Abstract
Identifying biomarkers is essential for early diagnosis of neurodegenerative diseases (NDs). Large (LEVs) and small extracellular vesicles (SEVs) are extracellular vesicles (EVs) of different sizes and biological functions transported in blood and they may be valid biomarkers for NDs. The aim of our study was to investigate common and different miRNA signatures in plasma derived LEVs and SEVs of Alzheimer’s disease (AD), Parkinson’s disease (PD), Amyotrophic Lateral Sclerosis (ALS) and Fronto-Temporal Dementia (FTD) patients. LEVs and SEVs were isolated from plasma of patients and healthy volunteers (CTR) by filtration and differential centrifugation and RNA was extracted. Small RNAs libraries were carried out by Next Generation Sequencing (NGS). MiRNAs discriminate all NDs diseases from CTRs and they can provide a signature for each NDs. Common enriched pathways for SEVs were instead linked to ubiquitin mediated proteolysis and Toll-like receptor signaling pathways and for LEVs to neurotrophin signaling and Glycosphingolipid biosynthesis pathway. LEVs and SEVs are involved in different pathways and this might give a specificity to their role in the spreading of the disease. The study of common and different miRNAs transported by LEVs and SEVs can be of great interest for biomarker discovery and for pathogenesis studies in neurodegeneration.
1. Introduction
Neurodegenerative disorders (NDs) are a group of diseases characterized by loss of neurons within the brain and/or spinal cord [1]. They include Alzheimer’s disease (AD), Parkinson’s disease (PD), Amyotrophic Lateral Sclerosis (ALS) and Fronto-Temporal Dementia (FTD) [2]. Each of these disorders is characterized by specific features, both clinical and pathological involving characteristic central nervous regions [3].
AD is characterized by the extracellular accumulation of beta amyloid (Aβ) peptide detectable as Aβ plaques and intracellular Tau protein in the parenchyma and cerebrovasculature of the brain [4]. In PD degeneration of dopaminergic pigmented neurons in the substantia nigra (SN) and accumulation of α-synuclein protein, which is the main component of Lewy bodies (LBs), are the main features [5]. ALS is a disease characterized by motor neurons death and one of the main pathological hallmarks is given by specific alterations of SOD1 [6,7,8,9,10], and aggregation of TDP-43 [11]. In FTD, there is a deregulation of RNA-binding proteins (RBPs) and aggregation of proteins in the frontal and temporal lobes with microvacuolation, neuronal loss and astrocytic gliosis [12]. TDP-43 and Tau aggregates are hallmarks of FTD [13].
The aberrant RNA metabolism processing converges as a common factor in the pathogenesis of these diseases [14,15]. Abnormal RNA metabolism is associated with disease-specific alterations in RNA-binding proteins (RBPs), and in non-coding RNAs, such as microRNAs (miRNA), transfer RNAs (tRNA) and long-noncoding RNAs (lncRNA) [16].
Several common and specific mechanisms of NDs are described in the literature, however, no biomarkers are available to identify the onset, progression and comorbidity of those diseases. Brain cells release extracellular vesicles (EVs), which can go through the brain barrier [17,18] and blood derived EVs can be used also to monitor disease processes occurring in the brain [19,20,21]. EVs, spherical vesicles heterogeneous in size (30 nm–1 µm in diameter) are transporters of receptors, bioactive lipids, proteins, and nucleic acids, such as mRNAs, lncRNAs and miRNAs [22,23,24,25]. EVs are classified as: exosomes (EXOs), microvesicles (MVs), and apoptotic bodies [22,23]. EXOs are secreted membrane vesicles (approximately 30–150 nm in diameter) formed intracellularly and released from exocytosis of multivesicular bodies, whereas apoptotic bodies (approximately 1000–4000 nm in diameter) are released by dying cells. MVs (approximately 100–1000 nm in diameter) are shed from cells by outward protrusion (or budding) of a plasma membrane followed by fission of their membrane stalk [22,23]. However, the guidelines of the International Society for the study of Extracellular Vesicle (ISEV) released in 2018 declare that MVs and EXOs cannot be distinguished on a particular biogenesis pathway and so they can be distinguished in small extracellular vesicles (SEVs) (30–130 nm) and large extracellular vesicles (LEVs) (130–1000 nm) mainly on their size [24].
Several studies on the role of EVs in NDs are available in the literature. Some studies have examined miRNAs and RNAs in EVs isolated from cultured cell media from the Central Nervous System (CNS) cells (e.g., neurons, astrocytes, microglia, and oligodendrocytes) and few have examined miRNAs in EVs in plasma of AD, PD and ALS [25,26,27]. However, none of these studies considered the differences between SEVs from LEVs. We previously described in ALS that SEVs and LEVs in plasma are different in dimensions and for loading of some pathological proteins for ALS (SOD1, TDP-43, p-TDP-43, and FUS) and lipids [28,29,30]. In this paper, we have investigated the miRNA cargo of EVs derived from plasma of patients affected by four neurodegenerative diseases (AD, PD, ALS and FTD). The aim was to identify common and specific small RNAs between the two subpopulation of EVs in the same NDs disease and in the four diseases in order to identify new biomarkers.
2. Results
2.1. miRNAs Selectively Traffic into SEVs and LEVs
We previously demonstrated significant differences between LEVs and SEVs derived from plasma for dimension, markers, protein loading (see Figure S1a–d) [28,29,30]. Cellular miRNAs can selectively traffic into LEVs and SEVs, so we first identified differentially expressed miRNAs (DE miRNAs) in SEVs and LEVs among the four groups of patients (AD, PD, ALS, FTD) and the healthy controls (CTRs) (Table 1, Table S1). We then moved to investigate the number of different and common deregulated miRNAs that sort into SEVs and LEVs in the same disease. In AD, of the 33 miRNAs found in SEVs and 13 in LEVs, 6 distribute to both (Figure 1a), 4 up-regulated and 2 down-regulated. In FTD, of the 88 miRNAs in SEVs and 130 in LEVs, 34 were in common, 32 up-regulated and 2 down-regulated (Figure 1b). Concerning miRNAs in ALS, of the 109 miRNAs in SEVs and 197 in LEVs, 67 were in common, (Figure 1c), 45 up-regulated and 22 down-regulated. In PD, of the 104 miRNAs found in SEVs and 109 in LEVs, 34 distribute to both (Figure 1d), 30 up-regulated and 4 down-regulated. The percentage of common miRNAs between SEVs and LEVs are shown in Table 2.

Table 1.
Statistically significant number of differentially expressed miRNAs in SEVs and LEVs from ALS, FTD, AD, PD patients. Up-regulated transcripts, down-regulated transcripts and total compared to CTRs were reported. Transcripts were considered as differentially expressed when |log2(disease sample/healthy controls)| ≥ 1 and a FDR ≤ 0.1.
