Secondary Metabolism and Defense Responses Are Differently Regulated in Two Grapevine Cultivars during Ripening
Abstract
1. Introduction
2. Results and Discussion
2.1. Genotype Is the Main Factor that Controls the Berry Anthocyanin Profiling
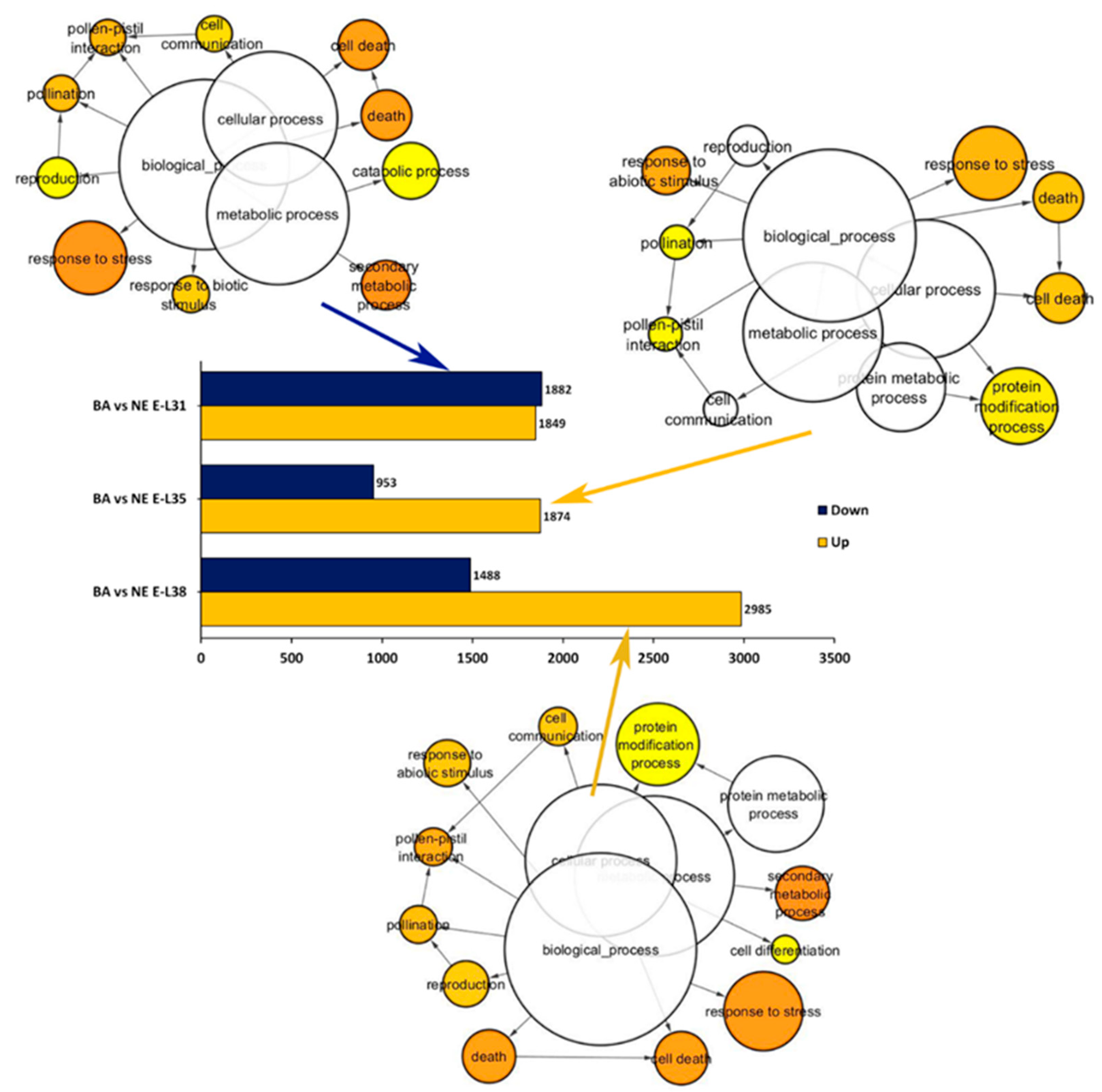
2.2. Overview of the Whole Transcriptome Changes Occurring in ‘Nebbiolo’ and ‘Barbera’ Grapes over Ripening
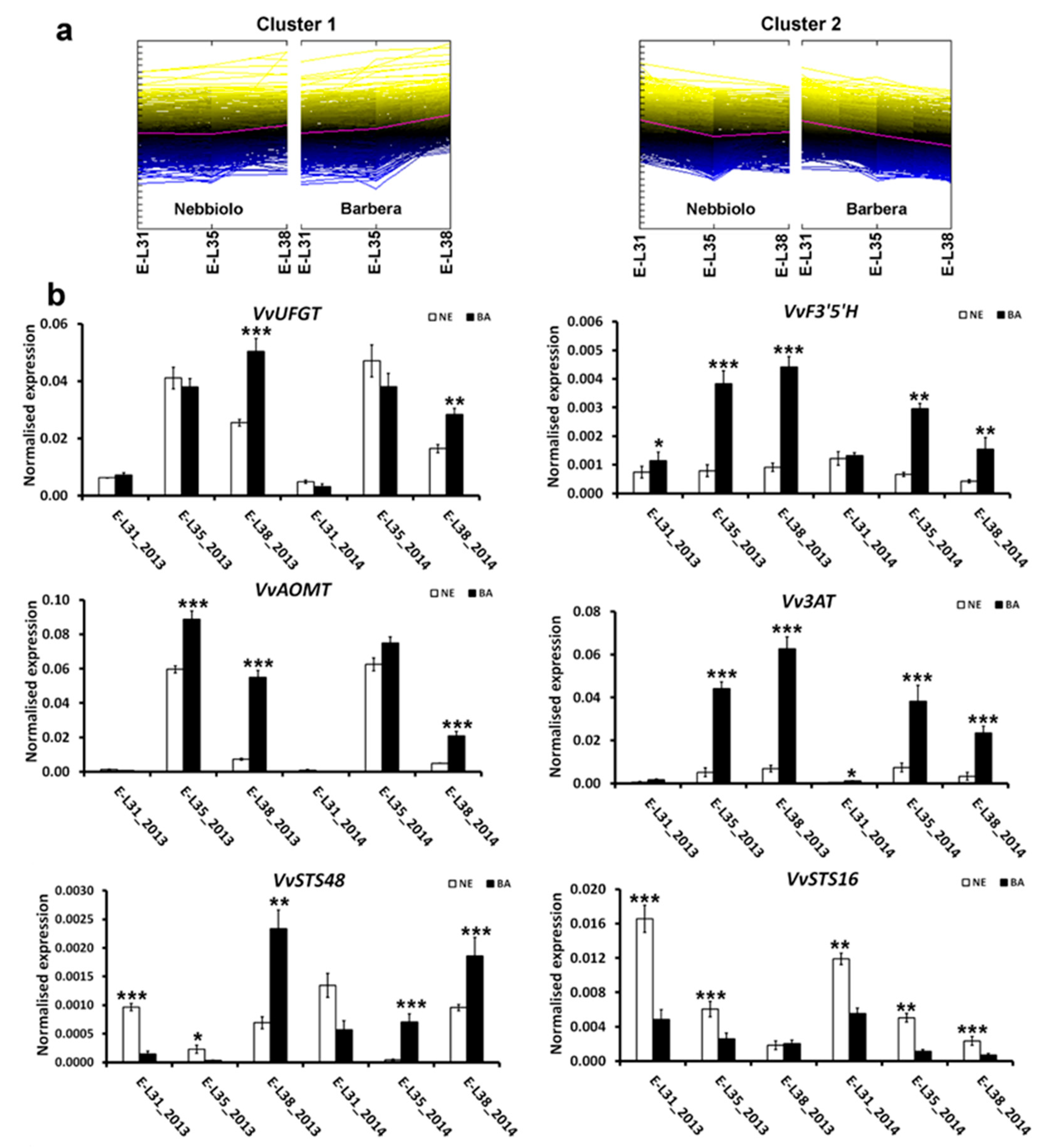
2.3. Genes Associated with Secondary Metabolism Are Differently Reprogrammed in ‘Nebbiolo’ and ‘Barbera’ Grapes
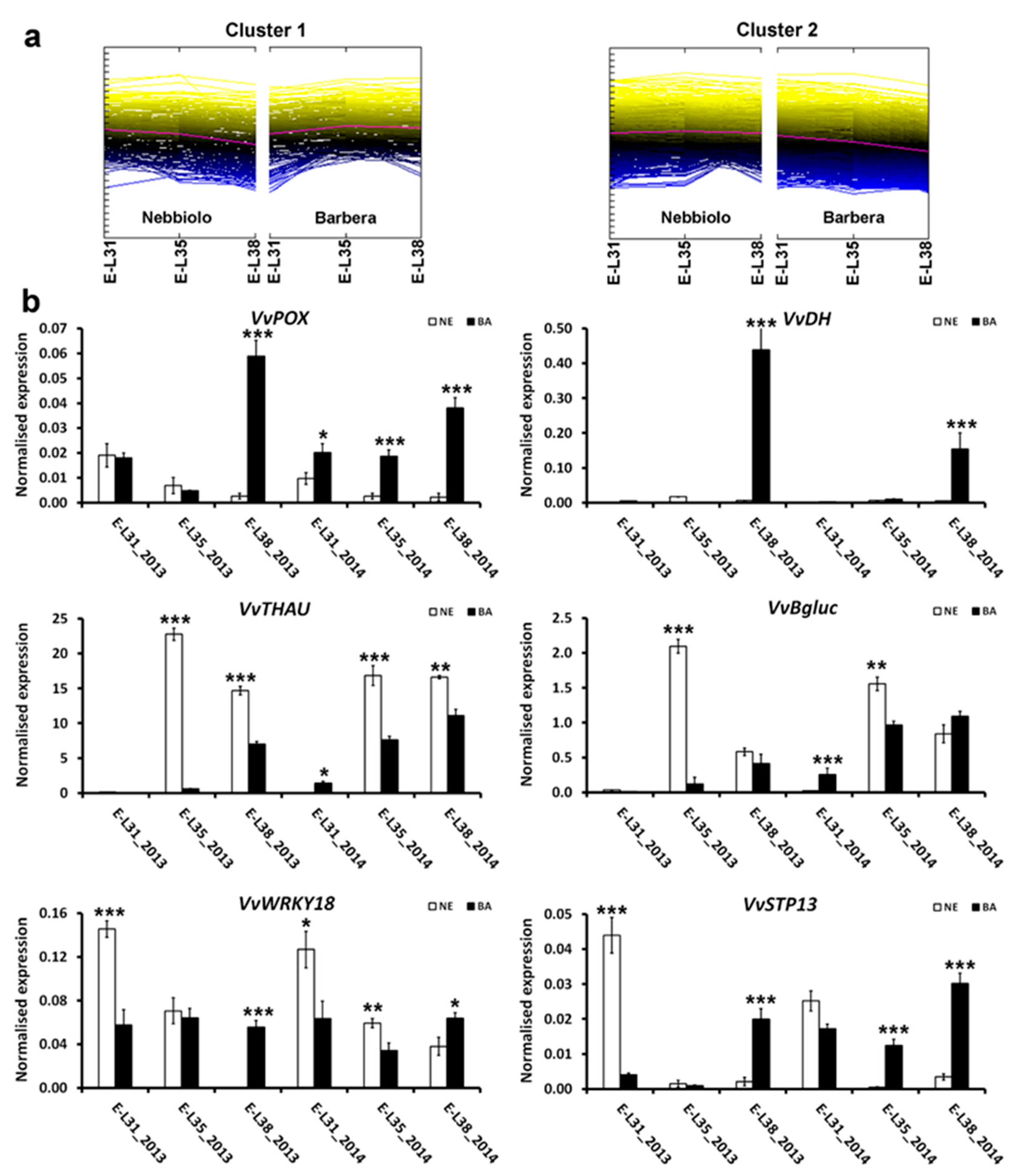
2.4. Regulation of Defense Metabolism Is Tightly Controlled by the Genotype Intrinsic Features
3. Material and Methods
3.1. Plant Material and Experimental Field
3.2. Determination of Anthocyanin Profiles
3.3. RNA Sequencing and Elaboration of the Data
3.4. Quantitative Real-Time PCR (RT-qPCR) Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morales-Castilla, I.; García de Cortázar-Atauri, P.; Cook, B.I.; Lacombe, T.; Parker, A.; van Leeuwen, C.; Nicholas, K.A.; Wolkovich, E.M. Diversity buffers winegrowing regions from climate change losses. Proc. Natl. Acad. Sci. USA 2020, 117, 2864–2869. [Google Scholar] [CrossRef]
- Sultan, S.E. Phenotypic plasticity for plant development, function and life history. Trends Plant Sci. 2010, 5, 537–542. [Google Scholar] [CrossRef]
- Dai, Z.W.; Ollat, N.; Gomes, E.; Decroocq, S.; Tandonnet, J.P.; Bordenave, L.; Pieri, P.; Hilbert, G.; Kappel, C.; van Leeuwen, C.; et al. Ecophysiological, genetic, and molecular causes of variation in grape berry weight and composition, a review. Am. J. Enol. Vitic. 2011, 62, 413–425. [Google Scholar] [CrossRef]
- van Leeuwen, C.; Schultz, H.R.; Garcia de Cortazar-Atauri, I.; Duchêne, E.; Ollat, N.; Pieri, P.; Bois, B.; Goutouly, J.-P.; Quénol, H.; Touzard, J.-M.; et al. Why climate change will not dramatically decrease viticultural suitability in main wine-producing areas by 2050. Proc. Natl. Acad. Sci. USA 2013, 110, E3051–E3052. [Google Scholar] [CrossRef] [PubMed]
- Dal Santo, S.; Tornielli, G.B.; Zenoni, S.; Fasoli, M.; Farina, L.; Anesi, A.; Guzzo, F.; Delledonne, M.; Pezzotti, M. The plasticity of the grapevine berry transcriptome. Genome Biol. 2013, 14, R54. [Google Scholar] [CrossRef]
- Anesi, A.; Stocchero, M.; Dal Santo, S.; Commisso, M.; Zenoni, S.; Ceoldo, S.; Tornielli, G.B.; Siebert, T.E.; Herderich, M.; Pezzotti, M.; et al. Towards a scientific interpretation of the terroir concept, plasticity of the grape berry metabolome. BMC Plant Biol. 2015, 15, 191. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Cramer, G.R.; Ghan, R.; Schlauch, K.A.; Tillett, R.L.; Heymann, H.; Ferrarini, A.; Delledonne, M.; Zenoni, S.; Fasoli, M.; Pezzotti, M. Transcriptomic analysis of the late stages of grapevine Vitis vinifera cv. Cabernet Sauvignon berry ripening reveals significant induction of ethylene signaling and flavor pathways in the skin. BMC Plant Biol. 2014, 14, 370. [Google Scholar] [CrossRef] [PubMed]
