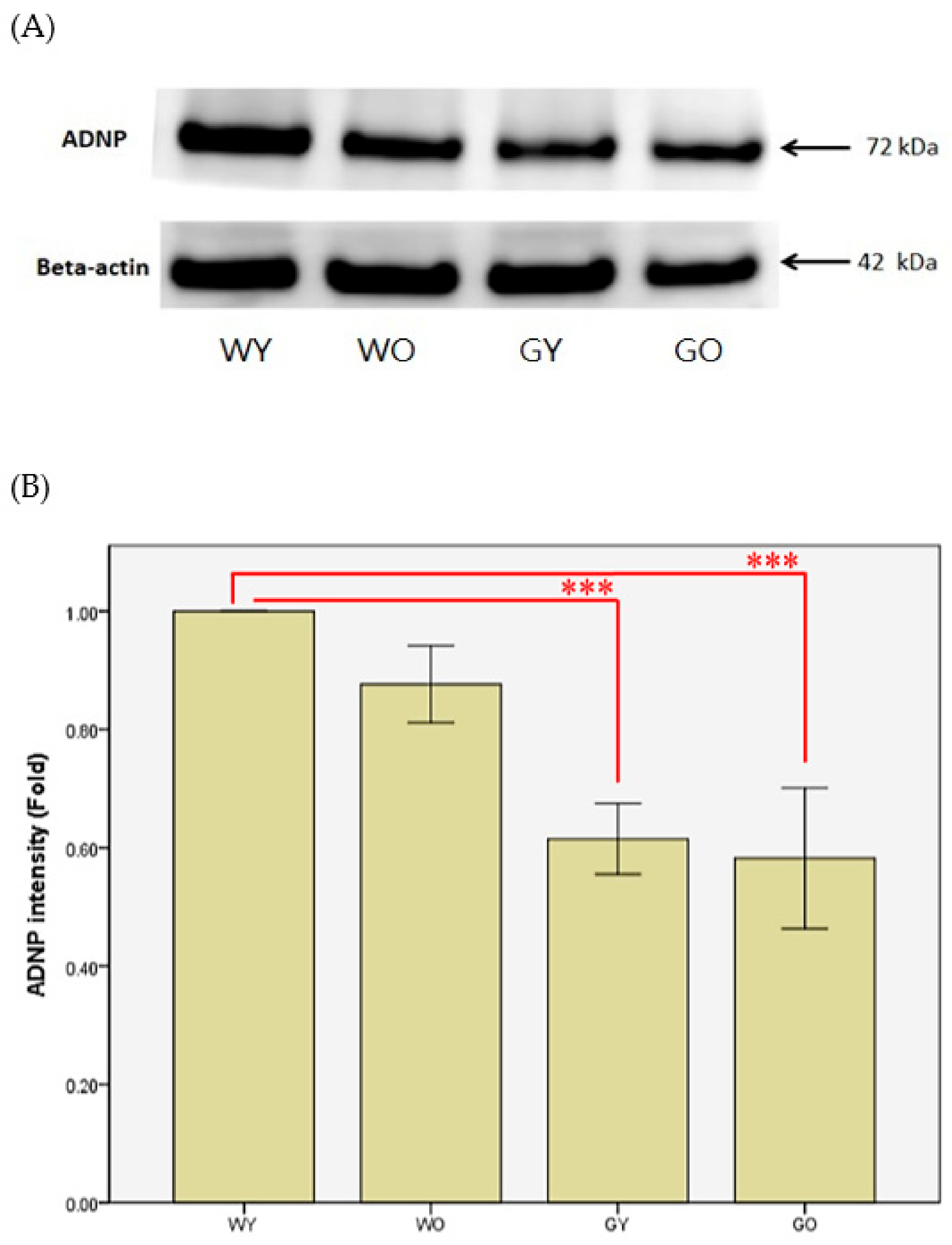
3.3. Activity-Dependent Neuroprotector (ADNP)
3.4. Receptor-Interacting Protein 1 (RIPK1)
3.5. Caspase 3
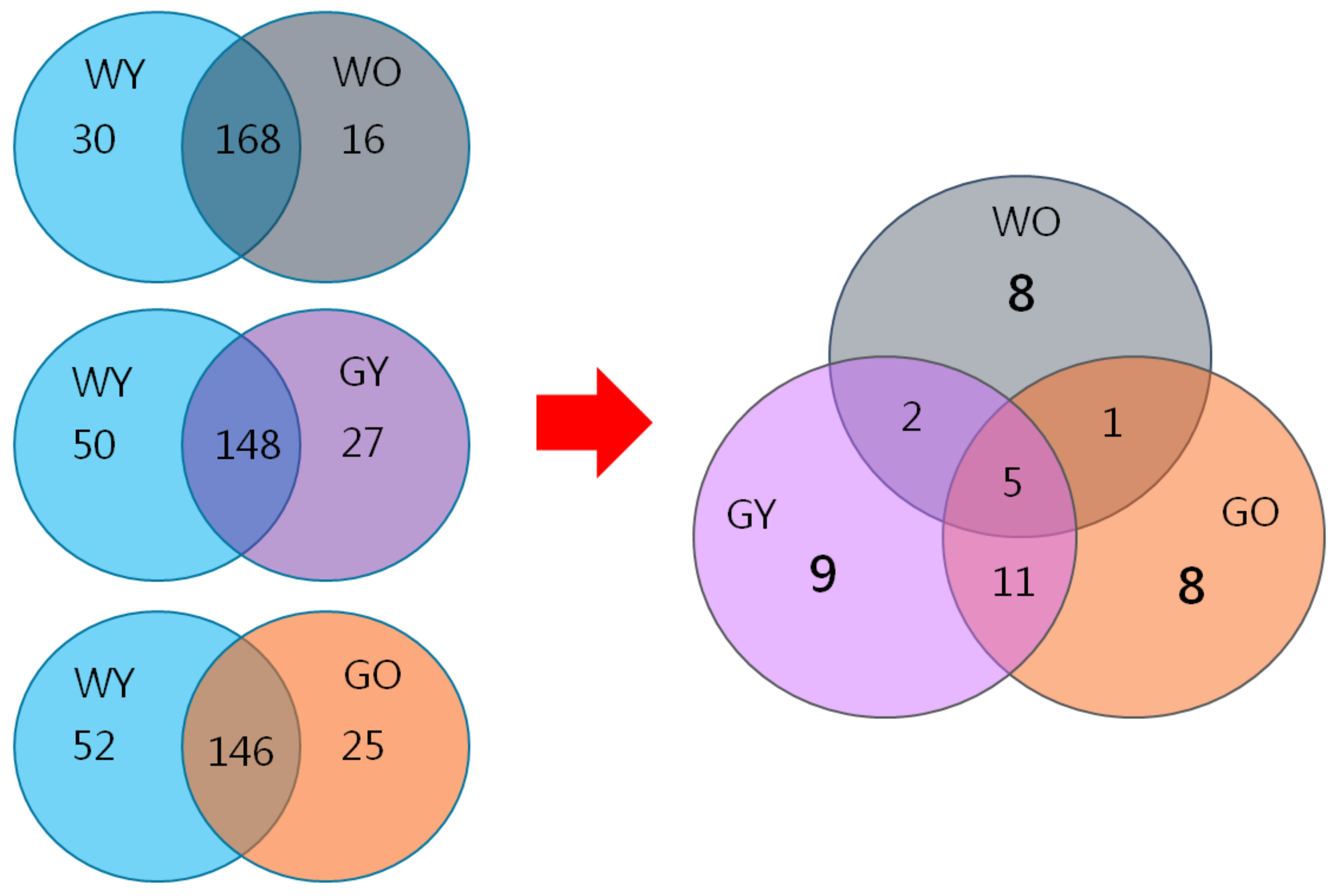
3.6. Biomarkers in Aged Mice- Proteome Database Search
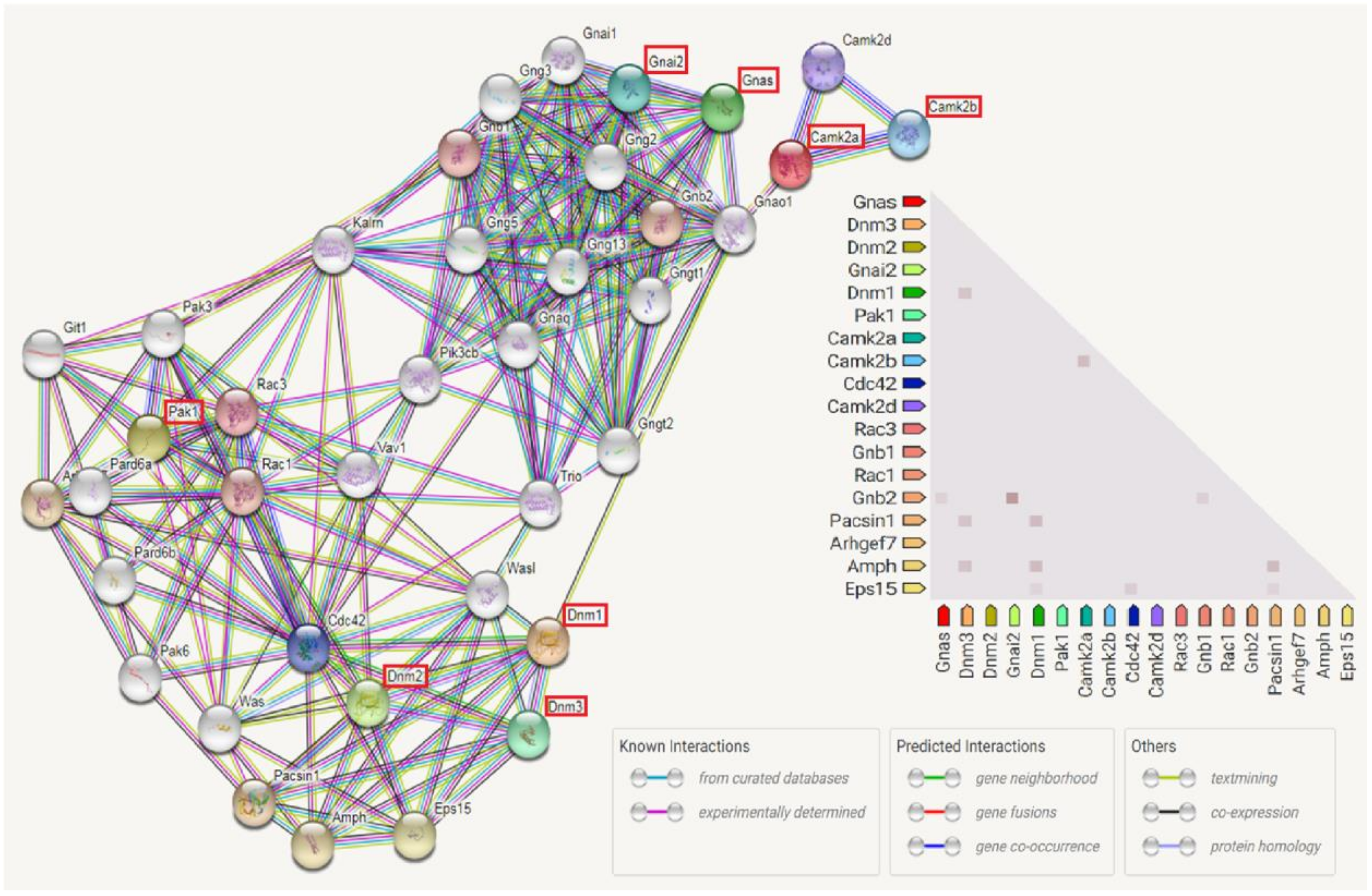
3.7. Enriched Biological Pathways by IPA Analysis
3.8. GNMT and Aging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Wt | wild-type mice |
| GNMT | glycine N-methyltransferase |
| WY | young wild-type mice |
| WO | older wild-type mice |
| GY | young GNMT−/− mice |
| GO | old GNMT−/− mice |
References
- Santos, A.L.; Lindner, A.B. Protein Posttranslational Modifications: Roles in Aging and Age-Related Disease. Oxid. Med. Cell. Longev. 2017, 5716409. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Walters, R.O.; Arias, E.; Diaz, A.; Burgos, E.S.; Guan, F.; Tiano, S.; Mao, K.; Green, C.L.; Qiu, Y.; Shah, H.; et al. Sarcosine Is Uniquely Modulated by Aging and Dietary Restriction in Rodents and Humans. Cell Rep. 2018, 25, 663–676.e6. [Google Scholar] [CrossRef]
- Obata, F.; Miura, M. Enhancing S-adenosyl-methionine catabolism extends Drosophila lifespan. Nat. Commun. 2015, 6, 8332. [Google Scholar] [CrossRef]
- Kerr, S.J. Competing methyltransferase systems. J. Biol. Chem. 1972, 247, 4248–4252. [Google Scholar] [CrossRef]
- Luka, Z.; Mudd, S.H.; Wagner, C. Glycine N-methyltransferase and regulation of S-adenosylmethionine levels. J. Biol. Chem. 2009, 284, 22507–22511. [Google Scholar] [CrossRef]
- Yeo, E.J.; Wagner, C. Purification and properties of pancreatic glycine N-methyltransferase. J. Biol. Chem. 1992, 267, 24669–24674. [Google Scholar] [CrossRef]
- DebRoy, S.; Kramarenko, I.I.; Ghose, S.; Oleinik, N.V.; Krupenko, S.A.; Krupenko, N.I. A novel tumor suppressor function of glycine N-methyltransferase is independent of its catalytic activity but requires nuclear localization. PLoS ONE 2013, 8, e70062. [Google Scholar] [CrossRef]
- Horgusluoglu, E.; Nudelman, K.; Nho, K.; Saykin, A.J. Adult neurogenesis and neurodegenerative diseases: A systems biology perspective. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2017, 174, 93–112. [Google Scholar] [CrossRef] [PubMed]
- Seibert, V.; Ebert, M.P.; Buschmann, T. Advances in clinical cancer proteomics: SELDI-ToF-mass spectrometry and biomarker discovery. Brief Funct. Genomic. Proteomic 2005, 4, 16–26. [Google Scholar] [CrossRef]
- Yang, M.H.; Chen, M.; Mo, H.H.; Tsai, W.C.; Chang, Y.C.; Chang, C.C.; Chen, K.C.; Wu, H.Y.; Yuan, C.H.; Lee, C.H.; et al. Utilizing Experimental Mouse Model to Identify Effectors of Hepatocellular Carcinoma Induced by HBx Antigen. Cancers 2020, 12, 409. [Google Scholar] [CrossRef]
- Horvatovich, P.; Végvári, Á.; Saul, J.; Park, J.G.; Qiu, J.; Syring, M.; Pirrotte, P.; Petritis, K.; Tegeler, T.J.; Aziz, M.; et al. In Vitro Transcription/Translation System: A Versatile Tool in the Search for Missing Proteins. J. Proteome Res. 2015, 14, 3441–3451. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Lin, W.L.; Lin, Y.J.; Tang, F.Y.; Chen, Y.M.; Chiang, E.P. A novel role of the tumor suppressor GNMT in cellular defense against DNA damage. Int. J. Cancer 2014, 134, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.P.; Wang, H.A.; Tsai, T.H.; Fan, A.; Hsu, C.L.; Chen, C.J.; Hong, C.J.; Chen, Y.M. Characterization of the neuropsychological phenotype of glycine N-methyltransferase-/- mice and evaluation of its responses to clozapine and sarcosine treatments. Eur. Neuropsychopharmacol. 2012, 22, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Popa, L.S.; Streng, M.L.; Ebner, T.J. Purkinje Cell Representations of Behavior: Diary of a Busy Neuron. Neuroscientist 2019, 25, 241–257. [Google Scholar] [CrossRef] [PubMed]
- Sassoè-Pognetto, M.; Patrizi, A. The Purkinje cell as a model of synaptogenesis and synaptic specificity. Brain Res. Bull. 2017, 129, 12–17. [Google Scholar] [CrossRef]