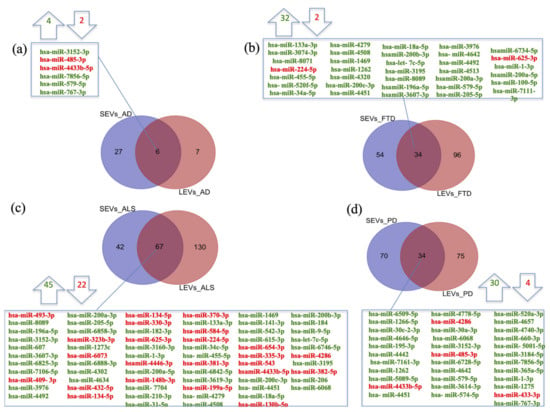
Figure 1.
Common packaging of deregulated miRNAs into SEVs and LEVs of NDs. (a) In AD, of the 33 miRNAs found in SEVs and 13 in LEVs, 6 distribute to both, 4 up-regulated-in green and 2 down-regulated-in red; (b) In FTD, of the 88 miRNAs in SEVs and 130 in LEVs, 34 were in common (32 up-regulated and 2 down-regulated); (c) for ALS, of the 109 miRNAs in SEVs and 197 in LEVs, 67 were in common, 45 up-regulated and 22 down-regulated; (d) in PD, of the 104 miRNAs found in SEVs and 109 in LEVs, 34 distribute to both, 30 up-regulated and 4 down-regulated. Differential miRNA expression analysis by DESeq2 (log2FC > 1, p-value < 0.05). AD = Alzheimer Disease; FTD = Fronto-Temporal Dementia; ALS = Amyotrophic Lateral Sclerosis; PD = Parkinson Disease; CTRs = controls; SEVs = small extracellular vesicles; LEVs = large extracellular vesicles.

Table 2.
Percentage of common miRNAs in SEVs and LEVs in AD, FTD, ALS and PD.
2.2. miRNAs Expression Profiles and Common Pathways in SEVs and LEVs of NDs
miRNAs detected as differentially expressed in SEVs were pooled together and analyzed by principal component analysis (PCA) (Figure 2a). The same approach was applied to LEVs.
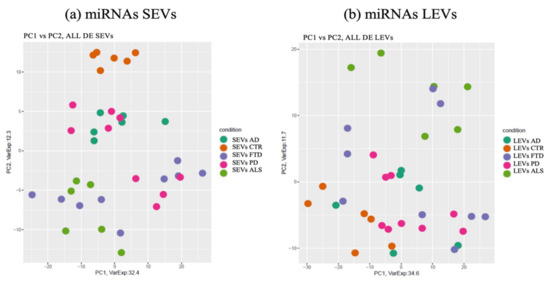
Figure 2.
Principal component Analysis (PCA) of miRNAs differentially expressed in SEVs (a) and LEVs (b) of ALS, FTD, AD and PD patients and healthy controls (CTRs). PCA is performed using as predictors all the miRNAs identified as differentially expressed in at least one disease in the comparison of each disease to the control state. Each dot represents a sample and each color represents a disease. AD = Alzheimer Disease; FTD = Fronto-Temporal Dementia; ALS = Amyotrophic Lateral Sclerosis; PD = Parkinson Disease; CTRs = controls; SEVs = small extracellular vesicles; LEVs = large extracellular vesicles; miRNA = microRNA.
As shown in Figure 2a, deregulated miRNAs in SEVs of the four NDs did not overlap with the CTRs (orange). Interestingly, clusters related to ALS (light green) and AD (dark green) patients could be identified in separate components, while PD (violet) overlapped with some ALS specimens. The miRNA cargo of SEVs were well differentiated in all four diseases.
The PCA of the differential expressed miRNAs detected in LEVs did not provide instead a clear separation of the four diseases (Figure 2b).
However, miRNAs in LEVs of ALS patients could be distinguished from CTRs and from the other three diseases, with some overlap with FTD.
In order to detect common miRNAs of the four diseases, we calculated the intersection of deregulated miRNAs compared to CTRs. We observed that 6 miRNAs were in common among the four diseases in SEVs (hsa-miR-133a-3, hsa-miR-543, hsa-miR-4451, hsa-miR-6889-5p, hsa-miR-4781-3p, hsa-miR-323b-3p) (Figure 3a) and 7 miRNAs (hsa-miR-1262, hsa-miR-3152-3p, hsa-miR-7856-5p, hsa-miR-365a-5p, hsa-miR-4433b-5p, hsa-miR-6068, hsa-miR-767-3p) in LEVs (Figure 3b). Enriched pathways found with MiRWalk included Ubiquitin mediated proteolysis, MAPK signaling pathway, Toll-like receptor signaling pathways for SEVs and Neurotrophin signaling pathway, MAPK signaling pathway, Glycosphingolipid biosynthesis, Ras signaling pathway for LEVs (Figure 3a,b).
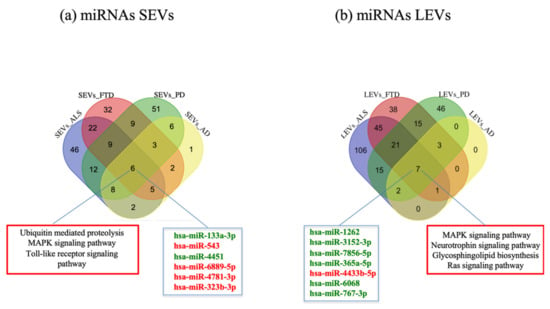
Figure 3.
Venn diagram showing numbers of common and unique miRNA and RNA in SEVs (a) and LEVs (b) from plasma of AD, FTD, ALS and PD patients. Common miRNAs and pathways are listed. Differential miRNA expression analysis by edgeR (log2FC > 1, p-value < 0.05). AD = Alzheimer Disease; FTD = Fronto-Temporal Dementia; ALS = Amyotrophic Lateral Sclerosis; PD = Parkinson Disease; CTRs = controls; SEVs = small extracellular vesicles; LEVs = large extracellular vesicles; miRNA = microRNA.
2.3. Specific miRNAs Pathway Analysis in SEVs of NDs
Enriched pathways targeted by the miRNAs that were differentially expressed between disease samples and CTRs were investigated by MiRWalk (Table S2).
Reactome and Gene Ontology (GO) are listed in Table S2.