- Dal Santo, S.; Fasoli, M.; Negri, S.; D’Incà, E.; Vicenzi, N.; Guzzo, F.; Tornielli, G.B.; Pezzotti, M.; Zenoni, S. Plasticity of the berry ripening program in a white grape variety. Front. Plant Sci. 2016, 7, 970. [Google Scholar] [CrossRef]
- Dal Santo, S.; Zenoni, S.; Sandri, M.; De Lorenzis, G.; Magris, G.; De Paoli, E.; Di Gaspero, G.; Del Fabbro, C.; Morgante, M.; Brancadoro, L.; et al. Grapevine field experiments reveal the contribution of genotype, the influence of environment and the effect of their interaction G×E on the berry transcriptome. Plant J. 2018, 93, 1143–1159. [Google Scholar] [CrossRef]
- Pagliarani, C.; Boccacci, P.; Chitarra, W.; Cosentino, E.; Sandri, M.; Perrone, I.; Mori, A.; Cuozzo, D.; Nerva, L.; Rossato, M.; et al. Distinct metabolic signals underlie clone by environment interplay in ‘Nebbiolo’ grapes over ripening. Front. Plant Sci. 2019, 10, 1575. [Google Scholar] [CrossRef]
- Degu, A.; Hochberg, U.; Sikron, N.; Venturini, L.; Buson, G.; Ghan, R.; Plaschkes, I.; Batushansky, A.; Chalifa-Caspi, V.; Mattivi, F.; et al. Metabolite and transcript profiling of berry skin during fruit development elucidates differential regulation between Cabernet Sauvignon and Shiraz cultivars at branching points in the polyphenol pathway. BMC Plant Biol. 2014, 14, 188. [Google Scholar] [CrossRef]
- Massonnet, M.; Fasoli, M.; Tornielli, G.B.; Altieri, M.; Sandri, M.; Zuccolotto, P.; Paci, P.; Gardiman, M.; Zenoni, S.; Pezzotti, M. Ripening transcriptomic program in red and white grapevine varieties correlates with berry skin anthocyanin accumulation. Plant Physiol. 2017, 174, 2376–2396. [Google Scholar] [CrossRef] [PubMed]
- Chitarra, W.; Perrone, I.; Avanzato, C.G.; Minio, A.; Boccacci, P.; Santini, D.; Gilardi, G.; Siciliano, I.; Gullino, M.L.; Delledonne, M.; et al. Grapevine grafting: Scion transcript profiling and defense-related metabolites induced by rootstocks. Front. Plant Sci. 2017, 8, 654. [Google Scholar] [CrossRef] [PubMed]
- Catacchio, C.R.; Alagna, F.; Perniola, R.; Bergamini, C.; Rotunno, S.; Calabrese, F.M.; Crupi, P.; Antonacci, D.; Ventura, M.; Cardone, M.F. Transcriptomic and genomic structural variation analyses on grape cultivars reveal new insights into the genotype-dependent responses to water stress. Sci. Rep. 2019, 9, 2809. [Google Scholar] [CrossRef] [PubMed]
- Pagliarani, C.; Moine, A.; Chitarra, W.; Meloni, G.R.; Abbà, S.; Nerva, L.; Pugliese, M.; Gullino, M.L.; Gambino, G. The molecular priming of defense responses is differently regulated in grapevine genotypes following elicitor application against powdery mildew. Int. J. Mol. Sci. 2020, 21, 6776. [Google Scholar] [CrossRef] [PubMed]