- Son, T.G.; Park, H.R.; Kim, S.J.; Kim, K.; Kim, M.S.; Ishigami, A.; Handa, S.; Maruyama, N.; Chung, H.Y.; Lee, J. Senescence marker protein 30 is up-regulated in kainate-induced hippocampal damage through ERK-mediated astrocytosis. J. Neurosci. Res. 2009, 87, 2890–2897. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Shirasawa, T.; Uchida, K.; Maruyama, N. Gene regulation of senescence marker protein-30 (SMP30): Coordinated up-regulation with tissue maturation and gradual down-regulation with aging. Mech. Ageing Dev. 1996, 87, 219–229. [Google Scholar] [CrossRef]
- Son, T.G.; Zou, Y.; Jung, K.J.; Yu, B.P.; Ishigami, A.; Maruyama, N.; Lee, J. SMP30 deficiency causes increased oxidative stress in brain. Mech. Ageing Dev. 2006, 127, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Gennet, N.; Herden, C.; Bubb, V.J.; Quinn, J.P.; Kipar, A. Expression of activity-dependent neuroprotective protein in the brain of adult rats. Histol. Histopathol. 2008, 23, 309–317. [Google Scholar] [CrossRef]
- Cappuyns, E.; Huyghebaert, J.; Vandeweyer, G.; Kooy, R.F. Mutations in ADNP affect expression and subcellular localization of the protein. Cell Cycle 2018, 17, 1068–1075. [Google Scholar] [CrossRef]
- Mollinedo, P.; Kapitansky, O.; Gonzalez-Lamuño, D.; Zaslavsky, A.; Real, P.; Gozes, I.; Gandarillas, A.; Fernandez-Luna, J.L. Cellular and animal models of skin alterations in the autism-related ADNP syndrome. Sci. Rep. 2019, 9, 736. [Google Scholar] [CrossRef]
- Furman, S.; Steingart, R.A.; Mandel, S.; Hauser, J.M.; Brenneman, D.E.; Gozes, I. Subcellular localization and secretion of activity-dependent neuroprotective protein in astrocytes. Neuron Glia Biol. 2004, 1, 193–199. [Google Scholar] [CrossRef]
- Pinhasov, A.; Mandel, S.; Torchinsky, A.; Giladi, E.; Pittel, Z.; Goldsweig, A.M.; Servoss, S.J.; Brenneman, D.E.; Gozes, I. Activity-dependent neuroprotective protein: A novel gene essential for brain formation. Brain Res. Dev. Brain Res. 2003, 144, 83–90. [Google Scholar] [CrossRef]
- Vulih-Shultzman, I.; Pinhasov, A.; Mandel, S.; Grigoriadis, N.; Touloumi, O.; Pittel, Z.; Gozes, I. Activity-dependent neuroprotective protein snippet NAP reduces tau hyperphosphorylation and enhances learning in a novel transgenic mouse model. J. Pharmacol. Exp. Ther. 2007, 323, 438–449. [Google Scholar] [CrossRef]
- Yang, M.H.; Chen, S.C.; Lin, Y.F.; Lee, Y.C.; Huang, M.Y.; Chen, K.C.; Wu, H.Y.; Lin, P.C.; Gozes, I.; Tyan, Y.C. Reduction of aluminum ion neurotoxicity through a small peptide application—NAP treatment of Alzheimer’s disease. J. Food Drug Anal. 2019, 27, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Mifflin, L.; Ofengeim, D.; Yuan, J. Receptor-interacting protein kinase 1 (RIPK1) as a therapeutic target. Nat. Rev. Drug Discov. 2020, 19, 553–571. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Wang, X.; Chen, G.; Quan, C.; Qu, S.; Tong, J. Inhibiting RIPK1 Limits Neuroinflammation and Alleviates Postoperative Cognitive Impairments in D-Galactose-Induced Aged Mice. Front. Behav. Neurosci. 2018, 12, 138. [Google Scholar] [CrossRef] [PubMed]
- Ofengeim, D.; Mazzitelli, S.; Ito, Y.; DeWitt, J.P.; Mifflin, L.; Zou, C.; Das, S.; Adiconis, X.; Chen, H.; Zhu, H.; et al. RIPK1 mediates a disease-associated microglial response in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, E8788–E8797. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yang, M.H.; Chang, M.M.; Tyan, Y.C.; Chen, Y.A. Tumor suppressor gene glycine N-methyltransferase and its potential in liver disorders and hepatocellular carcinoma. Toxicol. Appl. Pharmacol. 2019, 378, 114607. [Google Scholar] [CrossRef]