In SEVs from ALS patients, Reactome analysis showed miRNAs involved in MECP2 (methyl CpG binding protein 2) expression and activity, Intracellular signaling by second messengers and negative regulators of DDX58/IFIH1. IFIH1 and DDX58 encode retinoic acid-inducible gene I (RIG-I) (Table S2), cytosolic pattern recognition receptors function in viral RNA detection initiating an innate immune response through independent pathways that promote interferon expression [31]. Ingenuity pathway analysis (Figure 4a) confirmed regulation of the immune system by highlighting how miR31-5p and miR615-5p control CXCL8, C-X-C Motif Chemokine Ligand 8 activated in viral infections and miR584-5p/miR148-5p control TMEM9, which enhances production of proinflammatory cytokines induced by TNF, IL1B, and TLR ligands [31]. GO-BPs found negative regulation of Ras protein signal transduction and cell proliferation in forebrain.
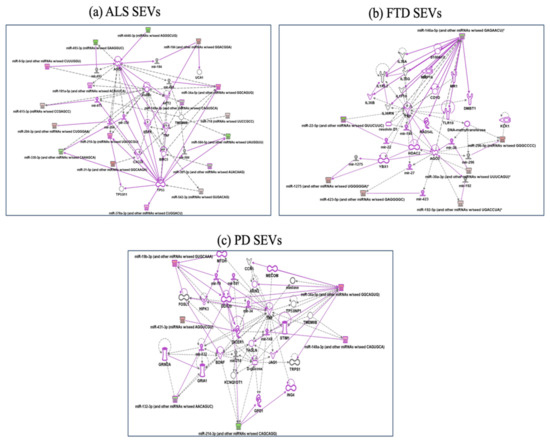
Figure 4.
IPA networks among deregulated miRNAs in SEVs from ALS (a), FTD (b), PD (c). For ALS miRNAs involvement in regulation of DDX58/IFIH1, through miR31-5p and miR615-5p (a); for FTD IL1F10 and TLR10 regulation by miR146a-5p (b); for PD, regulation of GRIA1, Glutamate receptors, by miR-132-3p, involvement of JAG1 and TNF regulated by miR-34 (c). Pink color indicates activation while green color indicate suppression. No pathway could be calculated for AD disease for the few miRNAs targets. FTD = Fronto-Temporal Dementia; ALS = Amyotrophic Lateral Sclerosis; PD = Parkinson Disease; CTRs = controls; SEVs = small extracellular vesicles.
In SEVs from FTD patients Reactome classified deregulated miRNAs in MyD88 cascade initiated on plasma membrane, diseases of signal transduction and axon guidance (Table S2). GO-BPs classified deregulated miRNAs in SEVs from FTD in pathway release of cytochrome c from mitochondria, mitotic G1 DNA damage checkpoint and DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest. IPA analysis (Figure 4b) confirmed involvement in MyD88 cascade (Adapter protein involved in the Toll-like receptor and IL-1 receptor signaling pathway in the innate immune response) and DNA damage response by regulation of miR146a-5p on RAD54, involved in homologous recombination and repair of DNA and on IL1F10 and TLR10 [32,33] (Figure 4b).
In SEVS from PD patients Reactome analysis identified signaling by TGF-beta family members, SLC-mediated transmembrane transport (regulation of GRIA1, subunit of Glutamate receptors, by miR-132-3p, Metabolism of lipids, MyD88 cascade initiated on plasma membrane (involvement of JAG1 and TNF regulated by miR-34-Sup IPA), while GeneOntology BP fibroblast migration, mRNA polyadenylation and positive regulation of gene silencing by miRNA (Figure 4c).
For AD, there were very few deregulated miRNAs to be classified in pathways.
2.4. Specific miRNAs Pathway Analysis in LEVs of NDs
Reactome underlined the role of miRNAs in LEVs (Table S2): (1) from ALS patients in intracellular signaling by second messengers (DAG, cAMP, cGMP, IP3, Ca2+ and phosphatidylinositols), signaling by TGF-beta family members, MyD88 cascade initiated on plasma membrane and metabolism of lipids; (2) from PD patients in intracellular signaling by second messengers, SLC-mediated transmembrane transport (solute carrier superfamily), some of which mediate neurotransmitter uptake in the CNS and peripheral nervous system (PNS) and metabolism of lipids. No significative pathways were detected for miRNAs of LEVs from FTD patients.
Gene ontology analysis found also pathways involved: (1) in regulation of G0 to G1 transition in LEVs from ALS patients; (2) in regulation of type I interferon-mediated signaling pathway and positive regulation of gene and posttranscriptional silencing in LEVs from FTD patients; (3) protein insertion into membrane, fibroblast apoptotic process, regulation of steroid biosynthetic process in LEVs from PD patients. Pathways are reported below in order of most significant p values and further details are shown in Table S2. IPA analysis confirmed the pathway found by both Reactome and Gene Ontology. (1) for LEVs from ALS regulation of p53 by mir-379, 378, 584-5p, 1207 emerged. hsa-miR-199a-5p and hsa-miR-329a-5p regulate LCN2, lipocalin2, inducible factor secreted by reactive astrocytes in transgenic rats with neuronal expression of mutant human TDP-43 or RNA-binding protein FUS and that is selectively toxic to neurons [34] (Figure 5a); (2) for LEVs from FTD patients regulation of genes like TP53 by miR-296 and RNA Polymerase II by miR-615 as well as Cyclin E regulated miR-615-5p, that might be part of gene expression and regulation of apoptosis [35] (Figure 5b); (3) for LEVs from PD patients regulation of ADAM9 (ADAM Metallopeptidase Domain 9), important in mediating cell–cell and cell–matrix interactions, by up-regulation of miR-291, miRNA that regulates cell proliferation and resolvin, a metabolic byproduct of omega-3 fatty acids, regulated by miR-302 [36,37] (Figure 5c).
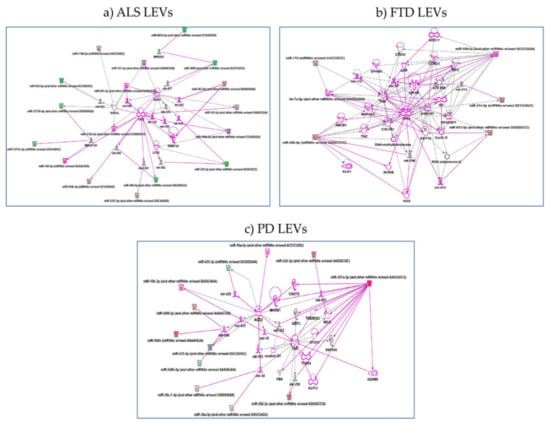
Figure 5.