- Zombardo, A.; Crosatti, C.; Bagnaresi, P.; Bassolino, L.; Reshef, N.; Puccioni, S.; Faccioli, P.; Tafuri, A.; Delledonne, M.; Fait, A.; et al. Transcriptomic and biochemical investigations support the role of rootstock-scion interaction in grapevine berry quality. BMC Genom. 2020, 21, 468. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Fischer, M.; Colot, V.; Bossdorf, O. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol. 2013, 197, 314–322. [Google Scholar] [CrossRef]
- Fortes, A.M.; Gallusci, P. Plant stress responses and phenotypic plasticity in the epigenomics era, perspectives on the grapevine scenario, a model for perennial crop plants. Front. Plant Sci. 2017, 8, 82. [Google Scholar] [CrossRef]
- Xie, H.; Konate, M.; Sai, N.; Tesfamicael, K.G.; Cavagnaro, T.; Gilliham, M.; Breen, J.; Metcalfe, A.; Stephen, J.R.; De Bei, R.; et al. Global DNA methylation patterns can play a role in defining terroir in grapevine Vitis vinifera cv. Shiraz. Front. Plant Sci. 2017, 8, 1860. [Google Scholar] [CrossRef] [PubMed]
- Pagliarani, C.; Gambino, G.; Ferrandino, A.; Chitarra, W.; Vrhovsek, U.; Cantù, D.; Palmano, S.; Marzachì, C.; Schubert, A. Molecular memory of Flavescence dorée phytoplasma in recovering grapevines. Hortic. Res. 2020, 7, 126. [Google Scholar] [CrossRef]
- Varela, A.; Ibañez, V.N.; Alonso, R.; Zavallo, D.; Asurmendi, S.; Gomez Talquenca, S.; Marfil, C.F.; Berli, F.J. Vineyard environments influence Malbec grapevine phenotypic traits and DNA methylation patterns in a clone-dependent way. Plant Cell Rep. 2020, 40, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Harding, J.; Vouillamoz, J. Wine Grapes: A complete Guide to 1368 Vine Varieties, Including Their Origins and Flavours; Penguin Books: London, UK, 2012; ISBN 13:9780062206367. [Google Scholar]
- Raimondi, S.; Tumino, G.; Ruffa, P.; Boccacci, P.; Gambino, G.; Schneider, A. DNA-based genealogy reconstruction of Nebbiolo, Barbera and other ancient grapevine cultivars from northwestern Italy. Sci. Rep. 2020, 10, 15782. [Google Scholar] [CrossRef]
- Schneider, A.; Boccacci, P.; Botta, R. Genetic relationships among grape cultivars from North-Western Italy. Acta Hort. 2003, 603, 229–235. [Google Scholar] [CrossRef]
- Gambino, G.; Dal Molin, A.; Boccacci, P.; Minio, A.; Chitarra, W.; Avanzato, C.G.; Tononi, P.; Perrone, I.; Raimondi, S.; Schneider, A.; et al. Whole-genome sequencing and SNV genotyping of ‘Nebbiolo’ (Vitis vinifera L.) clones. Sci. Rep. 2017, 7, 17294. [Google Scholar] [CrossRef]
- Guidoni, S.; Ferrandino, A.; Novello, V. Effects of seasonal and agronomical practices on skin anthocyanin profile of Nebbiolo grapes. Am. J. Enol. Vitic. 2008, 59, 22–29. [Google Scholar]
- Rio Segade, S.; Pace, C.; Torchio, F.; Giacosa, S.; Gerbi, V.; Rolle, L. Impact of maceration enzymes on skin softening and relationship with anthocyanin extraction in wine grapes with different anthocyanin profiles. Food Res. Int. 2015, 71, 50–57. [Google Scholar] [CrossRef]