- Yuan, J.; Amin, P.; Ofengeim, D. Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat. Rev. Neurosci. 2019, 20, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Snigdha, S.; Smith, E.D.; Prieto, G.A.; Cotman, C.W. Caspase-3 activation as a bifurcation point between plasticity and cell death. Neurosci. Bull. 2012, 28, 14–24. [Google Scholar] [CrossRef]
- Louneva, N.; Cohen, J.W.; Han, L.Y.; Talbot, K.; Wilson, R.S.; Bennett, D.A.; Trojanowski, J.Q.; Arnold, S.E. Caspase-3 is enriched in postsynaptic densities and increased in Alzheimer’s disease. Am. J. Pathol. 2008, 173, 1488–1495. [Google Scholar] [CrossRef] [PubMed]
- Shimohama, S.; Tanino, H.; Fujimoto, S. Changes in caspase expression in Alzheimer’s disease: Comparison with development and aging. Biochem. Biophys. Res. Commun. 1999, 256, 381–384. [Google Scholar] [CrossRef]
- Ghoumari, A.M.; Wehrlé, R.; Bernard, O.; Sotelo, C.; Dusart, I. Implication of Bcl-2 and Caspase-3 in age-related Purkinje cell death in murine organotypic culture: An in vitro model to study apoptosis. Eur. J. Neurosci. 2000, 12, 2935–2949. [Google Scholar] [CrossRef]
- Kim, K.; Lee, S.G.; Kegelman, T.P.; Su, Z.Z.; Das, S.K.; Dash, R.; Dasgupta, S.; Barral, P.M.; Hedvat, M.; Diaz, P.; et al. Role of excitatory amino acid transporter-2 (EAAT2) and glutamate in neurodegeneration: Opportunities for developing novel therapeutics. J. Cell. Physiol. 2011, 226, 2484–2493. [Google Scholar] [CrossRef]
- Woltjer, R.L.; Duerson, K.; Fullmer, J.M.; Mookherjee, P.; Ryan, A.M.; Montine, T.J.; Kaye, J.A.; Quinn, J.F.; Silbert, L.; Erten-Lyons, D.; et al. Aberrant detergent-insoluble excitatory amino acid transporter 2 accumulates in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2010, 69, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Thai, D.R. Excitatory amino acid transporter EAAT-2 in tangle-bearing neurons in Alzheimer’s disease. Brain Pathol. 2002, 12, 405–411. [Google Scholar] [CrossRef]
- Bodhinathan, K.; Kumar, A.; Foster, T.C. Intracellular redox state alters NMDA receptor response during aging through Ca2/calmodulin-dependent protein kinase II. J. Neurosci. 2010, 30, 1914–1924. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.R.; Cheng, X.R.; Zhou, W.X.; Zhang, Y.X. Age-related expression of calcium/calmodulin-dependent protein kinase II A in the hippocampus and cerebral cortex of senescence accelerated mouse prone/8 mice is modulated by anti-Alzheimer’s disease drugs. Neuroscience 2009, 159, 308–315. [Google Scholar] [CrossRef]
- Warner, H.R. Superoxide dismutase, aging, and degenerative disease. Free Radic. Biol. Med. 1994, 17, 249–258. [Google Scholar] [CrossRef]
- De Haan, J.B.; Newman, J.D.; Kola, I. Cu/Zn superoxide dismutase mRNA and enzyme activity, and susceptibility to lipid peroxidation, increases with aging in murine brains. Brain Res. Mol. Brain Res. 1992, 13, 179–187. [Google Scholar] [CrossRef]
- Shen, J.; Wang, C.; Ying, J.; Xu, T.; McAlinden, A.; O’Keefe, R.J. Inhibition of 4-aminobutyrate aminotransferase protects against injury-induced osteoarthritis in mice. JCI Insight 2019, 4, e128568. [Google Scholar] [CrossRef]
- Wang, Z.; Lyons, B.; Truscott, R.J.; Schey, K.L. Human protein aging: Modification and crosslinking through dehydroalanine and dehydrobutyrine intermediates. Aging Cell 2014, 13, 226–234. [Google Scholar] [CrossRef]
- Sanz, R.L.; Ferraro, G.B.; Girouard, M.P.; Fournier, A.E. Ectodomain shedding of Limbic System-Associated Membrane Protein (LSAMP) by ADAM Metallopeptidases promotes neurite outgrowth in DRG neurons. Sci. Rep. 2017, 7, 7961. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Yu, J.; Yu, T.; Fu, Y.; Han, J. Acupuncture regulates the aging-related changes in gene profile expression of the hippocampus in senescence-accelerated mouse (SAMP10). Neurosci. Lett. 2006, 399, 11–16. [Google Scholar] [CrossRef]
