Exosomal Aβ-Binding Proteins Identified by “In Silico” Analysis Represent Putative Blood-Derived Biomarker Candidates for Alzheimer´s Disease
Abstract
1. Introduction
2. Results and Discussion
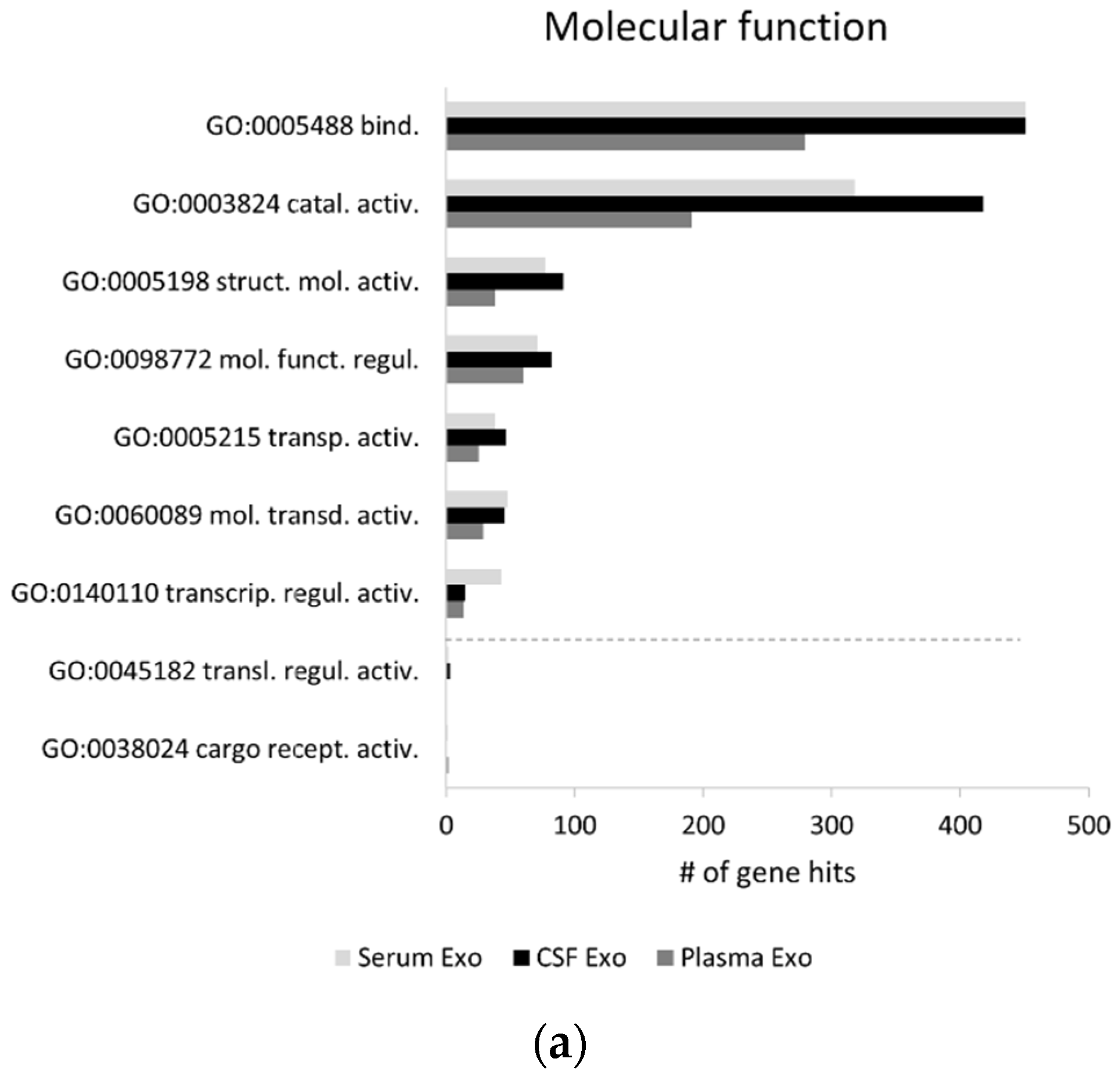
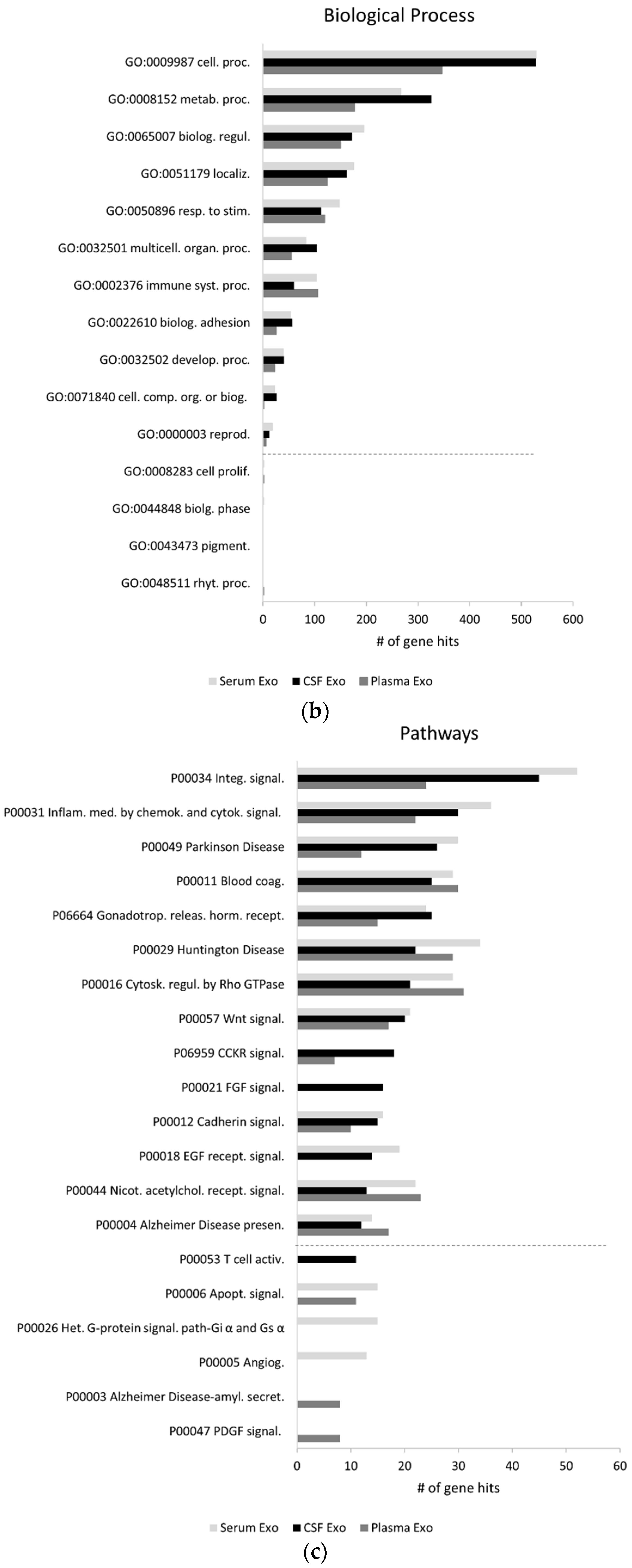
2.1. Exosomal Proteomes Gene Ontology (GO) Analysis
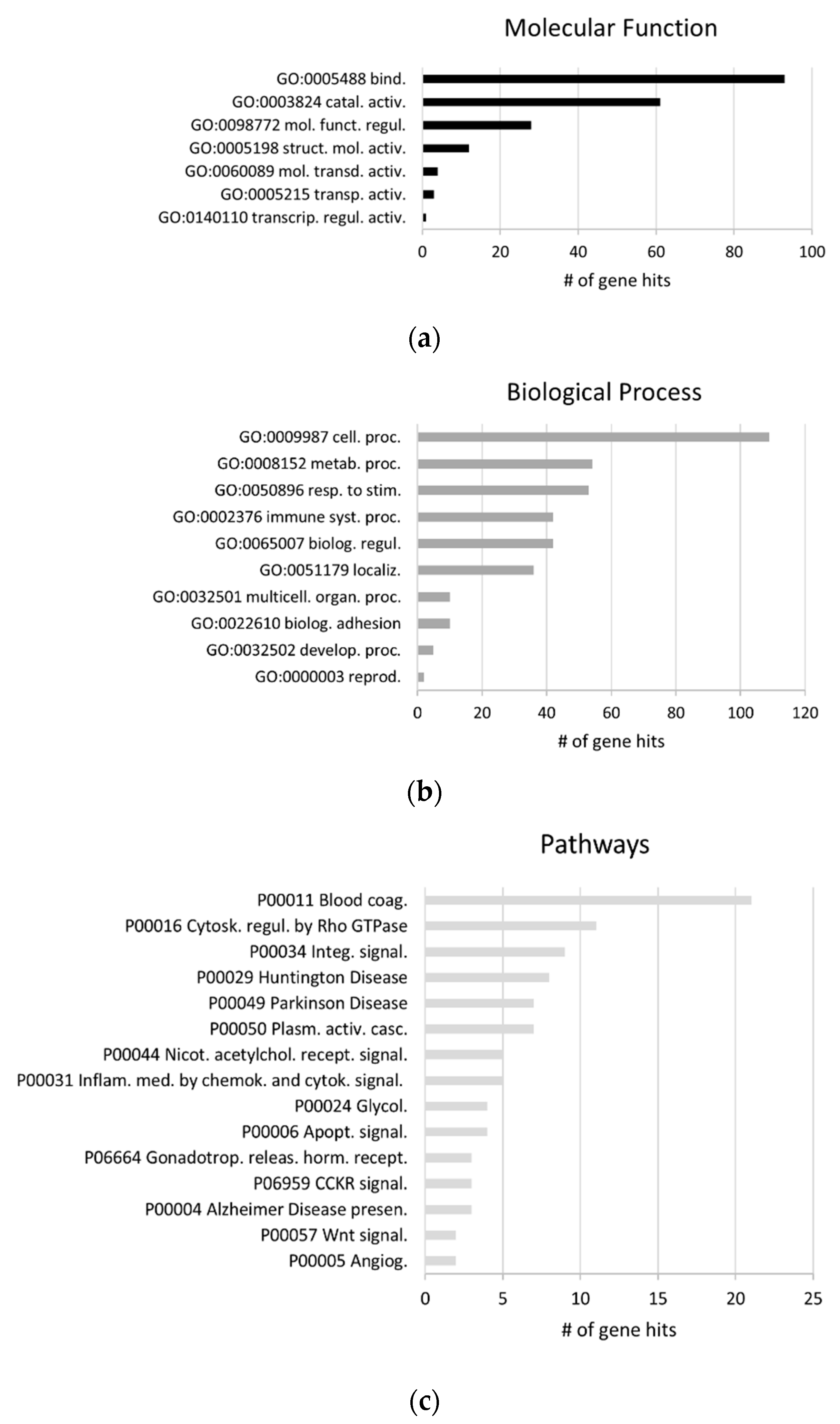
2.2. GO Analysis of the Biofluids Shared Exosomal Proteome
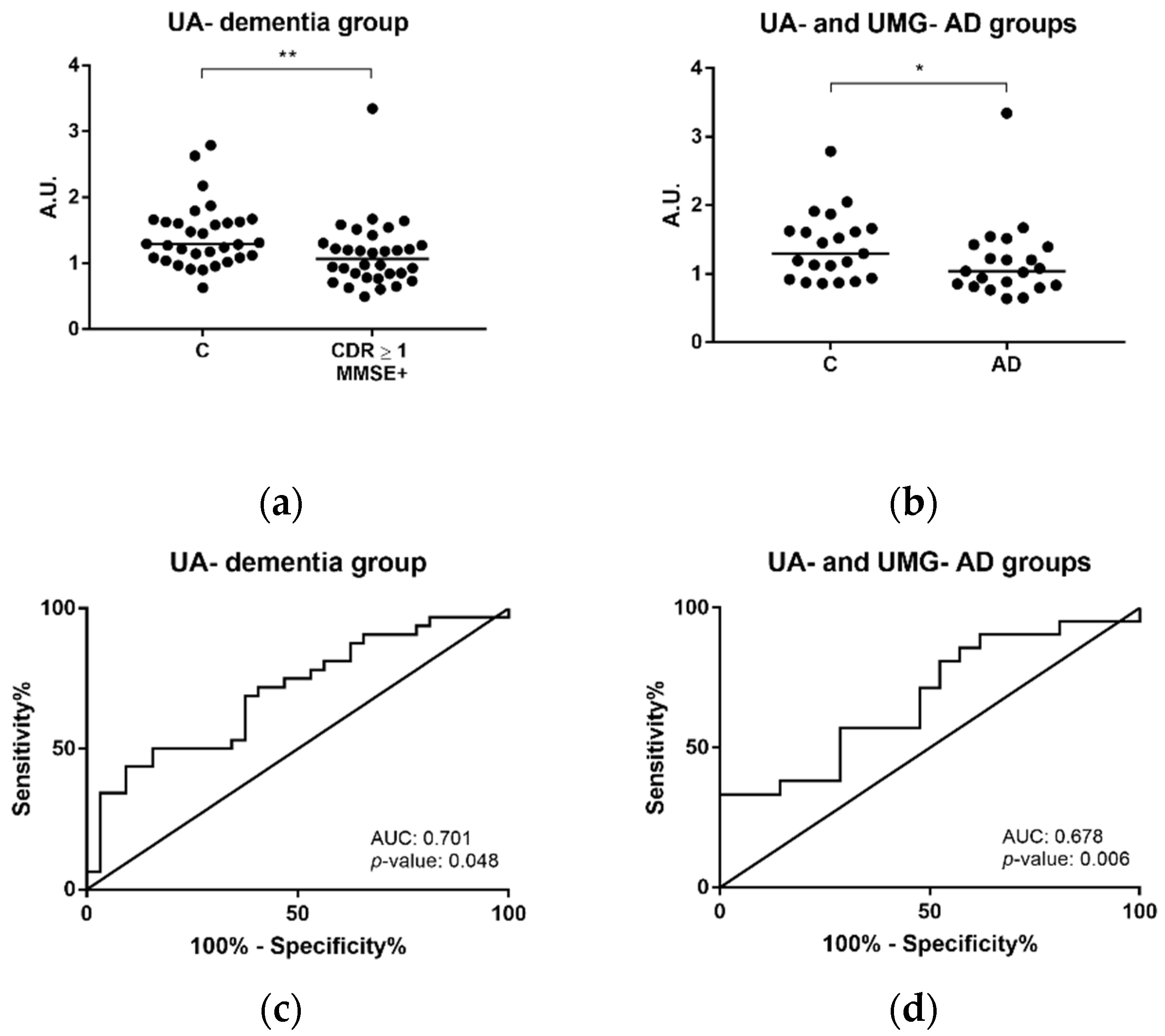
2.3. Potential Blood-Derived Exosomal Candidates for AD
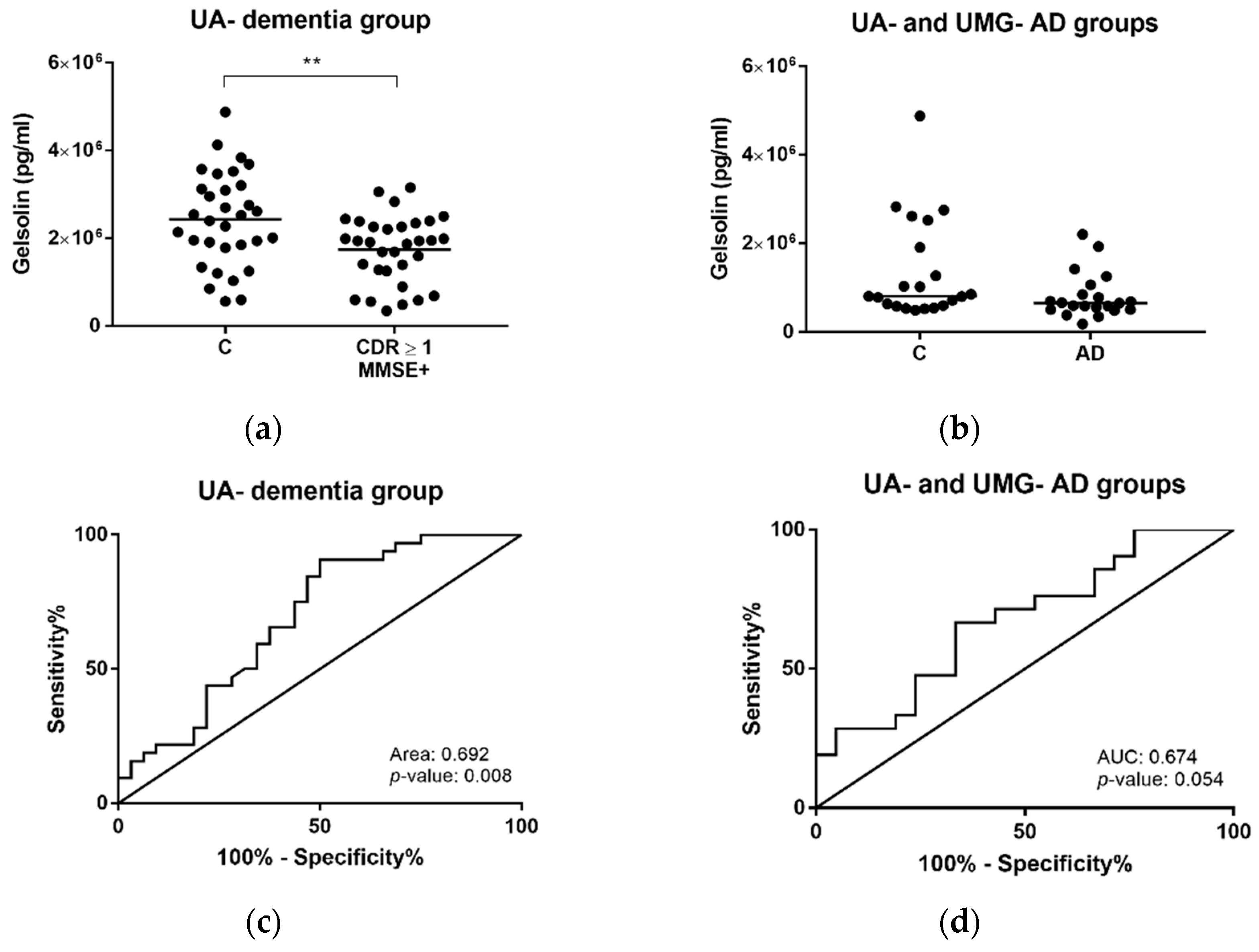
2.4. Gelsolin as Putative Exosomal Biomarker Candidate
3. Materials and Methods
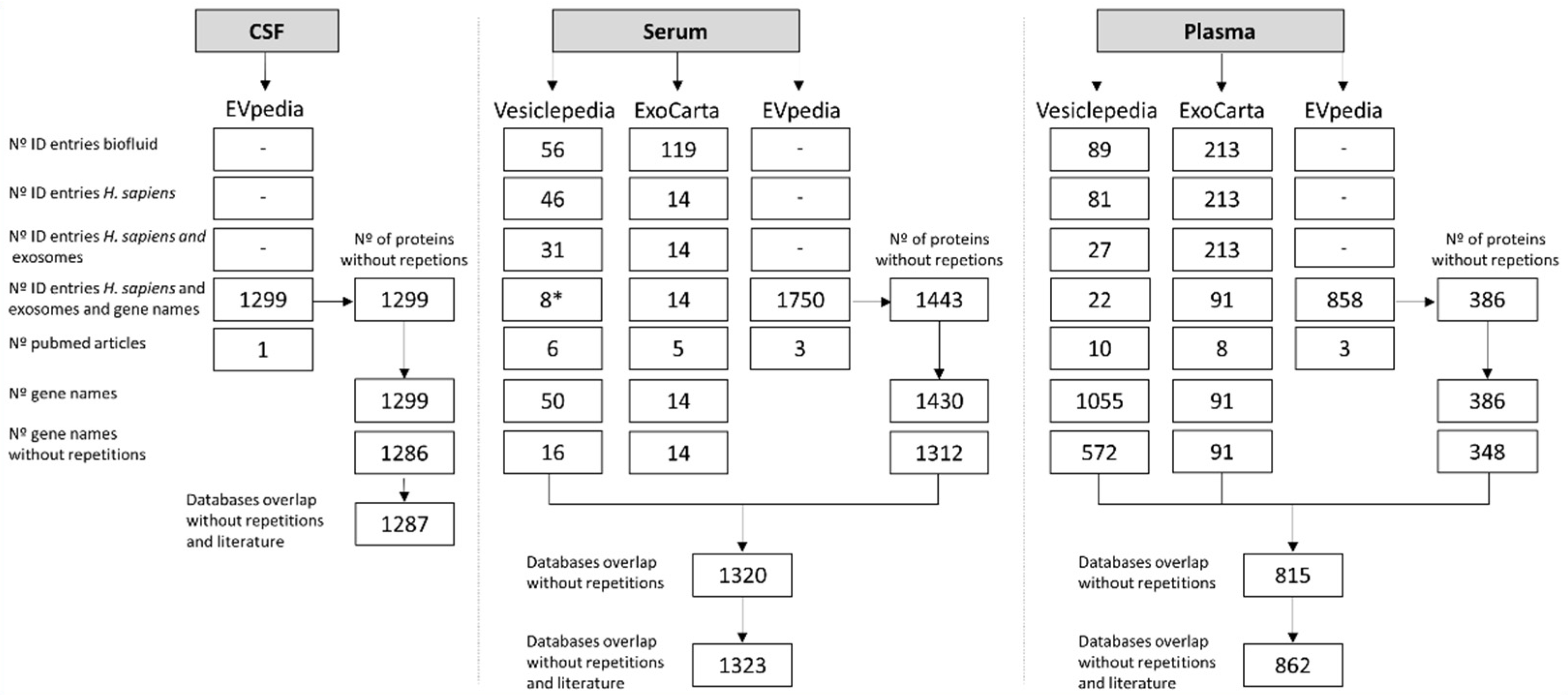
3.1. Exosomal Proteomes Construction
3.2. Proteome GO and Overlap Analysis
3.3. Aβ-binding Proteins Literature Search
3.4. EVs Isolation and Characterization
3.5. Evaluation of Gelsolin Levels
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeTure, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 1–18. [Google Scholar] [CrossRef]
- Marcus, C.; Mena, E.; Subramaniam, R.M. Brain PET in the diagnosis of Alzheimer’s disease. Clin. Nucl. Med. 2014, 39, e413–e426. [Google Scholar] [CrossRef]
- Vandenberghe, R.; Adamczuk, K.; Dupont, P.; Van Laere, K.; Chételat, G. Amyloid PET in clinical practice: Its place in the multidimensional space of Alzheimer’s disease. NeuroImage Clin. 2013, 2, 497–511. [Google Scholar] [CrossRef]
- Blennow, K.; Zetterberg, H. Cerebrospinal Fluid biomarkers for Alzheimer’s disease. J. Alzheimer’s Dis. 2009, 18, 413–417. [Google Scholar] [CrossRef]
- Welge, V.; Fiege, O.; Lewczuk, P.; Mollenhauer, B.; Esselmann, H.; Klafki, H.-W.; Wolf, S.; Trenkwalder, C.; Otto, M.; Kornhuber, J.; et al. Combined CSF tau, p-tau181 and amyloid-β 38/40/42 for diagnosing Alzheimer’s disease. J. Neural Transm. 2009, 116, 203–212. [Google Scholar] [CrossRef]
- Hampel, H.; O’Bryant, S.E.; Molinuevo, J.L.; Zetterberg, H.; Masters, C.L.; Lista, S.; Kiddle, S.J.; Batrla, R.; Blennow, K. Blood-based biomarkers for Alzheimer disease: Mapping the road to the clinic. Nat. Rev. Neurol. 2018, 14, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Martins, T.S.; Trindade, D.; Vaz, M.; Campelo, I.; Almeida, M.; Trigo, G.; Silva, O.A.B.D.C.E.; Henriques, A.G. Diagnostic and therapeutic potential of exosomes in Alzheimer’s disease. J. Neurochem. 2021, 156, 162–181. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.S.; Magalhães, S.; Rosa, I.M.; Vogelgsang, J.; Wiltfang, J.; Delgadillo, I.; Catita, J.; Silva, O.A.D.C.E.; Nunes, A.; Henriques, A.G. Potential of FTIR Spectroscopy applied to exosomes for Alzheimer’s disease discrimination: A pilot study. J. Alzheimer’s Dis. 2020, 74, 391–405. [Google Scholar] [CrossRef]
- Kapogiannis, D.; Mustapic, M.; Shardell, M.D.; Berkowitz, S.T.; Diehl, T.C.; Spangler, R.D.; Tran, J.; Lazaropoulos, M.P.; Chawla, S.; Gulyani, S.; et al. Association of extracellular vesicle biomarkers with Alzheimer disease in the baltimore longitudinal study of aging. JAMA Neurol. 2019, 76, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef] [PubMed]
- Sinha, M.S.; Ansell-Schultz, A.; Civitelli, L.; Hildesjö, C.; Larsson, M.; Lannfelt, L.; Ingelsson, M.; Hallbeck, M. Alzheimer’s disease pathology propagation by exosomes containing toxic amyloid-beta oligomers. Acta Neuropathol. 2018, 136, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.G.; Turola, E.; Ruiz, A.; Bergami, A.; Libera, D.D.; Benussi, L.; Giussani, P.; Magnani, G.; Comi, G.; Legname, G.; et al. Microglia convert aggregated amyloid-β into neurotoxic forms through the shedding of microvesicles. Cell Death Differ. 2014, 21, 582–593. [Google Scholar] [CrossRef]
- Ding, M.; Shen, Y.; Wang, P.; Xie, Z.; Xu, S.; Zhu, Z.; Wang, Y.; Lyu, Y.; Wang, D.; Xu, L.; et al. exosomes isolated from human umbilical cord mesenchymal stem cells alleviate neuroinflammation and reduce amyloid-beta deposition by modulating microglial activation in Alzheimer’s disease. Neurochem. Res. 2018, 43, 2165–2177. [Google Scholar] [CrossRef] [PubMed]
- Canales-Aguirre, A.A.; Reza-Zaldivar, E.E.; Hernández-Sapiéns, M.A.; Gutiérrez-Mercado, Y.K.; Sandoval-Ávila, S.; Gomez-Pinedo, U.; Márquez-Aguirre, A.L.; Vázquez-Méndez, E.; Padilla-Camberos, E. Mesenchymal stem cell-derived exosomes promote neurogenesis and cognitive function recovery in a mouse model of Alzheimer’s disease. Neural Regen. Res. 2019, 14, 1626–1634. [Google Scholar] [CrossRef] [PubMed]
- Yuyama, K.; Sun, H.; Usuki, S.; Sakai, S.; Hanamatsu, H.; Mioka, T.; Kimura, N.; Okada, M.; Tahara, H.; Furukawa, J.-I.; et al. A potential function for neuronal exosomes: Sequestering intracerebral amyloid-β peptide. FEBS Lett. 2015, 589, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Bulloj, A.; Leal, M.C.; Xu, H.; Castaño, E.M.; Morelli, L. Insulin-degrading enzyme sorting in exosomes: A secretory pathway for a key brain amyloid-β degrading protease. J. Alzheimer’s Dis. 2010, 19, 79–95. [Google Scholar] [CrossRef]
- Pacheco-Quinto, J.; Clausen, D.; Pérez-González, R.; Peng, H.; Meszaros, A.; Eckman, C.B.; Levy, E.; Eckman, E.A. Intracellular metalloprotease activity controls intraneuronal Aβ aggregation and limits secretion of Aβ via exosomes. FASEB J. 2018, 33, 3758–3771. [Google Scholar] [CrossRef]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L.; et al. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimer’s Dement. 2015, 11, 600–607.e1. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Schwartz, J.B.; Abner, E.L.; Jicha, G.A.; Kapogiannis, D. High complement levels in astrocyte-derived exosomes of Alzheimer disease. Ann. Neurol. 2018, 83, 544–552. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Kapogiannis, D.; Schwartz, J.B.; Lobach, I.V.; Goetzl, L.; Abner, E.L.; Jicha, G.A.; Karydas, A.M.; Boxer, A.; Miller, B.L. Decreased synaptic proteins in neuronal exosomes of frontotemporal dementia and Alzheimer’s disease. FASEB J. 2016, 30, 4141–4148. [Google Scholar] [CrossRef]