- Mattivi, F.; Guzzon, R.; Vrhovsek, U.; Stefanini, M.; Velasco, R. Metabolite profiling of grape: Flavonols and anthocyanins. J. Agric. Food Chem. 2006, 54, 7692–7702. [Google Scholar] [CrossRef]
- Falginella, L.; Castellarin, S.D.; Testolin, R.; Gambetta, G.A.; Morgante, M.; Di Gaspero, G. Expansion and subfunctionalisation of favonoid 3′,5′-hydroxylases in the grapevine lineage. BMC Genom. 2010, 11, 562. [Google Scholar] [CrossRef] [PubMed]
- Falginella, L.; Di Gaspero, G.; Castellarin, S.D. Expression of flavonoid genes in the red grape berry of ‘Alicante Bouschet’ varies with the histological distribution of anthocyanins and their chemical composition. Planta 2012, 236, 1037–1051. [Google Scholar] [CrossRef]
- Ferrandino, A.; Carra, A.; Rolle, L.; Schneider, A.; Schubert, A. Profiling of hydroxycinnamoyl tartrates and acylated anthocyanins in the skin of 34 Vitis vinifera genotypes. J. Agric. Food Chem. 2012, 60, 4931–4945. [Google Scholar] [CrossRef]
- Chorti, E.; Guidoni, S.; Ferrandino, A.; Novello, V. Effect of different cluster sunlight exposure levels on ripening and anthocyanin accumulation in Nebbiolo grapes. Am. J. Enol. Vitic. 2010, 61, 23–30. [Google Scholar]
- Ortega-Regules, A.; Romero-Cascales, I.; Lòpez-Roca, J.M.; Ros-García, J.M.; Gomez-Plaza, E. Anthocyanin fingerprint of grapes: Environmental and genetic variations. J. Sci. Food Agric. 2006, 86, 1460–1467. [Google Scholar] [CrossRef]
- González-Neves, G.; Gil, G.; Barreiro, L. Influence of grape variety on the extraction of anthocyanins during the fermentation on skins. Eur. Food Res. Technol. 2008, 226, 1349–1355. [Google Scholar] [CrossRef]
- Castellarin, S.D.; Pfeiffer, A.; Sivilotti, P.; Degan, M.; Peterlunger, E.; Di Gaspero, G. Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ. 2007, 30, 1381–1399. [Google Scholar] [CrossRef]
- Ferrandino, A.; Lovisolo, C. Abiotic stress effects on grapevine Vitis vinifera L., focus on abscisic acid-mediated consequences on secondary metabolism and berry quality. Environ. Exp. Bot. 2014, 103, 138–147. [Google Scholar] [CrossRef]
- Bergqvist, J.; Dokoozlian, N.; Ebisuda, N. Sunlight exposure and temperature effects on berry growth and composition of Cabernet Sauvignon and Grenache in the Central San Joaquin Valley of California. Am. J. Enol. Vitic. 2001, 52, 1–7. [Google Scholar]
- Pomar, F.; Novo, M.; Masa, A. Varietal differences among the anthocyanin profiles of 50 red table grape cultivars studied by high performance liquid chromatography. J. Chromatogr. A 2005, 1094, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Ferrandino, A.; Guidoni, S. Anthocyanins, flavonols and hydroxycinnamates, an attempt to use them to discriminate Vitis vinifera L. cv ‘Barbera’ clones. Eur. Food Res. Technol. 2010, 230, 417–427. [Google Scholar] [CrossRef]