- Fricker, L.D.; McKinzie, A.A.; Sun, J.; Curran, E.; Qian, Y.; Yan, L.; Patterson, S.D.; Courchesne, P.L.; Richards, B.; Levin, N.; et al. Identification and characterization of proSAAS, a granin-like neuroendocrine peptide precursor that inhibits prohormone processing. J. Neurosci. 2000, 20, 639–648. [Google Scholar] [CrossRef]
- Jarvela, T.S.; Lam, H.A.; Helwig, M.; Lorenzen, N.; Otzen, D.E.; McLean, P.J.; Maidment, N.T.; Lindberg, I. The neural chaperone proSAAS blocks α-synuclein fibrillation and neurotoxicity. Proc. Natl. Acad. Sci. USA 2016, 113, E4708–E4715. [Google Scholar] [CrossRef]
- Wada, M.; Ren, C.H.; Koyama, S.; Arawaka, S.; Kawakatsu, S.; Kimura, H.; Nagasawa, H.; Kawanami, T.; Kurita, K.; Daimon, M.; et al. A human granin-like neuroendocrine peptide precursor (proSAAS) immunoreactivity in tau inclusions of Alzheimer’s disease and parkinsonism-dementia complex on Guam. Neurosci. Lett. 2004, 356, 49–52. [Google Scholar] [CrossRef]
- Greenwood, M.P.; Greenwood, M.; Romanova, E.V.; Mecawi, A.S.; Paterson, A.; Sarenac, O.; Japundžić-Žigon, N.; Antunes-Rodrigues, J.; Paton, J.F.R.; Sweedler, J.V.; et al. The effects of aging on biosynthetic processes in the rat hypothalamic osmoregulatory neuroendocrine system. Neurobiol. Aging 2018, 65, 178–191. [Google Scholar] [CrossRef]
- Skillbäck, T.; Delsing, L.; Synnergren, J.; Mattsson, N.; Janelidze, S.; Nägga, K.; Kilander, L.; Hicks, R.; Wimo, A.; Winblad, B.; et al. CSF/serum albumin ratio in dementias: A cross-sectional study on 1861 patients. Neurobiol. Aging 2017, 59, 1–9. [Google Scholar] [CrossRef]
- Luo, L. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 2000, 1, 173–180. [Google Scholar] [CrossRef]
- Boettner, B.; Van Aelst, L. The role of Rho GTPases in disease development. Gene 2002, 286, 155–174. [Google Scholar] [CrossRef]
- Yu, J.; Gu, X.; Yi, S. Ingenuity Pathway Analysis of Gene Expression Profiles in Distal Nerve Stump following Nerve Injury: Insights into Wallerian Degeneration. Front. Cell Neurosci. 2016, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chadwick, W.; Park, S.S.; Zhou, Y.; Silver, N.; Martin, B.; Maudsley, S. Gonadotropin-releasing hormone receptor system: Modulatory role in aging and neurodegeneration. CNS Neurol. Disord. Drug Targets 2010, 9, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, C.A.; Manilall, A. Gonadotropin-Releasing Hormone (GnRH) Receptor Structure and GnRH Binding. Front. Endocrinol. 2017, 8, 274. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, B. Gonadotropin-Releasing Hormone and Its Role in the Enteric Nervous System. Front. Endocrinol. 2017, 8, 110. [Google Scholar] [CrossRef]
- Balasubramanian, R.; Dwyer, A.; Seminara, S.B.; Pitteloud, N.; Kaiser, U.B.; Crowley, W.F., Jr. Human GnRH deficiency: A unique disease model to unravel the ontogeny of GnRH neurons. Neuroendocrinology 2010, 92, 81–99. [Google Scholar] [CrossRef]
- Shaw, N.D.; Srouji, S.S.; Histed, S.N.; McCurnin, K.E.; Hall, J.E. Aging attenuates the pituitary response to gonadotropin-releasing hormone. J. Clin. Endocrinol. Metab. 2009, 94, 3259–3264. [Google Scholar] [CrossRef]
- Pastore, D.; Pacifici, F.; Dave, K.R.; Palmirotta, R.; Bellia, A.; Pasquantonio, G.; Guadagni, F.; Donadel, G.; Di Daniele, N.; Abete, P.; et al. Age-Dependent Levels of Protein Kinase Cs in Brain: Reduction of Endogenous Mechanisms of Neuroprotection. Int. J. Mol. Sci. 2019, 20, 3544. [Google Scholar] [CrossRef]
- Kim, H.; Oh, J.Y.; Choi, S.L.; Nam, Y.J.; Jo, A.; Kwon, A.; Shin, E.Y.; Kim, E.G.; Kim, H.K. Down-regulation of p21-activated serine/threonine kinase 1 is involved in loss of mesencephalic dopamine neurons. Mol. Brain 2016, 9, 45. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoo, D.Y.; Jung, H.Y.; Kim, J.W.; Yim, H.S.; Kim, D.W.; Nam, H.; Suh, J.G.; Choi, J.H.; Won, M.H.; Yoon, Y.S.; et al. Reduction of dynamin 1 in the hippocampus of aged mice is associated with the decline in hippocampal-dependent memory. Mol. Med. Rep. 2016, 14, 4755–4760. [Google Scholar] [CrossRef]