- Kapogiannis, D.; Boxer, A.; Schwartz, J.B.; Abner, E.L.; Biragyn, A.; Masharani, U.; Frassetto, L.; Petersen, R.C.; Miller, B.L.; Goetzl, E.J. Dysfunctionally phosphorylated type 1 insulin receptor substrate in neural-derived blood exosomes of preclinical Alzheimer’s disease. FASEB J. 2015, 29, 589–596. [Google Scholar] [CrossRef]
- Viola, K.L.; Klein, W.L. amyloid β oligomers in Alzheimer’s disease pathogenesis, treatment, and diagnosis. Acta Neuropathol. 2015, 129, 183–206. [Google Scholar] [CrossRef] [PubMed]
- Varma, V.R.; Varma, S.; An, Y.; Hohman, T.J.; Seddighi, S.; Casanova, R.; Beri, A.; Dammer, E.B.; Seyfried, N.T.; Pletnikova, O.; et al. Alpha-2 macroglobulin in Alzheimer’s disease: A marker of neuronal injury through the RCAN1 pathway. Mol. Psychiatry 2017, 22, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Scacchi, R.; Ruggeri, M.; Gambina, G.; Martini, M.C.; Corbo, R.M. α2-Macroglobulin Deletion Polymorphism and Plasma Levels in Late Onset Alzheimer’s disease. Clin. Chem. Lab. Med. 2002, 40, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Giometto, B.; Argentiero, V.; Sanson, F.; Ongaro, G.; Tavolato, B. Acute-Phase Proteins in Alzheimer’s disease. Eur. Neurol. 1988, 28, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Can, M.; Varlibas, F.; Guven, B.; Akhan, O.; Yuksel, G.A. Ischemia Modified Albumin and Plasma Oxidative Stress Markers in Alzheimer’s disease. Eur. Neurol. 2013, 69, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Schirinzi, T.; Di Lazzaro, G.; Sancesario, G.M.; Colona, V.L.; Scaricamazza, E.; Mercuri, N.B.; Martorana, A.; Sancesario, G. Levels of amyloid-beta-42 and CSF pressure are directly related in patients with Alzheimer’s disease. J. Neural Transm. 2017, 124, 1621–1625. [Google Scholar] [CrossRef] [PubMed]
- Miners, J.S.; Kehoe, P.G.; Love, S.; Zetterberg, H.; Blennow, K. CSF evidence of pericyte damage in Alzheimer’s disease is associated with markers of blood-brain barrier dysfunction and disease pathology. Alzheimer’s Res. Ther. 2019, 11, 1–6. [Google Scholar] [CrossRef]
- Crawford, J.R.; Bjorklund, N.L.; Taglialatela, G.; Gomer, R.H. Brain serum amyloid P levels are reduced in individuals that lack dementia while having Alzheimer’s disease neuropathology. Neurochem. Res. 2012, 37, 795–801. [Google Scholar] [CrossRef][Green Version]
- Yasojima, K.; Schwab, C.; McGeer, E.G.; McGeer, P.L. Human neurons generate C-reactive protein and amyloid P: Upregulation in Alzheimer’s disease. Brain Res. 2000, 887, 80–89. [Google Scholar] [CrossRef]
- Verwey, N.A.; Schuitemaker, A.; Van Der Flier, W.M.; Mulder, S.D.; Mulder, C.; Hack, C.E.; Scheltens, P.; Blankenstein, M.A.; Veerhuis, R. Serum amyloid p component as a biomarker in mild cognitive impairment and Alzheimer’s disease. Dement. Geriatr. Cogn. Disord. 2008, 26, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Poljak, A.; Crawford, J.; Kochan, N.A.; Wen, W.; Cameron, B.; Lux, O.; Brodaty, H.; Mather, K.; Smythe, G.A.; et al. Plasma Apolipoprotein Levels Are Associated with Cognitive Status and Decline in a Community Cohort of Older Individuals. PLoS ONE 2012, 7, e34078. [Google Scholar] [CrossRef] [PubMed]
- Kawano, M.; Kawakami, M.; Otsuka, M.; Yashima, H.; Yaginuma, T.; Ueki, A. Marked decrease of plasma apolipoprotein AI and AII in Japanese patients with late-onset non-familial Alzheimer’s disease. Clin. Chim. Acta 1995, 239, 209–211. [Google Scholar] [CrossRef]
- Song, F.; Poljak, A.; Kochan, N.A.; Raftery, M.; Brodaty, H.; Smythe, G.A.; Sachdev, P.S. Plasma protein profiling of Mild Cognitive Impairment and Alzheimer’s disease using iTRAQ quantitative proteomics. Proteome Sci. 2014, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Merched, A.; Xia, Y.; Visvikis, S.; Serot, J.; Siest, G. Decreased high-density lipoprotein cholesterol and serum apolipoprotein AI concentrations are highly correlated with the severity of Alzheimer’s disease. Neurobiol. Aging 2000, 21, 27–30. [Google Scholar] [CrossRef]
- Liu, H.-C.; Hu, C.-J.; Chang, J.-G.; Sung, S.-M.; Lee, L.-S.; Yuan, R.-Y.; Leu, S.-J. Proteomic Identification of Lower Apolipoprotein A-I in Alzheimer’s disease. Dement. Geriatr. Cogn. Disord. 2006, 21, 155–161. [Google Scholar] [CrossRef]
- Kuriyama, M.; Takahashi, K.; Yamano, T.; Hokezu, Y.; Togo, S.; Osame, M.; Igakura, T. Low Levels of Serum Apolipoprotein AI and AII in Senile Dementia. Psychiatry Clin. Neurosci. 1994, 48, 589–593. [Google Scholar] [CrossRef]
- Harr, S.D.; Uint, L.; Hollister, R.; Hyman, B.T.; Mendez, A.J. Brain Expression of Apolipoproteins E, J, and A-I in Alzheimer’s disease. J. Neurochem. 2002, 66, 2429–2435. [Google Scholar] [CrossRef]
- Song, H.; Saito, K.; Seishima, M.; Noma, A.; Urakami, K.; Nakashima, K. Cerebrospinal fluid apo E and apo A-I concentrations in early- and late-onset Alzheimer’s disease. Neurosci. Lett. 1997, 231, 175–178. [Google Scholar] [CrossRef]
- Kitamura, Y.; Usami, R.; Ichihara, S.; Kida, H.; Satoh, M.; Tomimoto, H.; Murata, M.; Oikawa, S. Plasma protein profiling for potential biomarkers in the early diagnosis of Alzheimer’s disease. Neurol. Res. 2017, 39, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.S.; He, Y.; Phan, K.; Ahmed, R.M.; Rye, K.-A.; Piguet, O.; Hodges, J.R.; Halliday, G.M. Altered High Density Lipoprotein Composition in Behavioral Variant Frontotemporal Dementia. Front. Neurosci. 2018, 12, 847. [Google Scholar] [CrossRef]