- Coombe, B.G. Adoption of a system for identifying grapevine growth stages. Aust. J. Grape Wine Res. 1995, 1, 104–110. [Google Scholar] [CrossRef]
- Lorenz, D.H.; Eichhorn, K.W.; Bleiholder, H.; Klose, R.; Meier, U.; Weber, E. Growth stages of the grapevine: Phenological growth stages of the grapevine (Vitis vinifera L. ssp. vinifera)-codes and descriptions according to the extended BBCH scale. Aust. J. Grape Wine Res. 1995, 1, 100–103. [Google Scholar] [CrossRef]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Retrotransposon induced mutations in grape skin color. Science 2004, 304, 982. [Google Scholar] [CrossRef]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.; Thomas, M.R.; Robinson, S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Castellarin, S.D.; Di Gaspero, G. Transcriptional control of anthocyanin biosynthetic genes in extreme phenotypes for berry pigmentation of naturally occurring grapevines. BMC Plant Biol. 2007, 7, 46. [Google Scholar] [CrossRef]
- Hugheney, P.; Provenzano, S.; Verriès, C.; Ferrandino, A.; Meudec, E.; Batelli, G.; Merdinoglu, D.; Cheynier, V.; Schubert, A.; Ageorges, A. A novel cation-dependent O-methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiol. 2009, 150, 2057–2070. [Google Scholar] [CrossRef]
- Rinaldo, A.R.; Cavallini, E.; Jia, Y.; Moss, S.M.; McDavid, D.A.; Hooper, L.C.; Robinson, S.P.; Tornielli, G.B.; Zenoni, S.; Ford, C.M.; et al. A grapevine anthocyanin acyltransferase, transcriptionally regulated by VvMYBA, can produce most acylated anthocyanins present in grape skins. Plant Physiol. 2015, 169, 1897–1916. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.R.; Nam, K.H.; D’Auria, J.C.; Pichersky, E. S-Adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch. Biochem. Biophys. 1999, 367, 9–16. [Google Scholar] [CrossRef]
- Negre, F.; Kolosova, N.; Knoll, J.M.; Kish, C.M.; Dudareva, N. Novel S-adenosyl-Lmethionine: Salicylic acid carboxyl methyltransferase, an enzyme responsible for biosynthesis of methyl salicylate and methyl benzoate, is not involved in floral scent production in snapdragon flowers. Arch. Biochem. Biophys. 2002, 406, 261–270. [Google Scholar] [CrossRef]
- Guillaumie, S.; Fouquet, R.; Kappel, C.; Camps, C.; Terrier, N.; Moncomble, D.; Dunlevy, J.D.; Davies, C.; Boss, P.K.; Delrot, S. Transcriptional analysis of late ripening stages of grapevine berry. BMC Plant Biol. 2011, 11, 165. [Google Scholar] [CrossRef] [PubMed]
- Vannozzi, A.; Dry, I.B.; Fasoli, M.; Zenoni, S.; Lucchin, M. Genome-wide analysis of the grapevine stilbene synthase multigenic family: Genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol. 2012, 12, 130. [Google Scholar] [CrossRef] [PubMed]