- Jiang, S.; Shao, C.; Tang, F.; Wang, W.; Zhu, X. Dynamin-like protein 1 cleavage by calpain in Alzheimer’s disease. Aging Cell 2019, 18, e12912. [Google Scholar] [CrossRef] [PubMed]
- Young, L.T.; Warsh, J.J.; Li, P.P.; Siu, K.P.; Becker, L.; Gilbert, J.; Hornykiewicz, O.; Kish, S.J. Maturational and aging effects on guanine nucleotide binding protein immunoreactivity in human brain. Brain Res. Dev. Brain Res. 1991, 61, 243–248. [Google Scholar] [CrossRef]
- De Oliveira, P.G.; Ramos, M.L.S.; Amaro, A.J.; Dias, R.A.; Vieira, S.I. Gi/o-Protein Coupled Receptors in the Aging Brain. Front. Aging Neurosci. 2019, 11, 89. [Google Scholar] [CrossRef]
- Rose, A.J.; Kiens, B.; Richter, E.A. Ca2-calmodulin-dependent protein kinase expression and signalling in skeletal muscle during exercise. J. Physiol. 2006, 574, 889–903. [Google Scholar] [CrossRef]
- Ghosh, A.; Giese, K.P. Calcium/calmodulin-dependent kinase II and Alzheimer’s disease. Mol. Brain 2015, 8, 78. [Google Scholar] [CrossRef]
- Oka, M.; Fujisaki, N.; Maruko-Otake, A.; Ohtake, Y.; Shimizu, S.; Saito, T.; Hisanaga, S.I.; Iijima, K.M.; Ando, K. Ca2/calmodulin-dependent protein kinase II promotes neurodegeneration caused by tau phosphorylated at Ser262/356 in a transgenic Drosophila model of tauopathy. J. Biochem. 2017, 162, 335–342. [Google Scholar] [CrossRef]
- Liu, S.P.; Li, Y.S.; Chen, Y.J.; Chiang, E.P.; Li, A.F.; Lee, Y.H.; Tsai, T.F.; Hsiao, M.; Huang, S.F.; Chen, Y.M. Glycine N-methyltransferase−/− mice develop chronic hepatitis and glycogen storage disease in the liver. Hepatology 2007, 46, 1413–1425. [Google Scholar] [CrossRef]
- Carrasco, M.; Rabaneda, L.G.; Murillo-Carretero, M.; Ortega-Martínez, S.; Martínez-Chantar, M.L.; Woodhoo, A.; Luka, Z.; Wagner, C.; Lu, S.C.; Mato, J.M.; et al. Glycine N-methyltransferase expression in the hippocampus and its role in neurogenesis and cognitive performance. Hippocampus 2014, 24, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Quadri, P.; Fragiacomo, C.; Pezzati, R.; Zanda, E.; Forloni, G.; Tettamanti, M.; Lucca, U. Homocysteine, folate, and vitamin B-12 in mild cognitive impairment, Alzheimer disease, and vascular dementia. Am. J. Clin. Nutr. 2004, 80, 114–122. [Google Scholar] [CrossRef] [PubMed]



| Swiss-Port TrEMBL Accession Number | Protein Name | MW | Score | Match Queries | pI | Sequence Coverage (%) | Matched Peptide | Subcellular Location | Protein Function |
|---|---|---|---|---|---|---|---|---|---|
| WO mice | |||||||||
| P43006 | Excitatory amino acid transporter 2 | 61,990 | 43 | 4 | 6.24 | 8 | MASTEGANNMPK.Q+Deamidated (NQ) K.KNDEVSSLDAFLDLIR.N R.CLEDNLGIDK.R +Carboxymethyl (C) K.SELDTIDSQHR.M | Cell membrane; Multi-pass membrane protein | Functions as a symporter that transports one amino acid molecule together with two or three Na+ ions and one proton, in parallel with the counter-transport of one K+ ion. |
| P15864 | Histone H1.2 | 21,254 | 100 | 5 | 11 | 18 | K.ERSGVSLAALK.K +Phospho (ST) R.SGVSLAALKK.A K.KALAAAGYDVEKNNSR.I +Deamidated (NQ) K.ALAAAGYDVEKNNSR.I K.GTGASGSFKLNK.K +Phospho (ST) | Nucleus; Chromosome | Histones H1 are necessary for the condensation of nucleosome chains into higher-order structured fibers. Acts also as a regulator of individual gene transcription through chromatin remodeling, nucleosome spacing and DNA methylation. |
| P11798 | Calcium/calmodulin-dependent protein kinase type II subunit alpha | 54,081 | 31 | 2 | 6.61 | 7 | R.KQEIIKVTEQLIEAISNGDFESYTK.M+2 Phospho (ST) R.FYFENLWSR.N | Cytoplasm; Synapse | Calcium/calmodulin-dependent protein kinase that functions autonomously after Ca2/calmodulin-binding and autophosphorylation, and is involved in synaptic plasticity, neurotransmitter release and long-term potentiation. |