IPA networks among deregulated miRNAs in LEVs from ALS (a), FTD (b), PD (c). (a) for ALS, regulation of p53 by mir-379, 378, 584-5p, 1207 emerged or hsa-miR-199a-5p and hsa-miR-329a-5p regulate LCN2, lipocalin2; (b) for FTD, regulation of genes like TP53 by miR-296 and RNA Polymerase II by miR-615 as well as Cyclin E regulated miR-615-5p; (c) for LEVs from PD patients, regulation of ADAM9 by up-regulation of miR-291. Pink color indicates activation while green color indicate suppression. FTD = Fronto-Temporal Dementia; ALS = Amyotrophic Lateral Sclerosis; PD = Parkinson Disease; LEVs = large extracellular vesicles.
3. Discussion
Identifying biomarkers is essential for early diagnosis of NDs. Extracellular vesicles (SEVs and LEVs) transported in blood might play this role. In this study, we have analyzed SEVs and LEVs cargo in order to detect specific and common miRNAs acting as novel, easily accessible biomarkers for AD, PD, ALS and FTD.
We first compared miRNA expression profiles of SEVs and LEVs in the same disease. We found a variable range of overlap between LEVs and SEVs: for miRNAs in SEVs the percentage was between 18.2–61.5% and in LEVs 25.2–46.2% (Table 2). Although there is some overlap between the two EVs subtypes, there is a significant difference that may justify, as we already described for dimension, protein and lipid loading [28,29,30], the different functions of LEVs and SEVs in plasma of ND patients. Conley et al. characterized protein coding transcripts in SEVs and LEVs from breast cancer patients by RNA-Seq and identified a small fraction of transcripts that were expressed at significantly different levels in large oncosomes and exosomes, suggesting they may mediate specialized functions [38]. SEVs are more enriched in deregulated miRNAs compared to LEVs. This is in agreement with the literature demonstrating that SEVs contain primarily small RNA [39]. Moreover, EVs from AD patients transport very few DE miRNAs compared to the other three NDs.
Regarding common deregulated miRNAs in LEVs and SEVs in ALS, our data showed an important deregulation in a small group of specific miRNAs already described in the literature (hsa-miR-206, hsa-miR-205-5p, miR-1-3p, hsa-miR-205-5p, hsa-miR-200b-3p, hsa-miR-200c-3p, hsa-miR-6888-3p, hsa-miR-31-5p, hsa-miR-141-3p, hsa-miR-210-3p). MiR-1 and miR-206 has already been described to be deregulated in ALS patients [40,41,42], while in previous studies miR-141 and miR-200 were reported as related to ALS since they bind a sequence in FUS promoter and in turn have an impact on FUS protein synthesis [43,44]. Additionally, miR-210 is already known to be up-regulated in neurodegenerative diseases [45]. To our knowledge, remaining miRNAs deregulation was never reported as associated to ALS.
We then analyzed deregulated miRNAs among the four NDs by principal component analysis (PCA). We found that deregulated miRNAs cargo of the four NDs was different from CTRs, in particular in SEVs, while in LEVs, the only group that did not overlap with CTRs was ALS. On the other hand, miRNAs split the group of PD patients in two, one overlapped with AD and the other with FTD patients. Additionally, FTD patients showed two subgroups, one overlapping with PD and the other with ALS. Multiple studies [46,47,48] already demonstrated that the observed overlap between FTD and ALS is due to common mechanisms contributing to the onset and development of the disease. Parkinsonism is found in approximately 20–30% of patients in FTLD; in particular, it is frequently observed in familial FTD, with mutations linked to microtubule associated protein Tau (MAPT), progranulin (GRN or PGRN), and chromosome 9 open reading frame 72 (C9ORF72) repeat expansion [49]. From the clinical point of view, in our cohort, only one FTD patient presented parkinsonism, which, however, clustered in the ALS group and did not present any mutation in the canonical genes associated to familial FTD. All FTD patients showed variable amounts of Tau, βamyloid 1-42 and only two patients presented ALS-FTD disease (Table 3).

Table 3.
Clinical characteristics of FTD subjects.
Common miRNAs in the four NDs were different between the two groups of EVs. Enriched pathways of common miRNAs found with MiRWalk showed pathway like Ubiquitin mediated proteolysis, MAPK signaling pathway, Toll-like receptor signaling pathways for SEVs and TGF-beta signaling pathway, Neurotrophin signaling pathway, MAPK signaling pathway, Glycosphingolipid biosynthesis, Ras signaling pathway for LEVs. Common miRNAs both in LEVs and SEVs are involved in signal transduction. For example, it is known that Phosphorylated Akt expression is augmented in AD and PD patients [50]. However, although recent reports have implicated EVs in intercellular signaling [51], their influence in modulating signaling pathways in NDs has not been clearly understood.
For the common enriched pathway of SEVs in the four NDs, it is known that many NDs are related to inflammation, which can increase cell injury and cause neuronal death. Toll like receptors (TLR) are innate immune receptors that, when activated, can induce the downstream signal molecules through the MyD88-dependent and TRIF signal adaptor proteins, which activate downstream kinases including IkB kinases and MAP kinases. In general, TLRs are expressed in the CNS, in neurons (TLR3, 4, 7, and 9), in human oligodendrocytes (TLR2), in human astrocytes (TLR3-4), and in human microglia (TLR1-4). Activation of both endosomal and plasma membrane receptors like TLRs can activate microglia and control the evolution of neurodegenerative processes [52].
Ubiquitin mediated proteolysis is the process of degradation of a protein via the ubiquitin system [53] and it is one of the common pathways of SEVs from the four NDs that we found. NDs are characterized by intraneuronal inclusions containing ubiquitynated filamentous protein aggregates, given by loss of function or mutations in enzymes of the ubiquitin conjugation/deconjugation pathway [54]. If there is an impairment of the ubiquitin mediated proteolysis, SEVs, which originate in the endocytic pathway, (differently from LEVs, which are shed from the budding of the cell membrane) might be the affected key part of the machinery as already suggested. In fact, the incorporation of ubiquitinated proteins into intraluminal vesicles (ILVs) is controlled through the Endosomal Sorting Complexes Required for Transport, ESCRT complex, key part of SEVs [55].
For LEVs, Neurotrophin signaling pathway, also called nerve growth factor (NGF) family members pathway, has multiple functions in both developing and mature neurons and it is connected to the downstream MAPK and Ras signaling pathway. On activation by BDNF, trkB initiates intracellular signaling through Shc and PLCγ binding sites. The Shc binding site plays major roles in neuronal survival and axonal outgrowth [56]. Another common deregulated pathway is glycosphingolipid biosynthesis pathway. Increasing evidence underlines the activation of ceramide-dependent pro-apoptotic signaling and reduction of neuroprotective S1P in neurodegeneration course. One hypothesis is the link between altered ceramide/S1P and the production, secretion, and aggregation of pathological proteins in NDs. Sphingolipids regulate EVs and the spread or release of neurotoxic proteins and/or regulatory miRNAs between brain cells [57].