- Gatto, P.; Vrhovsek, U.; Muth, J.; Segala, C.; Romualdi, C.; Fontana, P.; Pruefer, D.; Stefanini, M.; Moser, C.; Mattivi, F.; et al. Ripening and genotype control stilbene accumulation in healthy grapes. J. Agric. Food Chem. 2008, 56, 11773–11785. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, V.; Matus, J.T.; Pinto-Carnide, O.; Carrasco, D.; Arroyo-García, R.; Castro, I. Genetic analysis of a white-to-red berry skin color reversion and its transcriptomic and metabolic consequences in grapevine (Vitis vinifera cv. ‘Moscatel Galego’). BMC Genom. 2019, 20, 952. [Google Scholar] [CrossRef]
- Vincenzi, S.; Tomasi, D.; Gaiotti, F.; Lovat, L.; Giacosa, S.; Torchio, F.; Río Segade, S.; Rolle, L. Comparative study of the resveratrol content of twenty-one Italian red grape varieties. S. Afr. J. Enol. Viticult. 2013, 34, 30–35. [Google Scholar] [CrossRef]
- Chitarra, W.; Balestrini, R.; Vitali, M.; Pagliarani, C.; Perrone, I.; Schubert, A.; Lovisolo, C. Gene expression in vessel-associated cells upon xylem embolism repair in Vitis vinifera L. petioles. Planta 2014, 239, 887–899. [Google Scholar] [CrossRef]
- Wang, M.; Vannozzi, A.; Wang, G.; Liang, Y.H.; Tornielli, G.B.; Zenoni, S.; Cavallini, E.; Pezzotti, M.; Cheng, Z.-M. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic. Res. 2014, 1, 16. [Google Scholar] [CrossRef]
- Gilardi, G.; Chitarra, W.; Moine, A.; Mezzalama, M.; Boccacci, P.; Pugliese, M.; Gullino, M.L.; Gambino, G. Biological and molecular interplay between two viruses and powdery and downy mildews in two grapevine cultivars. Hortic. Res. 2020, 7, 188. [Google Scholar] [CrossRef]
- Gòmez-Ariza, J.; Campo, S.; Rufat, M.; Estopà, M.; Messeguer, J.; San Segundo, B.; Coca, M. Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRms Protein in rice plants. Mol. Plant Microbe Interact. 2007, 20, 832–842. [Google Scholar] [CrossRef] [PubMed]
- Roggia, C.; Caciagli, P.; Galetto, L.; Pacifico, D.; Veratti, F.; Bosco, D.; Marzachì, C. Flavescence dorée phytoplasma titre in field-infected Barbera and Nebbiolo grapevines. Plant Pathol. 2014, 63, 31–41. [Google Scholar] [CrossRef]
- Margaria, P.; Ferrandino, A.; Caciagli, P.; Kedrina, O.; Schubert, A.; Palmano, S. Metabolic and transcript analysis of the flavonoid pathway in diseased and recovered Nebbiolo and Barbera grapevines (Vitis vinifera L.) following infection by Flavescence dorée phytoplasma. Plant Cell Environ. 2014, 37, 2183–2200. [Google Scholar] [CrossRef]
- Marzachì, C.; Alma, A.; d’Aquilio, M.; Minuto, G.; Boccardo, G. Detection and identification of phytoplasmas infecting cultivated and wild plants in Liguria Italian Riviera. J. Plant Pathol. 1999, 81, 127–136. [Google Scholar] [CrossRef]
- Gambino, G. Multiplex RT-PCR method for the simultaneous detection of nine grapevine viruses. Methods Mol. Biol. 2015, 1236, 39–47. [Google Scholar] [CrossRef]
- Ferrandino, A.; Pagliarani, C.; Carlomagno, A.; Schubert, A.; Agati, G. Improved fluorescence-based evaluation of flavonoid in red and white wine grape cultivars. Aust. J. Grape Wine Res. 2017, 23, 207–214. [Google Scholar] [CrossRef]