| Q923T9 | Calcium/calmodulin-dependent protein kinase type II subunit gamma | 59,569 | 55 | 4 | 7.32 | 9 | MATTATCTR.F +Carboxymethyl (C); 2 Phospho (ST) K.IINTKKLSAR.D +Deamidated (NQ); 2 Phospho (ST) R.DLKPENLLLASK.C K.AGAYDFPSPEWDTVTPEAK.N | Sarcoplasmic reticulum membrane; Peripheral membrane protein; Cytoplasmic side | Calcium/calmodulin-dependent protein kinase in the central nervous system, it is involved in the regulation of neurite formation and arborization. It may participate in the promotion of dendritic spine and synapse formation and maintenance of synaptic plasticity which enables long-term potentiation (LTP) and hippocampus-dependent learning. |
| Q8C650 | Septin-10 | 52,388 | 40 | 2 | 6.17 | 4 | R.AQTYELQESNVR.L +3 Deamidated (NQ); Phospho (ST) K.VNIIPLIAK.A | Cytoskeleton; Cytoplasm | May play a role in cytokinesis. |
| P08228 | Superoxide dismutase [Cu-Zn] | 15,933 | 69 | 2 | 6.02 | 16 | K.GDGPVQGTIHFEQK.A R.HVGDLGNVTAGK.D | Nucleus; Cytoplasm | Destroys radicals which are normally produced within the cells and which are toxic to biological systems. |
| Q62277 | Synaptophysin | 34,002 | 37 | 3 | 4.82 | 7 | K.MATDPENIIK.E K.MATDPENIIK.E +Deamidated (NQ); Oxidation (M); Phospho (ST) K.EMPMCRQTGNTCK.E +Carboxymethyl (C); Deamidated (NQ) | Synaptic vesicle Membrane; Membrane protein; Synaptosome | Involved in the regulation of short-term and long-term synaptic plasticity. |
| Q9R0P9 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | 24,822 | 71 | 4 | 5.14 | 24 | MQLKPMEINPEMLNK.V+Deamidated (NQ); 2 Oxidation (M) K.LEFEDGSVLK.Q K.QFLSETEK.L R.MPFPVNHGASSEDSLLQDAAK.V | Endoplasmic reticulum Membrane; Cytoplasm | Ubiquitin-protein hydrolase was bound to free monoubiquitin and may prevent its degradation in lysosomes. The homodimer may have ATP-independent ubiquitin ligase activity. |
| GY mice | |||||||||
| P61922 | 4-aminobutyrate aminotransferase, mitochondrial | 56,416 | 27 | 1 | 8.35 | 3 | K.AIHKIDIPSFDWPIAPFPR.L | Mitochondrion matrix | Catalyzes the conversion of gamma-aminobutyrate and L-beta-aminoisobutyrate to succinate semialdehyde and methylmalonate semialdehyde, respectively. Can also convert delta-aminovalerate and beta-alanine. |
| P08752 | Guanine nucleotide-binding protein G(i) subunit alpha-2 | 40,463 | 41 | 3 | 5.28 | 10 | K.LLLLGAGESGK.S K.STIVKQMK.I +Oxidation (M); Phospho (ST) R.QYRAVVYSNTIQSIMAIVK.A +2 Deamidated (NQ); Oxidation (M) | Plasma membrane; Cell membrane; Cytoskeleton | The G(i) proteins are involved in hormonal regulation of adenylate cyclase: they inhibit the cyclase in response to beta-adrenergic stimuli. May play a role in cell division. |
| Q8BLK3 | Limbic system-associated membrane protein | 38,063 | 25 | 5 | 6.21 | 18 | R.GTDNITVR.Q +Phospho (ST) R.HALEYSLR.I K.AANEVSSADVK.Q +Deamidated (NQ) K.AANEVSSADVKQVK.V K.VTVNYPPTITESKSNEATTGRQASLK.C +2 Deamidated (NQ); 6 Phospho (ST); Phospho (Y) | Cell membrane | Mediates selective neuronal growth and axon targeting. Contributes to the guidance of developing axons and remodeling of mature circuits in the limbic system. Essential for normal growth of the hyppocampal mossy fiber projection. |
| P35486 | Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial | 43,204 | 45 | 2 | 8.49 | 4 | K.ADQLYK.Q +Deamidated (NQ); Phospho (Y) R.SKSDPIMLLKDR.M | Mitochondrion matrix | The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO2, and thereby links the glycolytic pathway to the tricarboxylic cycle. |
| Q8CGM2 | Retinitis pigmentosa 1-like 1 protein | 199,562 | 26 | 4 | 6.06 | 3 | K.VCMNEDGSLSVEMK.V +Carboxymethyl (C); 2 Phospho (ST) K.VRLEESPKYQEMLR.L +Oxidation (M) R.MLSQEDLGTVQGADEK.Q K.GEDEGSAGSLACTQVGGK.V +3 Phospho (ST) | Cytoskeleton | Plays a role in the organization of outer segment of rod and cone photoreceptors. |