- Shih, Y.-H.; Tsai, K.-J.; Lee, C.-W.; Shiesh, S.-C.; Chen, W.-T.; Pai, M.-C.; Kuo, Y.-M. Apolipoprotein C-III is an amyloid-β-Binding Protein and an Early Marker for Alzheimer’s disease. J. Alzheimer’s Dis. 2014, 41, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Muenchhoff, J.; Poljak, A.; Song, F.; Raftery, M.; Brodaty, H.; Duncan, M.; McEvoy, M.; Attia, J.; Schofield, P.W.; Sachdev, P.S. Plasma Protein Profiling of Mild Cognitive Impairment and Alzheimer’s disease Across Two Independent Cohorts. J. Alzheimer’s Dis. 2014, 43, 1355–1373. [Google Scholar] [CrossRef] [PubMed]
- Petit-Turcotte, C.; Stohl, S.M.; Beffert, U.; Cohn, J.S.; Aumont, N.; Tremblay, M.; Dea, D.; Yang, L.; Poirier, J.; Shachter, N.S. Apolipoprotein C-I Expression in the Brain in Alzheimer’s disease. Neurobiol. Dis. 2001, 8, 953–963. [Google Scholar] [CrossRef]
- Rezeli, M.; Zetterberg, H.; Blennow, K.; Brinkmalm, A.; Laurell, T.; Hansson, O.; Marko-Varga, G. Quantification of total apolipoprotein E and its specific isoforms in cerebrospinal fluid and blood in Alzheimer’s disease and other neurodegenerative diseases. EuPA Open Proteom. 2015, 8, 137–143. [Google Scholar] [CrossRef]
- Bertrand, P.; Poirier, J.; Oda, T.; Finch, C.E.; Pasinetti, G.M. Association of apolipoprotein E genotype with brain levels of apolipoprotein E and apolipoprotein J (clusterin) in Alzheimer disease. Mol. Brain Res. 1995, 33, 174–178. [Google Scholar] [CrossRef]
- Marksteiner, J.; Pirchl, M.; Ullrich, C.; Oberbauer, H.; Blasko, I.; Lederer, W.; Hinterhuber, H.; Humpel, C. Analysis of Cerebrospinal Fluid of Alzheimer Patients. Pharmacology 2008, 82, 214–220. [Google Scholar] [CrossRef]
- Zhuravin, I.A.; Nalivaeva, N.N.; Kozlova, D.I.; Kochkina, E.G.; Fedorova, Y.B.; Gavrilova, S.I. The activity of blood serum cholinesterases and neprilysin as potential biomarkers of mild-cognitive impairment and Alzheimer’s disease. Zhurnal Nevrol. Psikhiatrii Im. Korsakova 2015, 115, 110–117. [Google Scholar] [CrossRef]
- Macdonald, I.R.; Maxwell, S.P.; Reid, G.A.; Cash, M.K.; DeBay, D.R.; Darvesh, S. Quantification of Butyrylcholinesterase Activity as a Sensitive and Specific Biomarker of Alzheimer’s disease. J. Alzheimer’s Dis. 2017, 58, 491–505. [Google Scholar] [CrossRef]
- Smyth, M.D.; Cribbs, D.H.; Tenner, A.J.; Shankle, W.; Dick, M.; Kesslak, J.; Cotman, C.W. Decreased levels of C1q in cerebrospinal fluid of living Alzheimer patients correlate with disease state. Neurobiol. Aging 1994, 15, 609–614. [Google Scholar] [CrossRef]
- Khoonsari, P.E.; Häggmark, A.; Lönnberg, M.; Mikus, M.; Kilander, L.; Lannfelt, L.; Bergquist, J.; Ingelsson, M.; Nilsson, P.; Kultima, K.; et al. Analysis of the Cerebrospinal Fluid Proteome in Alzheimer’s disease. PLoS ONE 2016, 11, e0150672. [Google Scholar] [CrossRef] [PubMed]
- Winston, C.N.; Goetzl, E.J.; Schwartz, J.B.; Elahi, F.M.; Rissman, R.A. Complement protein levels in plasma astrocyte-derived exosomes are abnormal in conversion from mild cognitive impairment to Alzheimer’s disease dementia. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2019, 11, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Dejanovic, B.; Gandham, V.D.; Gogineni, A.; Edmonds, R.; Schauer, S.; Srinivasan, K.; Huntley, M.A.; Wang, Y.; Wang, T.-M.; et al. Complement C3 is activated in human AD brain and is required for neurodegeneration in mouse models of amyloidosis and tauopathy. Cell Rep. 2019, 28, 2111–2123.e6. [Google Scholar] [CrossRef]
- Litvinchuk, A.; Wan, Y.-W.; Cole, A.; Chen, F.; Liu, Z.; Zheng, H. The role of complement c3 and c3ar receptor in tau pathology in Alzheimer’s disease. Alzheimer’s Dement. 2017, 13, P229. [Google Scholar] [CrossRef]
- Trouw, L.A.; Nielsen, H.M.; Minthon, L.; Londos, E.; Landberg, G.; Veerhuis, R.; Janciauskiene, S.; Blom, A.M. C4b-binding protein in Alzheimer’s disease: Binding to Aβ1–42 and to dead cells. Mol. Immunol. 2008, 45, 3649–3660. [Google Scholar] [CrossRef]
- Puertas, M.; Martínez-Martos, J.; Cobo, M.; Carrera, M.; Mayas, M.; Ramírez-Expósito, M. Plasma oxidative stress parameters in men and women with early stage Alzheimer type dementia. Exp. Gerontol. 2012, 47, 625–630. [Google Scholar] [CrossRef]
- Zabel, M.; Nackenoff, A.; Kirsch, W.M.; Harrison, F.E.; Perry, G.; Schrag, M. Markers of oxidative damage to lipids, nucleic acids and proteins and antioxidant enzymes activities in Alzheimer’s disease brain: A meta-analysis in human pathological specimens. Free Radic. Biol. Med. 2018, 115, 351–360. [Google Scholar] [CrossRef]
- Lovell, M.A.; Ehmann, W.D.; Butler, S.M.; Markesbery, W.R. Elevated thiobarbituric acid-reactive substances and antioxidant enzyme activity in the brain in Alzheimer’s disease. Neurology 1995, 45, 1594–1601. [Google Scholar] [CrossRef]
- Lu, G.; Liu, W.; Huang, X.; Zhao, Y. Complement factor H levels are decreased and correlated with serum C-reactive protein in late-onset Alzheimer’s disease. Arq. Neuro-Psiquiatr. 2020, 78, 76–80. [Google Scholar] [CrossRef]
- Gezen-Ak, D.; Dursun, E.; Hanağası, H.; Bilgiç, B.; Lohman, E.; Araz, Ö.S.; Atasoy, I.L.; Alaylıoğlu, M.; Önal, B.; Gürvit, H.; et al. BDNF, TNFα, HSP90, CFH, and IL-10 Serum levels in patients with early or late onset alzheimer’s disease or mild cognitive impairment. J. Alzheimer’s Dis. 2013, 37, 185–195. [Google Scholar] [CrossRef]
- Williams, M.A.; Haughton, D.; Stevenson, M.; Craig, D.; Passmore, A.P.; Silvestri, G. Plasma Complement factor H in Alzheimer’s disease. J. Alzheimer’s Dis. 2015, 45, 369–372. [Google Scholar] [CrossRef] [PubMed]