The expression profile of some miRNAs has already been described in the brain or in blood of some NDs, but with an opposite regulatory pattern to the one found in EVs of our study.
For example, miR-133b expression is down-regulated in the midbrain of PD patients and in an animal model of PD, while in SEVs is up-regulated [58]. miR-4781–3p was instead found up-regulated in blood of AD patients by RNAseq, while in SEVs of the four NDs, this miRNA is down-regulated [59]. miR-323–3p associated with inflammatory responses, has been proposed as a target for therapy in AD and it is found down-regulated in SEVs [60]. Some of these miRNAs have not been found in neurodegeneration, and some only in one of the NDs studied in this work. Nevertheless, this study was carried out on a limited number of patients and controls; moreover, healthy donors age matched only two diseases of the four examined. These limitations should be overcome with further studies in which specific miRNAs and pathways will be investigated for each disease.
4. Materials and Methods
4.1. Study Subjects
Participants were recruited at the IRCCS Mondino Foundation, Pavia (Italy). Plasma isolated from 6 AD, 9 PD, 6 sporadic ALS (SALS), 9 FTD patients were used (Table 4). All patients were screened for mutations using a customized panel of 176 genes associated to neurodegenerative and neuromuscular diseases by Next Generation Sequencing (Sure Select QXT Target Enrichment, Agilent Technology, Santa Clara, CA, USA). ALS and FTD patients were screened for C9orf72 using the FastStart Taq DNA Polymerase Kit (Roche, Basel, Switzerland).

Table 4.
Baseline characteristics of recruited subjects for this study. Age is reported as mean ± SD. The percentage of male and female subjects is also indicated.
Diagnosis of AD was based on criteria expressed by Aging-Alzheimer’s Association workgroups [61]. For PD and FTD patients Movement Disorder Society (MDS) clinical diagnostic criteria were used [62,63]. ALS diagnosis was made according to the revised El Escorial Criteria [64].
A total of six age-matched healthy volunteers free from any pharmacological treatment were recruited at the Immunohematological and Transfusional Service IRCCS Foundation “San Matteo”, Pavia (Italy) and used as healthy controls (CTRs). All the subjects were assayed to rule out the presence of inflammatory diseases by white blood cell counts and subjects with WBCs >11 × 109 were excluded from the study. Patients’ characteristics are reported in Table 4, Table 5, Table 6 and Table 7.

Table 5.
Clinical characteristics of AD subjects.

Table 6.
Clinical characteristics of ALS subjects.

Table 7.
Clinical characteristics of PD subjects.
4.2. LEVs and SEVs Isolation
Venous blood (15 mL) was collected in sodium citrate tubes from all patients and controls and processed as previously described [28,29,30]. Briefly, platelet-free plasma was centrifuged at 20,000× g for 1 h. The pellet was washed in 0.2 µm filter filtered 1X PBS (Sigma-Aldrich, Milan, Italy). The supernatant of LEVs was filtered through a 0.2 µm filter and spun in an Optima MAX-TL Ultracentrifuge at 100,000× g for 1 h at 4 °C and SEVS pellet was washed with 1 mL of filtered 1X PBS. Western blot analysis for LEVs markers (Annexin V, Abcam, Cambridge, UK) and for SEVs markers (Alix-Abcam, Cambridge, UK), Transmission Electron Microscopy (TEM) and Nanoparticle-tracking analysis (NTA) were run to confirm LEVs and SEVs purity as we previously described [28,29].
4.3. RNA Extraction
RNA was extracted from LEVs and SEVs fractions using Qiagen miRNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
4.4. RNA Libraries Preparations
Small RNA libraries were constructed with NEBNext® kit (New England Biolabs, Ipswich, MA, USA). Individually-barcoded libraries were mixed. Pools were size selected on Novex 10% TBE gels (Life Technologies, Carlsbad, CA, USA) to enrich for miRNAs fraction. Sequencing (75 nts single-end) was performed on Illumina NextSeq500 (Illumina, San Diego, CA, USA).
4.5. Bioinformatic Data Analysis
The raw bcl files were converted into demultiplexed fastq files with bcl2fastq (Illumina, San Diego, CA, USA) implemented in docker4seq package [65]. For the row count analysis, only transcripts with counts above five were considered. No relevant difference between count in SEVs and LEVs in four diseases emerged.
Quantification of miRNAs was done as described in the literature [66]. The workflow, including quality control filter, trimming of adapters, reads mapping against miRNA precursors, is implemented in docker4seq package [65,66]. Differential expression analysis was performed with the R package DESeq2, implemented in docker4seq package. We imposed a minimum |Log2FC| of 1 and a FDR lower than 0.1 as thresholds to detect differentially expressed miRNAs.
In order to understand common miRNAs of the four diseases, we calculated the intersection of deregulated miRNAs compared to CTRs with http://bioinformatics.psb.ugent.be/webtools/Venn/ accessed on 11 December 2019). The datasets generated and analysed during the current study are available in the NCBI GEO repository [GSE155700].
4.6. Pathways Analysis
miRNA-targets analysis was done with miRWalk web tool (http://mirwalk.umm.uni-heidelberg.de, accessed on 11 December 2019).
In addition, Ingenuity pathway analysis (IPA, v. 2019 summer release, Qiagen, Germany) was performed to identify miRNAs and biological networks associated with the differentially expressed circulating miRNAs in SEVs and LEVs in each of the neurodegenerative disorders.
5. Conclusions
We found different deregulation of miRNAs between SEVs and LEVs from plasma of patients in four NDs.
We found a common signature of miRNAs in SEVs and LEVs among the four NDs and those miRNAs are involved in pathways already known in neurodegeneration.
For each NDs, we also found new DE miRNAs involved in different pathways and this might give a specificity to the role of SEVs and LEVs in the spreading/protection of each disease.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/22/5/2737/s1, Figure S1: LEVs and SEVs characterization; Table S1. Differentially expressed miRNAs in ALS, FTD, PD and AD groups respect to healthy controls. miRNA ID, measured log2FC and p-value are reported for each transcript; Table S2. miRWalk specific miRNAs pathways in ALS, FTD, PD and AD groups. Deregulated miRNAs specific of each disease were analysed with miRWalk web tool. Reactome Analysis and Gene Ontology Biological Processes with a p value < 0.05 are listed. Only pathways with p value < 0.05 are listed. BH and Pop Total stand for Benjamini–Hochberg and Population Total.