- Torchio, F.; Cagnasso, E.; Gerbi, V.; Rolle, L. Mechanical properties, phenolic composition and extractability indices of Barbera grapes of different soluble solids contents from several growing areas. Anal. Chim. Acta 2010, 660, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Rolle, L.; Torchio, F.; Ferrandino, A.; Guidoni, S. Influence of winegrape skin hardness on the kinetics of anthocyanin extraction. Int. J. Food Prop. 2012, 15, 249–261. [Google Scholar] [CrossRef][Green Version]
- Grimplet, J.; Van Hemert, J.; Carbonell-Bejerano, P.; Díaz-Riquelme, J.; Dickerson, J.; Fennell, A.; Pezzotti, M.; Martínez-Zapater, J.M. Comparative analysis of grapevine whole-genome gene predictions, functional annotation, categorization and integration of the predicted gene sequences. BMC Res. Notes 2012, 5, 213. [Google Scholar] [CrossRef]
- Grimplet, J.; Cramer, G.R.; Dickerson, J.A.; Mathiason, K.; Van Hemert, J.; Fennell, A.Y. VitisNet, “Omics” integration through grapevine molecular networks. PLoS ONE 2009, 4, e8365. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Margaria, P.; Palmano, S. Response of the Vitis vinifera L. cv. ‘Nebbiolo’ proteome to Flavescence dorée phytoplasma infection. Proteomics 2011, 11, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Cuozzo, D.; Fasoli, M.; Pagliarani, C.; Vitali, M.; Boccacci, P.; Pezzotti, M.; Mannini, F. Co-evolution between Grapevine rupestris stem pitting-associated virus and Vitis vinifera L. leads to decreased defense responses and increased transcription of genes related to photosynthesis. J. Exp. Bot. 2012, 63, 5919–5933. [Google Scholar] [CrossRef] [PubMed]
| 2013 | 2014 | Significance | |||||
|---|---|---|---|---|---|---|---|
| Nebbiolo | Barbera | Nebbiolo | Barbera | Genotype | Year | Genotype x Year | |
| Total Soluble Solids (TSS, °Brix) | 23.95 ± 0.18 | 27.28 ± 0.50 | 23.07 ± 0.46 | 23.18 ± 0.55 | ** | *** | ** |
| Total flavonoids (mg kg−1) | 2692 ± 741 | 4206 ± 210 | 3756 ± 224 | 2585 ± 96 | NS | * | *** |
| Total anthocyanins (mg kg−1) | 749 ± 26.96 | 1719 ± 125.83 | 904 ± 25.98 | 1108 ± 59.17 | *** | *** | *** |
| Anthocyanin extractability (%) | 61.53 ± 5.22 | 49.38 ± 0.88 | 76 ± 1.15 | 62 ± 2.74 | *** | *** | NS |
| Delphinidin-3-O-glucoside | 4.72 ± 0.33 | 21.16 ± 2.14 | 3.80 ± 0.10 | 18.5 ± 1.98 | *** | NS | NS |
| Cyanidin-3-O-glucoside | 15.61 ± 0.95 | 15.6 ± 1.48 | 14.73 ± 1.47 | 7.99 ± 1.14 | * | ** | * |
| Petunidin-3-O-glucoside | 4.10 ± 0.26 | 15.72 ± 1.38 | 3.43 ± 0.09 | 15.51 ± 0.29 | *** | NS | NS |
| Peonidin-3-O-glucoside | 51.96 ± 1.66 | 11.33 ± 1.07 | 57.60 ± 0.44 | 7.70 ± 0.75 | *** | NS | ** |
| Malvidin-3-O-glucoside | 18.28 ± 1.91 | 24.58 ± 1.5 | 15.50 ± 1.15 | 38.71 ± 0.99 | *** | ** | *** |
| Delphinidin-acetylglucoside | 0.09 ± 0.01 | 1.7 ± 0.16 | 0.07 ± 0.03 | 1.38 ± 0.48 | *** | NS | NS |