| P31650 | Sodium- and chloride-dependent GABA transporter 3 | 69,914 | 24 | 1 | 6.66 | 2 | K.VKGDGTISAITEKETHF. | Membrane | Terminates the action of GABA by its high affinity sodium-dependent reuptake into presynaptic terminals. Can also transport beta-alanine and taurine. |
| Q9CWF2 | Tubulin beta-2B chain | 49,921 | 362 | 14 | 4.78 | 41 | K.FWEVISDEHGIDPTGSYHGDSDLQLER.I R.INVYYNEATGNKYVPR.A R.AILVDLEPGTMDSVR.S R.AILVDLEPGTMDSVR.S +Oxidation (M) R.SGPFGQIFRPDNFVFGQSGAGNNWAK.G K.GHYTEGAELVDSVLDVVR.K K.GHYTEGAELVDSVLDVVRK.E K.LAVNMVPFPR.L R.LHFFMPGFAPLTSR.G R.LHFFMPGFAPLTSR.G +Oxidation (M) R.ALTVPELTQQMFDSK.N R.YLTVAAIFR.G K.NSSYFVEWIPNNVK.T K.MSATFIGNSTAIQELFKR.I | Cytoskeleton | Tubulin plays a critical role in proper axon guidance in both central and peripheral axon tracts. Implicated in neuronal migration. |
| P20029 | Endoplasmic reticulum chaperone BiP | 72,377 | 61 | 3 | 5.07 | 7 | R.IINEPTAAAIAYGLDK.R K.SQIFSTASDNQPTVTIK.V +2 Deamidated (NQ) K.DNHLLGTFDLTGIPPAPR.G +Phospho (ST) | Endoplasmic reticulum; Cytoplasm | Involved in the correct folding of proteins and degradation of misfolded proteins via its interaction with DNAJC10/ERdj5, probably to facilitate the release of DNAJC10/ERdj5 from its substrate. |
| Q9QXV0 | ProSAAS | 27,254 | 48 | 1 | 5.68 | 6 | R.AVPRGEAAGAVQELAR.A | Extracellular region or secreted; Golgi apparatus | Proposed be a specific endogenous inhibitor of PCSK1. ProSAAS and Big PEN-LEN, both containing the C-terminal inhibitory domain, but not the processed peptides reduce PCSK1 activity in the endoplasmic reticulum and Golgi. |
| GO mice | |||||||||
| P07724 | Serum albumin | 68,648 | 49 | 2 | 5.75 | 3 | K.CSSMQK.F +Carboxymethyl (C); Oxidation (M); Phospho (ST) R.YTQKAPQVSTPTLVEAAR.N | Extracellular region or secreted | The main function of serum albumin is the regulation of the colloidal osmotic pressure of blood. |
| O55143 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 114,784 | 38 | 2 | 5.23 | 3 | K.TVEEVLGHFGVNESTGLSLEQVKK.L K.TGTLTTNQMSVCR.M +Deamidated (NQ); Oxidation (M) | Endoplasmic reticulum | Acts as a regulator of TNFSF11-mediated Ca2 signaling pathways via its interaction with TMEM64 which is critical for the TNFSF11-induced CREB1 activation and mitochondrial ROS generation necessary for proper osteoclast generation. |
| Q3U319 | E3 ubiquitin-protein ligase BRE1B | 113,897 | 37 | 4 | 6.13 | 4 | R.RLQDLATQLQEK.H +Deamidated (NQ) R.TNERLKVALR.S R.EVQAEIGK.L K.ARLTCPCCNTRK.K +Carboxymethyl (C); Phospho (ST) | Nucleus | H2BK120ub1 gives a specific tag for epigenetic transcriptional activation and is also prerequisite for histone H3 ’Lys-4’ and ’Lys-79’ methylation. |
| Q99PT1 | Rho GDP-dissociation inhibitor 1 | 23,393 | 32 | 1 | 5.12 | 8 | K.SIQEIQELDKDDESLRK.Y | Cytoplasm | Regulates the GDP/GTP exchange reaction of the Rho proteins by inhibiting the dissociation of GDP from them, and the subsequent binding of GTP to them. Retains Rho proteins such as CDC42, RAC1 and RHOA in an inactive cytosolic pool, regulating their stability and protecting them from degradation. |
| P70168 | Importin subunit beta-1 | 97,122 | 33 | 1 | 4.68 | 1 | R.AAVENLPTFLVELSR.V | Nucleus; Cytoplasm | Functions in nuclear protein import, either in association with an adapter protein, like an importin-alpha subunit, which binds to nuclear localization signals (NLS) in cargo substrates, or by acting as autonomous nuclear transport receptor. |
| P60761 | Neurogranin | 7492 | 42 | 2 | 6.54 | 41 | MDCCTESACSKPDDDILDIPLDDPGANAAAAK.I +2 Carboxymethyl (C); Oxidation (M); Phospho (ST) MDCCTESACSKPDDDILDIPLDDPGANAAAAK.I +3 Carboxymethyl (C); Oxidation (M); Phospho (ST) | Cytoplasm; Synapse; Dendritic spine | Regulates the affinity of calmodulin for calcium. Involved in synaptic plasticity and spatial learning. |