- Strohmeyer, R.; Shen, Y.; Rogers, J. Detection of complement alternative pathway mRNA and proteins in the Alzheimer’s disease brain. Mol. Brain Res. 2000, 81, 7–18. [Google Scholar] [CrossRef]
- Nilselid, A.-M.; Davidsson, P.; Nägga, K.; Andreasen, N.; Fredman, P.; Blennow, K. Clusterin in cerebrospinal fluid: Analysis of carbohydrates and quantification of native and glycosylated forms. Neurochem. Int. 2006, 48, 718–728. [Google Scholar] [CrossRef]
- Schürmann, B.; Wiese, B.; Bickel, H.; Weyerer, S.; Riedel-Heller, S.G.; Pentzek, M.; Bachmann, C.; Williams, J.; Bussche, H.V.D.; Maier, W.; et al. Association of the Alzheimer’s disease clusterin risk allele with plasma clusterin concentration. J. Alzheimer’s Dis. 2011, 25, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Thambisetty, M.; Simmons, A.; Velayudhan, L.; Hye, A.; Campbell, J.; Zhang, Y.; Wahlund, L.-O.; Westman, E.; Kinsey, A.; Güntert, A.; et al. Association of plasma clusterin concentration with severity, pathology, and progression in Alzheimer disease. Arch. Gen. Psychiatry 2010, 67, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Liao, P.-C.; Yu, L.; Kuo, C.-C.; Lin, C.; Kuo, Y.-M. Proteomics analysis of plasma for potential biomarkers in the diagnosis of Alzheimer’s disease. Proteom. Clin. Appl. 2007, 1, 506–512. [Google Scholar] [CrossRef]
- Suidan, G.L.; Singh, P.K.; Patel-Hett, S.; Chen, Z.-L.; Volfson, D.; Yamamoto-Imoto, H.; Norris, E.H.; Bell, R.D.; Strickland, S. Abnormal clotting of the intrinsic/contact pathway in Alzheimer disease patients is related to cognitive ability. Blood Adv. 2018, 2, 954–963. [Google Scholar] [CrossRef]
- Tsai, C.W.; Tsai, C.F.; Lin, K.H.; Chen, W.J.; Lin, M.S.; Hsieh, C.C.; Lin, C.C. An investigation of the correlation between the S-glutathionylated GAPDH levels in blood and Alzheimer’s disease progression. PLoS ONE 2020, 15, e0233289. [Google Scholar] [CrossRef]
- Sultana, R.; Boyd-Kimball, D.; Cai, J.; Pierce, W.M.; Klein, J.B.; Merchant, M.; Butterfield, D.A. Proteomics Analysis of the Alzheimer’s disease Hippocampal Proteome. J. Alzheimer’s Dis. 2007, 11, 153–164. [Google Scholar] [CrossRef]
- Güntert, A.; Campbell, J.; Saleem, M.; O’Brien, D.P.; Thompson, A.J.; Byers, H.L.; Ward, M.A.; Lovestone, S. Plasma gelsolin is decreased and correlates with rate of decline in Alzheimer’s disease. J. Alzheimer’s Dis. 2010, 21, 585–596. [Google Scholar] [CrossRef]
- Peng, M.; Jia, J.; Qin, W. Plasma gelsolin and matrix metalloproteinase 3 as potential biomarkers for Alzheimer disease. Neurosci. Lett. 2015, 595, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Antequera, D.; Vargas, T.; Ugalde, C.; Spuch, C.; Molina, J.A.; Ferrer, I.; Bermejo-Pareja, F.; Carro, E. Cytoplasmic gelsolin increases mitochondrial activity and reduces Aβ burden in a mouse model of Alzheimer’s disease. Neurobiol. Dis. 2009, 36, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Zhang, K.; Zhang, Y.; Guo, Y.; Li, A.; Xiao, S.; Liu, Q.; Shen, L.; Ni, J. Identification of blood biomarkers for Alzheimer’s disease through computational prediction and experimental validation. Front. Neurol. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Elovaara, I.; Icen, A.; Palo, J.; Erkinjuntti, T. CSF in Alzheimer’s disease. J. Neurol. Sci. 1985, 70, 73–80. [Google Scholar] [CrossRef]
- Ayton, S.; Janelidze, S.; Roberts, B.; Palmqvist, S.; Kalinowski, P.; Diouf, I.; Belaidi, A.A.; Stomrud, E.; Bush, A.I.; Hansson, O. Acute phase markers in CSF reveal inflammatory changes in Alzheimer’s disease that intersect with pathology, APOE ε4, sex and age. Prog. Neurobiol. 2021, 198, 101904. [Google Scholar] [CrossRef]
- Song, I.-U.; Kim, Y.-D.; Chung, S.-W.; Cho, H.-J. Association between serum haptoglobin and the pathogenesis of Alzheimer’s disease. Intern. Med. 2015, 54, 453–457. [Google Scholar] [CrossRef]
- Zhu, C.-J.; Jiang, G.-X.; Chen, J.-M.; Zhou, Z.-M.; Cheng, Q. Serum haptoglobin in chinese patients with Alzheimer’s disease and mild cognitive impairment: A case-control study. Brain Res. Bull. 2018, 137, 301–305. [Google Scholar] [CrossRef]
- Björkdahl, C.; Sjögren, M.J.; Zhou, X.; Concha, H.; Avila, J.; Winblad, B.; Pei, J.-J. Small heat shock proteins Hsp27 or αB-crystallin and the protein components of neurofibrillary tangles: Tau and neurofilaments. J. Neurosci. Res. 2007, 86, 1343–1352. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Boxer, A.L.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Miller, B.L.; Kapogiannis, D. Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology 2015, 85, 40–47. [Google Scholar] [CrossRef]
- Zetterberg, H. Identification of a β-amyloid-binding plasma protein, LRP1: Implications for biomarker research and therapy in Alzheimer’s disease. Biomarker. Med. 2007, 1, 347–348. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.E.; Pietrzik, C.U.; Baum, L.; Chevallier, N.; Merriam, D.E.; Kounnas, M.Z.; Wagner, S.L.; Troncoso, J.C.; Kawas, C.H.; Katzman, R.; et al. Modulation of amyloid β-protein clearance and Alzheimer’s disease susceptibility by the LDL receptor–related protein pathway. J. Clin. Investig. 2000, 106, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, Y.; Faghihi, M.A.; Magistri, M.; Alvarez-Garcia, O.; Lotz, M.; Wahlestedt, C. Antisense RNA controls LRP1 sense transcript expression through interaction with a chromatin-associated protein, HMGB2. Cell Rep. 2015, 11, 967–976. [Google Scholar] [CrossRef]
- Matsui, T.; Ingelsson, M.; Fukumoto, H.; Ramasamy, K.; Kowa, H.; Frosch, M.P.; Irizarry, M.C.; Hyman, B.T. Expression of APP pathway mRNAs and proteins in Alzheimer’s disease. Brain Res. 2007, 1161, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Dranovsky, A.; Vincent, I.; Gregori, L.; Schwarzman, A.; Colflesh, D.; Enghild, J.; Strittmatter, W.; Davies, P.; Goldgaber, D. Cdc2 phosphorylation of nucleolin demarcates mitotic stages and Alzheimer’s disease pathology. Neurobiol. Aging 2001, 22, 517–528. [Google Scholar] [CrossRef]