| Cyanidin-acetylglucoside | 0.27 ± 0.02 | 1.15 ± 0.09 | 0.23 ± 0.03 | 0.47 ± 0.15 | *** | *** | *** |
| Petunidin-acetylglucoside | 0.06 ± 0.01 | 1.48 ± 0.10 | 0.03 ± 0.03 | 1.23 ± 0.50 | *** | NS | NS |
| Peonidin-acetylglucoside | 1.14 ± 0.07 | 0.72 ± 0.06 | 0.90 ± 0.00 | 0.38 ± 0.13 | *** | ** | NS |
| Malvidin-acetylglucoside | 0.55 ± 0.05 | 2.3 ± 0.08 | 0.37 ± 0.03 | 2.98 ± 1.26 | *** | NS | NS |
| Peonidin-caffeoylglucoside | 0.11 ± 0.01 | 0.89 ± 0.18 | 0.10 ± 0.00 | 0.98 ± 0.16 | *** | NS | NS |
| Malvidin-caffeoylglucoside | 0.32 ± 0.01 | 0.03 ± 0.01 | 0.30 ± 0.00 | 0.03 ± 0.01 | *** | ** | * |
| Dephinidin p-coumaroylglucoside | 0.14 ± 0.01 | 0.08 ± 0.02 | 0.10 ± 0.00 | 0.13 ± 0.07 | NS | NS | NS |
| Cyanidin p-coumaroylglucoside | 0.40 ± 0.02 | 0.93 ± 0.18 | 0.40 ± 0.06 | 0.58 ± 0.07 | *** | * | * |
| Petunidin p-coumaroylglucoside | 0.09 ± 0.01 | 0.59 ± 0.07 | 0.10 ± 0.00 | 0.68 ± 0.12 | *** | NS | NS |
| Peonidin p-coumaroylglucoside | 1.62 ± 0.08 | 0.49 ± 0.08 | 1.90 ± 0.06 | 0.34 ± 0.11 | *** | NS | * |
| Malvidin p-coumaroylglucoside | 0.54 ± 0.06 | 1.25 ± 0.20 | 0.53 ± 0.03 | 2.41 ± 0.33 | *** | ** | *** |
| Total free trihydroxylated anthocyanins | 27.10 ± 2.43 | 61.46 ± 1.53 | 22.77 ± 1.16 | 72.72 ± 1.12 | *** | NS | ** |
| Total free dihydroxylated anthocyanins | 67.57 ± 2.43 | 26.93 ± 1.46 | 72.32 ± 1.09 | 15.69 ± 1.09 | *** | NS | ** |
| Total acylated anthocyanins | 5.33 ± 0.16 | 11.61 ± 0.45 | 4.90 ± 0.07 | 11.59 ± 1.94 | *** | NS | NS |
| Trihydroxylated/dihydroxylated anthocyanins | 0.40 ± 0.05 | 2.30 ± 0.19 | 0.32 ± 0.02 | 4.67 ± 0.27 | *** | *** | *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gambino, G.; Boccacci, P.; Pagliarani, C.; Perrone, I.; Cuozzo, D.; Mannini, F.; Gribaudo, I. Secondary Metabolism and Defense Responses Are Differently Regulated in Two Grapevine Cultivars during Ripening. Int. J. Mol. Sci. 2021, 22, 3045. https://doi.org/10.3390/ijms22063045
Gambino G, Boccacci P, Pagliarani C, Perrone I, Cuozzo D, Mannini F, Gribaudo I. Secondary Metabolism and Defense Responses Are Differently Regulated in Two Grapevine Cultivars during Ripening. International Journal of Molecular Sciences. 2021; 22(6):3045. https://doi.org/10.3390/ijms22063045
Chicago/Turabian StyleGambino, Giorgio, Paolo Boccacci, Chiara Pagliarani, Irene Perrone, Danila Cuozzo, Franco Mannini, and Ivana Gribaudo. 2021. "Secondary Metabolism and Defense Responses Are Differently Regulated in Two Grapevine Cultivars during Ripening" International Journal of Molecular Sciences 22, no. 6: 3045. https://doi.org/10.3390/ijms22063045
APA StyleGambino, G., Boccacci, P., Pagliarani, C., Perrone, I., Cuozzo, D., Mannini, F., & Gribaudo, I. (2021). Secondary Metabolism and Defense Responses Are Differently Regulated in Two Grapevine Cultivars during Ripening. International Journal of Molecular Sciences, 22(6), 3045. https://doi.org/10.3390/ijms22063045