| P32848 | Parvalbumin alpha | 11,923 | 59 | 1 | 5.02 | 23 | K.TLLAAGDKDGDGKIGVEEFSTLVAES. | Cytoskeleton Cytosol Nucleus | In muscle, parvalbumin is thought to be involved in relaxation after contraction. It binds two calcium ions. |
| Q8R0A5 | Transcription elongation factor A protein-like 3 | 22,455 | 45 | 2 | 5.33 | 6 | R.AAEKRPAEDYVPR.K K.RPAEDYVPR.K | Nucleus | May be involved in transcriptional regulation. |
| Ingenuity Canonical Pathways | -log(p-Value) | Ratio | Molecules |
|---|---|---|---|
| GnRH Signaling | 5.92 | 0.0729 | PAK1, DNM1, GNAI2, CAMK2A, GNAS, DNM3, DNM2, CAMK2B |
| Clathrin-mediated Endocytosis Signaling | 5.6 | 0.0654 | DNM1, SYNJ1, ACTB, PPP3R1, DNM3, PPP3CA, DNM2 |
| Huntington’s Disease Signaling | 4.73 | 0.0483 | DNM1, NSF, PACSIN1, HSPA1A/HSPA1B, DNM3, SNCA, DNM2 |
| Remodeling of Epithelial Adherens Junctions | 4.57 | 0.133 | DNM1, ACTB, DNM3, DNM2 |
| Role of NFAT in Cardiac Hypertrophy | 4.02 | 0.0469 | GNAI2, CAMK2A, GNAS, PPP3R1, PPP3CA, CAMK2B |
| GM-CSF Signaling | 3.68 | 0.08 | CAMK2A, PPP3R1, PPP3CA, CAMK2B |
| cAMP-mediated signaling | 3.46 | 0.037 | GNAI2, CAMK2A, GNAS, PPP3R1, PPP3CA, CAMK2B |
| B Cell Receptor Signaling | 3.4 | 0.0467 | CAMK2A, SYNJ1, PPP3R1, PPP3CA, CAMK2B |
| Breast Cancer Regulation by Stathmin1 | 3.36 | 0.0459 | STMN1, GNAI2, CAMK2A, GNAS, CAMK2B |
| iCOS-iCOSL Signaling in T Helper Cells | 3.34 | 0.0656 | CAMK2A, PPP3R1, PPP3CA, CAMK2B |
| nNOS Signaling in Neurons | 3.22 | 0.107 | CAMK2A, PPP3R1, PPP3CA |
| PKCθ Signaling in T Lymphocytes | 3.21 | 0.0606 | CAMK2A, PPP3R1, PPP3CA, CAMK2B |
| Gap Junction Signaling | 3.18 | 0.042 | GNAI2, GNAS, ACTB, PPP3R1, PPP3CA |
| Protein Kinase A Signaling | 3.17 | 0.0328 | GNAI2, CAMK2A, GNAS, PPP3R1, PPP3CA, CAMK2B |
| fMLP Signaling in Neutrophils | 3.09 | 0.0563 | GNAI2, GNAS, PPP3R1, PPP3CA |
| Crosstalk between Dendritic Cells and Natural Killer Cells | 3.01 | 0.0909 | CAMK2A, ACTB, CAMK2B |
| PI3K Signaling in B Lymphocytes | 2.98 | 0.0526 | CAMK2A, PPP3R1, PPP3CA, CAMK2B |
| Synaptic Long Term Potentiation | 2.96 | 0.0519 | CAMK2A, PPP3R1, PPP3CA, CAMK2B |
| Chemokine Signaling | 2.83 | 0.0789 | GNAI2, CAMK2A, CAMK2B |
| Role of NFAT in Regulation of the Immune Response | 2.8 | 0.0471 | GNAI2, GNAS, PPP3R1, PPP3CA |
| Melatonin Signaling | 2.73 | 0.0732 | GNAI2, CAMK2A, CAMK2B |
| Dopamine-DARPP32 Feedback in cAMP Signaling | 2.57 | 0.0408 | GNAI2, GNAS, PPP3R1, PPP3CA |
| GABA Receptor Signaling | 2.56 | 0.0638 | DNM1, NSF, GNAS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.-H.; Chen, Y.-M.A.; Tu, S.-C.; Chi, P.-L.; Chuang, K.-P.; Chang, C.-C.; Lee, C.-H.; Chen, Y.-L.; Lee, C.-H.; Yuan, C.-H.; et al. Utilizing an Animal Model to Identify Brain Neurodegeneration-Related Biomarkers in Aging. Int. J. Mol. Sci. 2021, 22, 3278. https://doi.org/10.3390/ijms22063278
Yang M-H, Chen Y-MA, Tu S-C, Chi P-L, Chuang K-P, Chang C-C, Lee C-H, Chen Y-L, Lee C-H, Yuan C-H, et al. Utilizing an Animal Model to Identify Brain Neurodegeneration-Related Biomarkers in Aging. International Journal of Molecular Sciences. 2021; 22(6):3278. https://doi.org/10.3390/ijms22063278
Chicago/Turabian StyleYang, Ming-Hui, Yi-Ming Arthur Chen, Shan-Chen Tu, Pei-Ling Chi, Kuo-Pin Chuang, Chin-Chuan Chang, Chiang-Hsuan Lee, Yi-Ling Chen, Che-Hsin Lee, Cheng-Hui Yuan, and et al. 2021. "Utilizing an Animal Model to Identify Brain Neurodegeneration-Related Biomarkers in Aging" International Journal of Molecular Sciences 22, no. 6: 3278. https://doi.org/10.3390/ijms22063278
APA StyleYang, M.-H., Chen, Y.-M. A., Tu, S.-C., Chi, P.-L., Chuang, K.-P., Chang, C.-C., Lee, C.-H., Chen, Y.-L., Lee, C.-H., Yuan, C.-H., & Tyan, Y.-C. (2021). Utilizing an Animal Model to Identify Brain Neurodegeneration-Related Biomarkers in Aging. International Journal of Molecular Sciences, 22(6), 3278. https://doi.org/10.3390/ijms22063278