- Ijsselstijn, L.; Dekker, L.J.M.; Stingl, C.; Van Der Weiden, M.M.; Hofman, A.; Kros, J.M.; Koudstaal, P.J.; Smitt, P.A.E.S.; Ikram, M.A.; Breteler, M.M.B.; et al. Serum levels of pregnancy zone protein are elevated in presymptomatic Alzheimer’s disease. J. Proteome Res. 2011, 10, 4902–4910. [Google Scholar] [CrossRef] [PubMed]
- Nijholt, D.A.; Ijsselstijn, L.; Van Der Weiden, M.M.; Zheng, P.-P.; Smitt, P.A.E.S.; Koudstaal, P.J.; Luider, T.M.; Kros, J.M. Pregnancy zone protein is increased in the Alzheimer’s disease brain and associates with senile plaques. J. Alzheimer’s Dis. 2015, 46, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Valero, J.; Costell, M.; Sjögren, M.; Andreasen, N.; Blennow, K.; Luque, J.M. Altered levels of cerebrospinal fluid reelin in frontotemporal dementia and Alzheimer’s disease. J. Neurosci. Res. 2003, 72, 132–136. [Google Scholar] [CrossRef]
- Botella-López, A.; Burgaya, F.; Gavín, R.; García-Ayllón, M.S.; Gómez-Tortosa, E.; Peña-Casanova, J.; Ureña, J.M.; Del Río, J.A.; Blesa, R.; Soriano, E.; et al. Reelin expression and glycosylation patterns are altered in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2006, 103, 5573–5578. [Google Scholar] [CrossRef]
- Mata-Balaguer, T.; Cuchillo-Ibañez, I.; Calero, M.; Ferrer, I.; Sáez-Valero, J. Decreased generation of C-terminal fragments of ApoER2 and increased reelin expression in Alzheimer’s disease. FASEB J. 2018, 32, 3536–3546. [Google Scholar] [CrossRef]
- Shen, L.; Liao, L.; Chen, C.; Guo, Y.; Song, D.; Wang, Y.; Chen, Y.; Zhang, K.; Ying, M.; Li, S.; et al. Proteomics analysis of blood serums from Alzheimer’s disease patients using iTRAQ labeling technology. J. Alzheimer’s Dis. 2017, 56, 361–378. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.-A.; Shin, K.Y.; Nam, E.; Lee, Y.-B.; Moon, C.; Suh, Y.-H.; Lee, S.H. Plasma soluble neuregulin-1 as a diagnostic biomarker for Alzheimer’s disease. Neurochem. Int. 2016, 97, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Horvath, I.; Jia, X.; Johansson, P.; Wang, C.; Moskalenko, R.; Steinau, A.; Forsgren, L.; Wågberg, T.; Svensson, J.; Zetterberg, H.; et al. Pro-inflammatory S100A9 protein as a robust biomarker differentiating early stages of cognitive impairment in Alzheimer’s disease. ACS Chem. Neurosci. 2015, 7, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Rueli, R.H.; Parubrub, A.C.; Dewing, A.S.; Hashimoto, A.C.; Bellinger, M.T.; Weeber, E.J.; Uyehara-Lock, J.H.; White, L.R.; Berry, M.J.; Bellinger, F.P. Increased selenoprotein P in choroid plexus and cerebrospinal fluid in Alzheimer’s disease brain. J. Alzheimer’s Dis. 2015, 44, 379–383. [Google Scholar] [CrossRef]
- Nielsen, H.M.; Minthon, L.; Londos, E.; Blennow, K.; Miranda, E.; Perez, J.; Crowther, D.C.; Lomas, D.A.; Janciauskiene, S.M. Plasma and CSF serpins in Alzheimer disease and dementia with Lewy bodies. Neurology 2007, 69, 1569–1579. [Google Scholar] [CrossRef]
- Shinohara, Y.; Ohsuga, H.; Yamamoto, M.; Ohsuga, S.; Akiyama, K.; Yoshii, F.; Tsuda, M.; Kamiguchi, H.; Yamamura, M. CSF α1-Antichymotrypsin and antitrypsin in multi-infarct dementia and Alzheimer’s disease. Cereb. Ischemia Dement. 1991, 334–337. [Google Scholar] [CrossRef]
- Abraham, C.R.; Selkoe, D.J.; Potter, H. Immunochemical identification of the serine protease inhibitor α1-antichymotrypsin in the brain amyloid deposits of Alzheimer’s disease. Cell 1988, 52, 487–501. [Google Scholar] [CrossRef]
- Furby, A.; Leys, D.; Delacourte, A.; Buee, L.; Soetaert, G.; Petit, H. Are alpha-1-antichymotrypsin and inter-alpha-trypsin inhibitor peripheral markers of Alzheimer’s disease? J. Neurol. Neurosurg. Psychiatry 1991, 54, 469. [Google Scholar] [CrossRef][Green Version]
- Pirttila, T.; Mehta, P.; Frey, H.; Wisniewski, H. α1-Antichymotrypsin and IL-1β are not increased in CSF or serum in Alzheimer’s disease. Neurobiol. Aging 1994, 15, 313–317. [Google Scholar] [CrossRef]
- Lanzrein, A.-S.; Johnston, C.M.; Perry, V.H.; Jobst, K.A.; King, E.M.; Smith, A.D. Longitudinal study of inflammatory factors in serum, cerebrospinal fluid, and brain tissue in Alzheimer disease. Alzheimer Dis. Assoc. Disord. 1998, 12, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, B.A.; Swanwick, G.R.; Feighery, C.; Walsh, J.B.; Coakley, D. Acute phase reactants in Alzheimer’s disease. Biol. Psychiatry 1996, 39, 1051–1052. [Google Scholar] [CrossRef]
- Matsubara, E.; Amari, M.; Shoji, M.; Harigaya, Y.; Yamaguchi, H.; Okamoto, K.; Hirai, S. Serum concentration of alpha 1-antichymotrypsin is elevated in patients with senile dementia of the Alzheimer type. Prog. Clin. Boil. Res. 1989, 317, 707–714. [Google Scholar]
- Brugge, K.; Katzman, R.; Hill, L.R.; Hansen, L.A.; Saitoh, T. Serological α1-Antichymotrypsin in Down’s syndrome and Alzheimer’s disease. Ann. Neurol. 1992, 32, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Altstiel, L.D.; Lawlor, B.; Mohs, R.C.; Schmeidler, J.; Dalton, A.J.; Mehta, P.D.; Davis, K.L. Elevated Alpha1-Antichymotrypsin serum levels in a subset of nondemented first-degree relatives of Alzheimer’s disease patients. Dement. Geriatr. Cogn. Disord. 1995, 6, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, J.; Schleissner, L.; Tachiki, K.H.; Kling, A.S. Serum α1-antichymotrypsin level as a marker for Alzheimer-type dementia. Neurobiol. Aging 1995, 16, 747–753. [Google Scholar] [CrossRef]