Author Contributions
Conceptualization, C.C. and R.A.C.; methodology, D.S., C.C., M.A. (Maddalena Arigoni), M.O. and R.A.C.; software, S.Z., R.A.C., M.D.O., N.I.P.-B., D.S.; validation, M.G. (Marta Giannini), M.G. (Maria Garofalo), D.S., S.G. and M.A. (Maddalena Arigoni); formal analysis, S.Z., S.G., D.S.; investigation, C.C.; resources, S.B.; M.A. (Micol Avenali); A.C.; B.M.; G.P.; L.D.; R.Z.; M.C.R.; M.C.; data curation, D.S., S.G. and S.Z.; writing—original draft preparation, S.G., D.S. and S.Z.; writing—review and editing C.C, D.S., O.P., N.I.P.-B., A.C., R.A.C.; visualization O.P., N.I.P.-B.; supervision, R.A.C. and C.C.; project administration, C.C. and O.P.; funding acquisition, C.C. and S.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Italian Ministry of Health (Grant N°5*1000 anno 2016, Ricerca Corrente 2018–2020, Young research project GR-2016-02361552); Fondazione Regionale per la Ricerca Biomedica for TRANS–ALS (FRRB 2015-0023); Fondazione Cariplo 2017 (Extracellular vesicles in the pathogenesis of Frontotemporal Dementia 2017-0747; Association between frailty trajectories and biological markers of aging 2017-0557).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board (for ALS patients Protocol n°-20180034329; for PD patients Protocol n°20170001758; for AD patients Protocol n° 20200045845; for FTD patients Protocol n°20180049077).
Informed Consent Statement
Before being enrolled, the subjects participating in the study signed an informed consent form (Code 375/04—version 07/01/2004).
Data Availability Statement
The datasets generated and analysed during the current study are available in the NCBI GEO repository [GSE155700].
Acknowledgments
We kindly thank Fabio Corsi and Raffaele Allevi (Dipartimento di Scienze Biomediche e Cliniche “L. Sacco”) for TEM images and analysis. We thank patients and family for contributing to this study.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AD | Alzheimer’s disease |
| ALS | Amyotrophic Lateral Sclerosis |
| CTRs | healthy controls |
| DE miRNAs | differentially expressed miRNAs |
| ESCRT | Endosomal Sorting Complexes Required for Transport |
| EVs | extracellular vesicles |
| EXOs | exosomes |
| FTD | frontotemporal dementia |
| GWAS | Genome-wide association studies |
| ILVs | intraluminal vesicles |
| IPA | Ingenuity pathway analysis |
| LBs | Lewy bodies |
| LEVs | large extracellular vesicles |
| lncRNA | long non coding RNA |
| miRNA | microRNA |
| MVs | microvesicles |
| NDs | neurodegenerative diseases |
| PCA | principal component analysis |
| PD | Parkinson’s disease |
| RBPs | RNA-binding proteins |
| SEVs | small extracellular vesicles |
| SN | substantia nigra |
| SNPs | single nucleotide polymorphisms |
| TLRs | Toll like receptors |
References
- Gitler, A.D.; Dhillon, P.; Shorter, J. Neurodegenerative disease: Models, mechanisms, and a new hope. Dis. Model. Mech. 2017, 10, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Habib, R.; Noureen, N.; Nadeem, N. Decoding Common Features of Neurodegenerative Disorders: From Differentially Expressed Genes to Pathways. Curr. Genom. 2018, 19, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Montie, H.L.; Durcan, T.M. The Cell and Molecular Biology of Neurodegenerative Diseases: An Overview. Front. Neurol. 2013, 4, 194. [Google Scholar] [CrossRef] [PubMed]
- García, J.C.; Bustos, R.H. The Genetic Diagnosis of Neurodegenerative Diseases and Therapeutic Perspectives. Brain Sci. 2018, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- Obeso, J.A.; Stamelou, M.; Goetz, C.G.; Poewe, W.; Lang, A.E.; Weintraub, D.; Burn, D.; Halliday, G.M.; Bezard, E. Past, present, and future of Parkinson’s disease: A special essay on the 200th Anniversary of the Shaking Palsy. Mov. Disord. 2017, 32, 1264–1310. [Google Scholar] [CrossRef]
- Cereda, C.; Leoni, E.; Milani, P.; Pansarasa, O.; Mazzini, G.; Guareschi, S.; Alvisi, E.; Ghiroldi, A.; Diamanti, L.; Bernuzzi, S.; et al. Altered intracellular localization of SOD1 in leukocytes from patients with sporadic amyotrophic lateral sclerosis. PLoS ONE 2013, 8, e75916. [Google Scholar] [CrossRef]
- Guareschi, S.; Cova, E.; Cereda, C.; Ceroni, M.; Donetti, E.; Bosco, D.A.; Trotti, D.; Pasinelli, P. An over-oxidized form of superoxide dismutase found in sporadic amyotrophic lateral sclerosis with bulbar onset shares a toxic mechanism with mutant SOD1. Proc. Natl. Acad. Sci. USA 2012, 109, 5074–5079. [Google Scholar] [CrossRef] [PubMed]
- Pansarasa, O.; Bordoni, M.; Diamanti, L.; Sproviero, D.; Gagliardi, S.; Cereda, C. SOD1 in Amyotrophic Lateral Sclerosis: “Ambivalent” Behavior Connected to the Disease. Int. J. Mol. Sci. 2018, 19, 1345. [Google Scholar] [CrossRef]
- Gagliardi, S.; Cova, E.; Davin, A.; Guareschi, S.; Abel, K.; Alvisi, E.; Laforenza, U.; Ghidoni, R.; Cashman, J.R.; Ceroni, M.; et al. SOD1 mRNA expression in sporadic amyotrophic lateral sclerosis. Neurobiol. Dis. 2010, 39, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Fogh, I.; Lin, K.; Tiloca, C.; Rooney, J.; Gellera, C.; Diekstra, F.P.; Ratti, A.; Shatunov, A.; Van Es, M.A.; Proitsi, P.; et al. Association of a Locus in theCAMTA1Gene With Survival in Patients with Sporadic Amyotrophic Lateral Sclerosis. JAMA Neurol. 2016, 73, 812–820. [Google Scholar] [CrossRef]
- Arai, T.; Hasegawa, M.; Akiyama, H.; Ikeda, K.; Nonaka, T.; Mori, H.; Mann, D.; Tsuchiya, K.; Yoshida, M.; Hashizume, Y.; et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006, 351, 602–611. [Google Scholar] [CrossRef]
- Bott, N.T.; Radke, A.; Stephens, M.L.; Kramer, J.H. Frontotemporal dementia: Diagnosis, deficits and management. Neurodegener. Dis. Manag. 2014, 4, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Götzl, J.K.; Lang, C.M.; Haass, C.; Capell, A. Impaired protein degradation in FTLD and related disorders. Ageing Res. Rev. 2016, 32, 122–139. [Google Scholar] [CrossRef] [PubMed]