- Licastro, F.; Pedrini, S.; Caputo, L.; Annoni, G.; Davis, L.J.; Ferri, C.; Casadei, V.; Grimaldi, L.M.E. Increased plasma levels of interleukin-1, interleukin-6 and α-1-antichymotrypsin in patients with Alzheimer’s disease: Peripheral inflammation or signals from the brain? J. Neuroimmunol. 2000, 103, 97–102. [Google Scholar] [CrossRef]
- Licastro, F.; Mallory, M.; Hansen, L.; Masliah, E. Increased levels of α-1-antichymotrypsin in brains of patients with Alzheimer’s disease correlate with activated astrocytes and are affected by APOE 4 genotype. J. Neuroimmunol. 1998, 88, 105–110. [Google Scholar] [CrossRef]
- Padmanabhan, J.; Levy, M.; Dickson, D.W.; Potter, H. Alpha1-antichymotrypsin, an inflammatory protein overexpressed in Alzheimer’s disease brain, induces tau phosphorylation in neurons. Brain 2006, 129, 3020–3034. [Google Scholar] [CrossRef]
- Vanni, S.; Moda, F.; Zattoni, M.; Bistaffa, E.; De Cecco, E.; Rossi, M.; Giaccone, G.; Tagliavini, F.; Haïk, S.; Deslys, J.P.; et al. Differential overexpression of SERPINA3 in human prion diseases. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Licastro, F.; Parnetti, L.; Morini, M.C.; Davis, L.J.; Cucinotta, D.; Gaiti, A.; Senin, U. Acute phase reactant α1Antichymotrypsin is increased in cerebrospinal fluid and serum of patients with probable Alzheimer disease. Alzheimer Dis. Assoc. Disord. 1995, 9, 112–118. [Google Scholar] [CrossRef]
- Porcellini, E.; Davis, E.; Chiappelli, M.; Ianni, E.; Di Stefano, G.; Forti, P.; Ravaglia, G.; Licastro, F. Elevated plasma levels of α -1-anti-chymotrypsin in age-related cognitive decline and Alzheimers disease: A potential therapeutic target. Curr. Pharm. Des. 2008, 14, 2659–2664. [Google Scholar] [CrossRef] [PubMed]
- DeKosky, S.T.; Ikonomovic, M.D.; Wang, X.; Farlow, M.; Wisniewski, S.; Lopez, O.L.; Becker, J.T.; Saxton, J.; Klunk, W.E.; Sweet, R.; et al. Plasma and cerebrospinal fluid α1-antichymotrypsin levels in Alzheimer’s disease: Correlation with cognitive impairment. Ann. Neurol. 2002, 53, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, E.; Hirai, S.; Amari, M.; Shoji, M.; Yamaguchi, H.; Okamoto, K.; Ishiguro, K.; Harigaya, Y.; Wakabayashi, K. α1-Antichymotrypsin as a possible biochemical marker for Alzheimer-type dementia. Ann. Neurol. 1990, 28, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Delamarche, C.; Berger, F.; Gallard, L.; Pouplard-Barthelaix, A. Aging and Alzheimer’s disease: Protease inhibitors in cerebrospinal fluid. Neurobiol. Aging 1991, 12, 71–74. [Google Scholar] [CrossRef]
- Loeffler, D.A.; DeMaggio, A.J.; Juneau, P.L.; Brickman, C.M.; Mashour, G.A.; Finkelman, J.H.; Pomara, N.; LeWitt, P.A. Ceruoplasmin is increased in cerebrospinal fluid in Alzheimer’s disease but not Parkinson’s disease. Alzheimer Dis. Assoc. Disord. 1994, 8, 190–197. [Google Scholar] [CrossRef]
- Hare, D.J.; Doecke, J.D.; Faux, N.G.; Rembach, A.; Volitakis, I.; Fowler, C.J.; Grimm, R.; Doble, P.A.; Cherny, R.A.; Masters, C.L.; et al. Decreased plasma iron in Alzheimer’s disease is due to transferrin desaturation. ACS Chem. Neurosci. 2015, 6, 398–402. [Google Scholar] [CrossRef]
- Son, S.M.; Nam, D.W.; Cha, M.-Y.; Kim, K.H.; Byun, J.; Ryu, H.; Mook-Jung, I. Thrombospondin-1 prevents amyloid beta–mediated synaptic pathology in Alzheimer’s disease. Neurobiol. Aging 2015, 36, 3214–3227. [Google Scholar] [CrossRef]
- Merched, A.; Serot, J.-M.; Visvikis, S.; Aguillon, D.; Faure, G.; Siest, G. Apolipoprotein E, transthyretin and actin in the CSF of Alzheimer’s patients: Relation with the senile plaques and cytoskeleton biochemistry. FEBS Lett. 1998, 425, 225–228. [Google Scholar] [CrossRef]
- Han, S.-H.; Jung, E.S.; Sohn, J.-H.; Hong, H.S.; Kim, J.W.; Na, D.L.; Kim, M.; Kim, H.; Ha, H.J.; Kim, Y.H.; et al. Human serum transthyretin levels correlate inversely with Alzheimer’s disease. J. Alzheimer’s Dis. 2011, 25, 77–84. [Google Scholar] [CrossRef]
- Schultz, K.; Nilsson, K.; Nielsen, J.E.; Lindquist, S.G.; Hjermind, L.E.; Andersen, B.B.; Wallin, A.; Petersén, Å. Transthyretin as a potential CSF biomarker for Alzheimer’s disease and dementia with Lewy bodies: Effects of treatment with cholinesterase inhibitors. Eur. J. Neurol. 2010, 17, 456–460. [Google Scholar] [CrossRef]
- Ribeiro, C.A.; Santana, I.; Oliveira, C.; Baldeiras, I.; Moreira, J.; Saraiva, M.J.; Cardoso, I. Transthyretin decrease in plasma of MCI and AD patients: Investigation of mechanisms for disease modulation. Curr. Alzheimer Res. 2012, 9, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Velayudhan, L.; Killick, R.; Hye, A.; Kinsey, A.; Güntert, A.; Lynham, S.; Ward, M.; Leung, R.; Lourdusamy, A.; To, A.W.; et al. Plasma transthyretin as a candidate marker for Alzheimer’s disease. J. Alzheimer’s Dis. 2012, 28, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Haas, C.; Cazorla, P.; De Miguel, C.; Valdivieso, F.; Vázquez, J. Apolipoprotein E forms stable complexes with recombinant Alzheimer’s disease β-amyloid precursor protein. Biochem. J. 1997, 325, 169–175. [Google Scholar] [CrossRef]
- Strittmatter, W.J.; Saunders, A.M.; Schmechel, D.; Pericak-Vance, M.; Enghild, J.; Salvesen, G.S.; Roses, A.D. Apolipoprotein E: High-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA 1993, 90, 1977–1981. [Google Scholar] [CrossRef] [PubMed]
- Vogelgsang, J.; Vukovich, R.; Wedekind, D.; Wiltfang, J. Higher level of mismatch in APOEε4 carriers for amyloid-beta peptide Alzheimer’s disease biomarkers in cerebrospinal fluid. ASN Neuro 2019, 11. [Google Scholar] [CrossRef]
- Liu, C.-C.; Zhao, N.; Fu, Y.; Wang, N.; Linares, C.; Tsai, C.-W.; Bu, G. ApoE4 Accelerates early seeding of amyloid pathology. Neuron 2017, 96, 1024–1032.e3. [Google Scholar] [CrossRef] [PubMed]
- Bachmeier, C.; Paris, D.; Beaulieu-Abdelahad, D.; Mouzon, B.; Mullan, M.; Crawford, F. A multifaceted role for apoE in the clearance of Beta-amyloid across the blood-brain barrier. Neurodegener. Dis. 2013, 11, 13–21. [Google Scholar] [CrossRef]
- Strittmatter, W.J.; Saunders, A.M.; Goedert, M.; Weisgraber, K.H.; Dong, L.M.; Jakes, R.; Huang, D.Y.; Pericak-Vance, M.; Schmechel, D.; Roses, A.D. Isoform-specific interactions of apolipoprotein E with microtubule-associated protein tau: Implications for Alzheimer disease. Proc. Natl. Acad. Sci. USA 1994, 91, 11183–11186. [Google Scholar] [CrossRef]
- Gollin, P.A.; Kalaria, R.N.; Eikelenboom, P.; Rozemuller, A.; Perry, G. α1-Antitrypsin and α1-antichymotrypsin are in the lesions of Alzheimer’s disease. NeuroReport 1992, 3, 201–203. [Google Scholar] [CrossRef]
- Eriksson, S.; Janciauskiene, S.; Lannfelt, L. Alpha 1-antichymotrypsin regulates Alzheimer beta-amyloid peptide fibril formation. Proc. Natl. Acad. Sci. USA 1995, 92, 2313–2317. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, E.; Fiorelli, T.; Norden, M.; Padmanabhan, J. Alpha 1-Antichymotrypsin, an inflammatory protein overexpressed in the brains of patients with Alzheimer’s disease, induces Tau hyperphosphorylation through c-Jun N-terminal kinase activation. Int. J. Alzheimer’s Dis. 2013, 2013, 1–11. [Google Scholar] [CrossRef]
- Nilsson, L.N.G.; Bales, K.R.; Dicarlo, G.; Gordon, M.N.; Morgan, D.; Paul, S.M.; Potter, H. α-1-Antichymotrypsin promotes β-sheet amyloid plaque deposition in a transgenic mouse model of Alzheimer’s disease. J. Neurosci. 2001, 21, 1444–1451. [Google Scholar] [CrossRef]
- Gold, M.; Dolga, A.M.; Koepke, J.; Mengel, D.; Culmsee, C.; Dodel, R.; Koczulla, A.R.; Bach, J.-P. α 1-antitrypsin modulates microglial-mediated neuroinflammation and protects microglial cells from amyloid-β-induced toxicity. J. Neuroinflamm. 2014, 11, 165. [Google Scholar] [CrossRef]
- Wyatt, A.R.; Constantinescu, P.; Ecroyd, H.; Dobson, C.M.; Wilson, M.R.; Kumita, J.R.; Yerbury, J.J. Protease-activated alpha-2-macroglobulin can inhibit amyloid formation via two distinct mechanisms. FEBS Lett. 2013, 587, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.D.; Sagare, A.P.; Friedman, A.E.; Bedi, G.S.; Holtzman, D.M.; Deane, R.; Zlokovic, B.V. Transport pathways for clearance of human alzheimer’s amyloid β-Peptide and apolipoproteins E and J in the mouse central nervous system. Br. J. Pharmacol. 2006, 27, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Storck, S.E.; Meister, S.; Nahrath, J.; Meißner, J.N.; Schubert, N.; Di Spiezio, A.; Baches, S.; Vandenbroucke, R.E.; Bouter, Y.; Prikulis, I.; et al. Endothelial LRP1 transports amyloid-β1–42 across the blood-brain barrier. J. Clin. Investig. 2015, 126, 123–136. [Google Scholar] [CrossRef]
- Tachibana, M.; Holm, M.-L.; Liu, C.-C.; Shinohara, M.; Aikawa, T.; Oue, H.; Yamazaki, Y.; Martens, Y.A.; Murray, M.E.; Sullivan, P.M.; et al. APOE4-mediated amyloid-β pathology depends on its neuronal receptor LRP1. J. Clin. Investig. 2019, 129, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.S.; Eira, J.; Ribeiro, C.A.; Oliveira, Â.; Sousa, M.M.; Cardoso, I.; Liz, M.A. Transthyretin neuroprotection in Alzheimer’s disease is dependent on proteolysis. Neurobiol. Aging 2017, 59, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Alemi, M.; Gaiteiro, C.; Ribeiro, C.A.; Santos, L.M.; Gomes, J.R.; Oliveira, S.M.; Couraud, P.-O.; Weksler, B.; Romero, I.; Saraiva, M.J.; et al. Transthyretin participates in beta-amyloid transport from the brain to the liver- involvement of the low-density lipoprotein receptor-related protein 1? Sci. Rep. 2016, 6, 20164. [Google Scholar] [CrossRef]
- Tenner, A.J. Complement-mediated events in Alzheimer’s disease: Mechanisms and potential therapeutic targets. J. Immunol. 2020, 204, 306–315. [Google Scholar] [CrossRef]
- Piktel, E.; Levental, I.; Durnaś, B.; Janmey, P.A.; Bucki, R. Plasma Gelsolin: Indicator of inflammation and its potential as a diagnostic tool and therapeutic target. Int. J. Mol. Sci. 2018, 19, 2516. [Google Scholar] [CrossRef] [PubMed]
- Hirko, A.C.; Meyer, E.M.; King, M.A.; Hughes, J. Peripheral transgene expression of plasma gelsolin reduces amyloid in transgenic mouse models of Alzheimer’s disease. Mol. Ther. 2007, 15, 1623–1629. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y.; Saito, M.; Lafrancois, J.; Saito, M.; Gaynor, K.; Olm, V.; Wang, L.; Casey, E.; Lu, Y.; Shiratori, C.; et al. Novel therapeutic approach for the treatment of Alzheimer’s disease by peripheral administration of agents with an affinity to β-amyloid. J. Neurosci. 2003, 23, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Corina, K.; Chow, E.P.; Miller, S.; Janmey, P.A.; Pepinsky, R.B. The plasma and cytoplasmic forms of human gelsolin differ in disulfide structure. Biochem. 1996, 35, 9700–9709. [Google Scholar] [CrossRef]
- Solomon, J.P.; Page, L.J.; Balch, W.E.; Kelly, J.W. Gelsolin amyloidosis: Genetics, biochemistry, pathology and possible strategies for therapeutic intervention. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Henriques, A.G.; Oliveira, J.M.; Gomes, B.; Ruivo, R.; Silva, E.F.D.C.E.; Silva, O.A.B.D.C.E. Complexing Aβ prevents the cellular anomalies induced by the Peptide alone. J. Mol. Neurosci. 2014, 53, 661–668. [Google Scholar] [CrossRef][Green Version]
- Martins, T.S.; Catita, J.; Rosa, I.M.; da Cruz e Silva, A.B.; Henriques, A.G. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE 2018, 13, e0198820. [Google Scholar] [CrossRef]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2011, 40, D1241–D1244. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef]