- Belzil, V.V.; Gendron, T.F.; Petrucelli, L. RNA-mediated toxicity in neurodegenerative disease. Mol. Cell. Neurosci. 2013, 56, 406–419. [Google Scholar] [CrossRef]
- Ling, S.C.; Polymenidou, M.; Cleveland, D.W. Converging mechanisms in ALS and FTD: Disrupted RNA and protein homestasis. Neuron 2013, 79, 416–438. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.Y.; Cali, C.P.; Lee, E.B. RNA metabolism in neurodegenerative disease. Dis. Model. Mech. 2017, 10, 509–518. [Google Scholar] [CrossRef]
- Iraci, N.; Leonardi, T.; Gessler, F.; Vega, B.; Pluchino, S. Focus on Extracellular Vesicles: Physiological Role and Signalling Properties of Extracellular Membrane Vesicles. Int. J. Mol. Sci. 2016, 17, 171. [Google Scholar] [CrossRef] [PubMed]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L.; et al. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally de-rived blood exosomes: A case-control study. Alzheimers Dement. 2015, 11, 600.e1–601.e1. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Boxer, A.L.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Miller, B.L.; Kapogiannis, D. Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology 2015, 85, 40–47. [Google Scholar] [CrossRef]
- Fatima, F.; Nawaz, M. Vesiculated Long Non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation between Cells, Cancer Progression and Resistance to Therapies. Non-Coding RNA 2017, 3, 10. [Google Scholar] [CrossRef]
- Matsumoto, J.; Stewart, T.; Banks, W.A.; Zhang, J. The Transport Mechanism of Extracellular Vesicles at the Blood-Brain Barrier. Curr. Pharm. Des. 2017, 23, 6206–6214. [Google Scholar] [CrossRef]
- Cocucci, E.; Meldolesi, J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015, 25, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Yao, Y.F.; Qu, M.W.; Li, G.C.; Zhang, F.B.; Rui, H.C. Circulating exosomal miRNAs as diagnostic biomarkers in Parkinson’s disease. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5278–5283. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Reñé, R.; Álvarez, R.; Armengol, M.P.; Borràs, F.E.; Beyer, K. Ex-ploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Transl. Neurodegener. 2019, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Katsu, M.; Hama, Y.; Utsumi, J.; Takashina, K.; Yasumatsu, H.; Mori, F.; Wakabayashi, K.; Shoji, M.; Sasaki, H. MicroRNA expression profiles of neuron-derived extracellular vesicles in plasma from patients with amyotrophic lateral sclerosis. Neurosci. Lett. 2019, 708, 134176. [Google Scholar] [CrossRef] [PubMed]
- Sproviero, D.; La Salvia, S.; Giannini, M.; Crippa, V.; Gagliardi, S.; Bernuzzi, S.; Diamanti, L.; Ceroni, M.; Pansarasa, O.; Poletti, A.; et al. Pathological Proteins Are Transported by Extracellular Vesicles of Sporadic Amyotrophic Lateral Sclerosis Patients. Front. Neurosci. 2018, 12, 487. [Google Scholar] [CrossRef]
- Morasso, C.F.; Sproviero, D.; Mimmi, M.C.; Giannini, M.; Gagliardi, S.; Vanna, R.; Diamanti, L.; Bernuzzi, S.; Piccotti, F.; Truffi, M.; et al. Raman spectroscopy reveals biochemical differences in plasma derived extra-cellular vesicles from sporadic Amyotrophic Lateral Sclerosis patients. Nanomedicine 2020, 29, 102249. [Google Scholar] [CrossRef] [PubMed]
- Sproviero, D.; La Salvia, S.; Colombo, F.; Zucca, S.; Pansarasa, O.; Diamanti, L.; Costa, A.; Lova, L.; Giannini, M.; Gagliardi, S.; et al. Leukocyte derived microvesicles as disease progres-sion biomarkers in slow progressing amyotrophic lateral sclerosis patients. Front. Neurosci. 2019, 13, 344. [Google Scholar] [CrossRef]
- Bowie, A.G.; Unterholzner, L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 2008, 8, 911–922. [Google Scholar] [CrossRef]
- Mazin, A.V.; Mazina, O.M.; Bugreev, D.V.; Rossi, M.J. Rad54, the motor of homologous recombination. DNA Repair 2010, 9, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Sato, S.; Ishii, K.J.; Coban, C.; Hemmi, H.; Yamamoto, M.; Terai, K.; Matsuda, M.; Inoue, J.-I.; Uematsu, S.; et al. Interferon-α induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 2004, 5, 1061–1068. [Google Scholar] [CrossRef]
- Bi, F.; Huang, C.; Tong, J.; Qiu, G.; Huang, B.; Wu, Q.; Li, F.; Xu, Z.; Bowser, R.; Xia, X.-G.; et al. Reactive astrocytes secrete lcn2 to promote neuron death. Proc. Natl. Acad. Sci. USA 2013, 110, 4069–4074. [Google Scholar] [CrossRef]
- Liu, C.; Yao, M.-D.; Li, C.-P.; Shan, K.; Yang, H.; Wang, J.-J.; Liu, B.; Li, X.-M.; Yao, J.; Jiang, Q.; et al. Silencing of Circular RNA-ZNF609 Ameliorates Vascular Endothelial Dysfunction. Theranostics 2017, 7, 2863–2877. [Google Scholar] [CrossRef]
- Wang, Y.; Baskerville, S.; Shenoy, A.; Babiarz, J.E.; Baehner, L.; Blelloch, R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008, 40, 1478–1483. [Google Scholar] [CrossRef]
- Recchiuti, A.; Krishnamoorthy, S.; Fredman, G.; Chiang, N.; Serhan, C.N. MicroRNAs in resolution of acute inflammation: Identification of novel resolvin D1-miRNA circuits. FASEB J. 2011, 25, 544–560. [Google Scholar] [CrossRef]
- Conley, A.; Minciacchi, V.R.; Lee, D.H.; Knudsen, B.S.; Karlan, B.Y.; Citrigno, L.; Viglietto, G.; Tewari, M.; Freeman, M.R.; Demichelis, F.; et al. High-throughput sequencing of two populations of extracellular vesicles provides an mRNA signature that can be detected in the circulation of breast cancer patients. RNA Biol. 2017, 14, 305–316. [Google Scholar] [CrossRef]
- Crescitelli, R.; Lässer, C.; Szabó, T.G.; AKittel, M.E.; Dianzani, I.; Buzás, E.I.; Lötvall, J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. JEVs 2013, 2, 20677. [Google Scholar] [CrossRef]
- Waller, R.; Goodall, E.F.; Milo, M.; Cooper-Knock, J.; Da Costa, M.; Hobson, E.; Kazoka, M.; Wollff, H.; Heath, P.R.; Shaw, P.J.; et al. Serum miRNAs miR-206, 143–3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol. Aging 2017, 55, 123–131. [Google Scholar] [CrossRef] [PubMed]
- De Andrade, H.M.T.; de Albuquerque, M.; Avansini, S.H.; Rocha, C.d.S.; Dogini, D.B.; Nucci, A.; Carvalho, B.; Lopes-Cendes, I.; França, M.C., Jr. MicroRNAs-424 and 206 are potential prognostic markers in spinal onset amyotrophic lateral sclerosis. J. Neurol. Sci. 2016, 368, 19–24. [Google Scholar] [CrossRef]
- Raheja, R.; Regev, K.; Healy, B.C.; Mazzola, M.A.; Beynon, V.; Von Glehn, F.; Paul, A.; Diaz-Cruz, C.; Gholipour, T.; Glanz, B.I.; et al. Correlating serum micrornas and clinical parameters in amyotrophic lateral sclerosis. Muscle Nerve 2018, 58, 261–269. [Google Scholar] [CrossRef]
- Apolloni, S.; Parisi, C.; Volonté, C. MicroRNAs: Newcomers into the ALS Picture. CNS Neurol. Disord. Drug Targets 2015, 14, 194–207. [Google Scholar] [CrossRef]
- Modigliani, S.D.; Morlando, M.; Errichelli, L.; Sabatelli, M.; Bozzoni, I. An ALS-associated Mutation in the FUS 3′-UTR Disrupts a microRNA-FUS Regulatory Circuitry. Nat. Commun. 2014, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Watts, M.E.; Williams, S.M.; Nithianantharajah, J.; Claudianos, C. Hypoxia-Induced MicroRNA-210 Targets Neurodegenerative Pathways. Non-Coding RNA 2018, 4, 10. [Google Scholar] [CrossRef]
- Ferrari, R.; Kapogiannis, D.; Huey, E.D.; Momeni, P. FTD and ALS: A Tale of Two Diseases. Curr. Alzheimer Res. 2011, 8, 273–294. [Google Scholar] [CrossRef]
- Achi, E.Y.; Rudnicki, S.A. ALS and Frontotemporal Dysfunction: A Review. Neurol. Res. Int. 2012, 2012, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liscic, R.M. Als and Ftd: Insights into the disease mechanisms and therapeutic targets. Eur. J. Pharmacol. 2017, 817, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Park, H.K.; Chung, S.J. New Perspective on Parkinsonism in Frontotemporal Lobar Degeneration. J. Mov. Disord. 2013, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Armentero, M.T.; Sinforiani, E.; Ghezzi, C.; Bazzini, E.; Levandis, G.; Ambrosi, G.; Zangaglia, R.; Pacchetti, C.; Cereda, C.; Cova, E.; et al. Peripheral expression of key regulatory kinases in Alzheimer’s disease and Parkinson’s disease. Neurobiol. Aging 2011, 32, 2142–2151. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Fiebich, B.L.; Batista, C.R.A.; Saliba, S.W.; Yousif, N.M.; De Oliveira, A.C.P. Role of Microglia TLRs in Neurodegeneration. Front. Cell. Neurosci. 2018, 12, 329. [Google Scholar] [CrossRef]
- Ciechanover, A.; Schwartz, A.L. The ubiquitin-mediated proteolytic pathway: Mechanisms of recognition of the proteolytic substrate and involvement in the degradation of native cellular proteins. FASEB J. 1994, 8, 182–191. [Google Scholar] [CrossRef]
- Layfield, R.; Cavey, J.R.; Lowe, J. Role of ubiquitin-mediated proteolysis in the pathogenesis of neurodegenerative disorders. Ageing Res. Rev. 2003, 2, 343–356. [Google Scholar] [CrossRef]
- Raiborg, C.; Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nat. Cell Biol. 2009, 458, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Numakawa, T.; Suzuki, S.; Kumamaru, E.; Adachi, N.; Richards, M.; Kunugi, H. BDNF function and intracellular signaling in neurons. Histol. Histopathol. 2010, 25, 2. [Google Scholar] [CrossRef]
- Czubowicz, K.; Jęśko, H.; Wencel, P.; Lukiw, W.J.; Strosznajder, R.P. The Role of Ceramide and Sphingosine-1-Phosphate in Alzheimer’s Disease and Other Neurodegenerative Disorders. Mol. Neurobiol. 2019, 56, 5436–5455. [Google Scholar] [CrossRef]
- Kim, J.; Inoue, K.; Ishii, J.; Vanti, W.B.; Voronov, S.V.; Murchison, E.; Hannon, G.; Abeliovich, A.A. MicroRNA feedback circuit in midbrain dopamine neurons. Science 2007, 317, 1220–1224. [Google Scholar] [CrossRef] [PubMed]
- Satoh, J.; Kino, Y.; Niida, S. MicroRNA-Seq data analysis pipeline to identify blood biomarkers for Alzheimer’s disease from public data. Biomark. Insights 2015, 10, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Li, L.; Huang, C.; Li, X.; Peng, Y.; Li, J. MicroRNA-323–3p with clinical potential in rheumatoid arthritis. Alzheimer’s disease and ectopic pregnancy. Expert. Opin. Ther. Targets 2014, 18, 153–158. [Google Scholar] [CrossRef] [PubMed]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the national institute on aging—Alzheimer’s association work groups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Postuma, R.B.; Berg, D.; Stern, M.; Poewe, W.; Olanow, C.W.; Oertel, W.; Obeso, J.; Marek, K.; Litvan, I.; Lang, A.E.; et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. 2015, 30, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Chare, L.; Hodges, J.R.; Leyton, C.E.; McGinley, C.; Tan, R.H.; Kril, J.K.; Halliday, G.M. New criteria for frontotemporal dementia syndromes: Clinical and pathological diagnostic implications. J. Neurol. Neurosurg. Psychiatry 2014, 85, 865–870. [Google Scholar] [CrossRef]
- Brooks, B.R.; Miller, R.G.; Swash, M.; Munsat, T.L. El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2000, 1, 293–299. [Google Scholar] [CrossRef]
- Cordero, F.; Beccuti, M.; Arigoni, M.; Donatelli, S.; Calogero, R.A. Optimizing a Massive Parallel Sequencing Workflow for Quantitative miRNA Expression Analysis. PLoS ONE 2012, 7, e31630. [Google Scholar] [CrossRef]
- Kulkarni, N.; Alessandrì, L.; Panero, R.; Arigoni, M.; Olivero, M.; Ferrero, G.; Cordero, F.; Beccuti, M.; Calogero, R.A. Reproducible bioinformatics project: A community for reproducible bioinformatics analysis pipelines. BMC Bioinform. 2018, 19, 5–13. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).