- Simpson, R.J.; Kalra, H.; Mathivanan, S. ExoCarta as a resource for exosomal research. J. Extracell. Vesicles 2012, 1. [Google Scholar] [CrossRef]
- Mathivanan, S.; Simpson, R.J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009, 9, 4997–5000. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.W.; Bond, V.C.; Borràs, F.E.; Breakefield, X.O.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-K.; Kang, B.; Kim, O.Y.; Choi, D.-S.; Lee, J.; Kim, S.R.; Go, G.; Yoon, Y.J.; Kim, J.H.; Jang, S.C.; et al. EVpedia: An integrated database of high-throughput data for systemic analyses of extracellular vesicles. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-K.; Lee, J.; Simpson, R.J.; Lötvall, J.; Gho, Y.S. EVpedia: A community web resource for prokaryotic and eukaryotic extracellular vesicles research. Semin. Cell Dev. Biol. 2015, 40, 4–7. [Google Scholar] [CrossRef]
- Rosa, I.M.; Henriques, A.G.; Carvalho, L.; Oliveira, J.; Silva, O.A.D.C.E. Screening younger individuals in a primary care setting flags putative dementia cases and correlates gastrointestinal diseases with poor cognitive performance. Dement. Geriatr. Cogn. Disord. 2016, 43, 15–28. [Google Scholar] [CrossRef]
- Rosa, I.M.; Henriques, A.G.; Wiltfang, J.; Silva, O.A.D.C.E. Putative dementia cases fluctuate as a function of mini-mental state examination cut-off points. J. Alzheimer’s Dis. 2017, 61, 157–167. [Google Scholar] [CrossRef]

| Gene | Protein Name | CSF | Serum | Plasma | Brain | Publication |
|---|---|---|---|---|---|---|
| A2M | Alpha-2-macroglobulin | ↑/= | ↑/= | [24,25,26] | ||
| ALB | Albumin | = | = | [26,27,28,29] | ||
| APCS | Serum amyloid P-component | = | = | ↑ | [30,31,32] | |
| APOA1 | Apolipoprotein A-I | = | ↓ | ↓/= | = | [33,34,35,36,37,38,39,40,41,42,43] |
| APOA2 | Apolipoprotein A-II | ↓ | ↓/= | [34,35,38,42] | ||
| APOA4 | Apolipoprotein A-IV | ↑ | [41] | |||
| APOB | Apolipoprotein B-100 | ↓/=/↑ | [35,43,44] | |||
| APOC1 | Apolipoprotein C-I | = | ↓ | [42,45] | ||
| APOC3 | Apolipoprotein C-III | ↓ | [43] | |||
| APOE | Apolipoprotein E | =/↑ | ↑/= | ↓/= | [33,39,40,43,46,47] | |
| BCHE | Cholinesterase | = | ↓ | ↑ | [48,49,50] | |
| C1QA, C1QB and C1QC | Complement C1q | ↓* | ↑** | [51,52,53] | ||
| C3 | Complement C3 | ↑ | ↑ | [54,55] | ||
| C4BPα | C4b-binding protein alpha chain | =* | =*/↓ | [35,56] | ||
| CAT | Catalase | = | ↑/= | [57,58,59] | ||
| CFH | Complement factor H | ↓ | = | = | [60,61,62,63] | |
| CFI | Complement factor I | = | [63] | |||
| CLU | Clusterin | ↑ | ↑/= | ↑/= | [33,35,39,46,47,64,65,66,67] | |
| CSTB | Cystatin-B | Not found | ||||
| F12 | Coagulation factor XII | ↑ | = | [68] | ||
| FGB | Fibrinogen beta chain | ↑ | [44] | |||
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | ↑** | ↑ | [69,70] | ||
| GC | Vitamin D-binding protein | ↓ | [35,44] | |||
| GSN | Gelsolin | ↓ | ↓/↑ | =/↓ | [35,71,72,73,74] | |
| HP | Haptoglobin | ↑ | ↑/= | [26,75,76,77,78] | ||
| HSPB1 | Heat shock protein beta-1 | ↑ | [79] | |||
| IGHM | Immunoglobulin heavy constant mu | ↑ | [35] | |||
| L1CAM | Neural cell adhesion molecule L1 | =*** | [19,80] | |||
| LRP1 | Prolow-density lipoprotein receptor-related protein 1 | ↓ | ↑/↓ | [81,82,83,84] | ||
| NCL | Nucleolin | ↓ | [85] | |||
| PFN1 | Profilin-1 | Not found | ||||
| PZP | Pregnancy zone protein | ↑ | ↑ | [86,87] | ||
| RELN | Reelin | ↑ | = | ↑ | [88,89,90] | |
| S100A8 | Protein S100-A8 | ↓ | [91] | |||
| S100A9 | Protein S100-A9 | ↓ | ↓ | = | [91,92,93] | |
| SELENOP | Selenoprotein P | ↑ | ↑ | [94] | ||
| SERPINA1 | Alpha-1-antitrypsin | ↑/= | ↑/= | ↑/= | [26,42,67,95,96] | |
| SERPINA3 | Alpha-1-antichymotrypsin, ACT | ↑/= | ↑/= | ↑ | ↑ | [95,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114] |
| TF | Serotransferrin | = | = | [115,116] | ||
| THBS1 | Thrombospondin-1 | ↓ | ↓ | [91,117] | ||
| TTR | Transthyretin | ↓/= | ↓ | ↓/↑ | [35,118,119,120,121,122] | |
| TUBB | Tubulin | ↓* | [70] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares Martins, T.; Marçalo, R.; Ferreira, M.; Vaz, M.; Silva, R.M.; Martins Rosa, I.; Vogelgsang, J.; Wiltfang, J.; da Cruz e Silva, O.A.B.; Henriques, A.G. Exosomal Aβ-Binding Proteins Identified by “In Silico” Analysis Represent Putative Blood-Derived Biomarker Candidates for Alzheimer´s Disease. Int. J. Mol. Sci. 2021, 22, 3933. https://doi.org/10.3390/ijms22083933
Soares Martins T, Marçalo R, Ferreira M, Vaz M, Silva RM, Martins Rosa I, Vogelgsang J, Wiltfang J, da Cruz e Silva OAB, Henriques AG. Exosomal Aβ-Binding Proteins Identified by “In Silico” Analysis Represent Putative Blood-Derived Biomarker Candidates for Alzheimer´s Disease. International Journal of Molecular Sciences. 2021; 22(8):3933. https://doi.org/10.3390/ijms22083933
Chicago/Turabian StyleSoares Martins, Tânia, Rui Marçalo, Maria Ferreira, Margarida Vaz, Raquel M. Silva, Ilka Martins Rosa, Jonathan Vogelgsang, Jens Wiltfang, Odete A. B. da Cruz e Silva, and Ana Gabriela Henriques. 2021. "Exosomal Aβ-Binding Proteins Identified by “In Silico” Analysis Represent Putative Blood-Derived Biomarker Candidates for Alzheimer´s Disease" International Journal of Molecular Sciences 22, no. 8: 3933. https://doi.org/10.3390/ijms22083933
APA StyleSoares Martins, T., Marçalo, R., Ferreira, M., Vaz, M., Silva, R. M., Martins Rosa, I., Vogelgsang, J., Wiltfang, J., da Cruz e Silva, O. A. B., & Henriques, A. G. (2021). Exosomal Aβ-Binding Proteins Identified by “In Silico” Analysis Represent Putative Blood-Derived Biomarker Candidates for Alzheimer´s Disease. International Journal of Molecular Sciences, 22(8), 3933. https://doi.org/10.3390/ijms22083933








