Hyalectanase Activities by the ADAMTS Metalloproteases
Abstract
1. Introduction
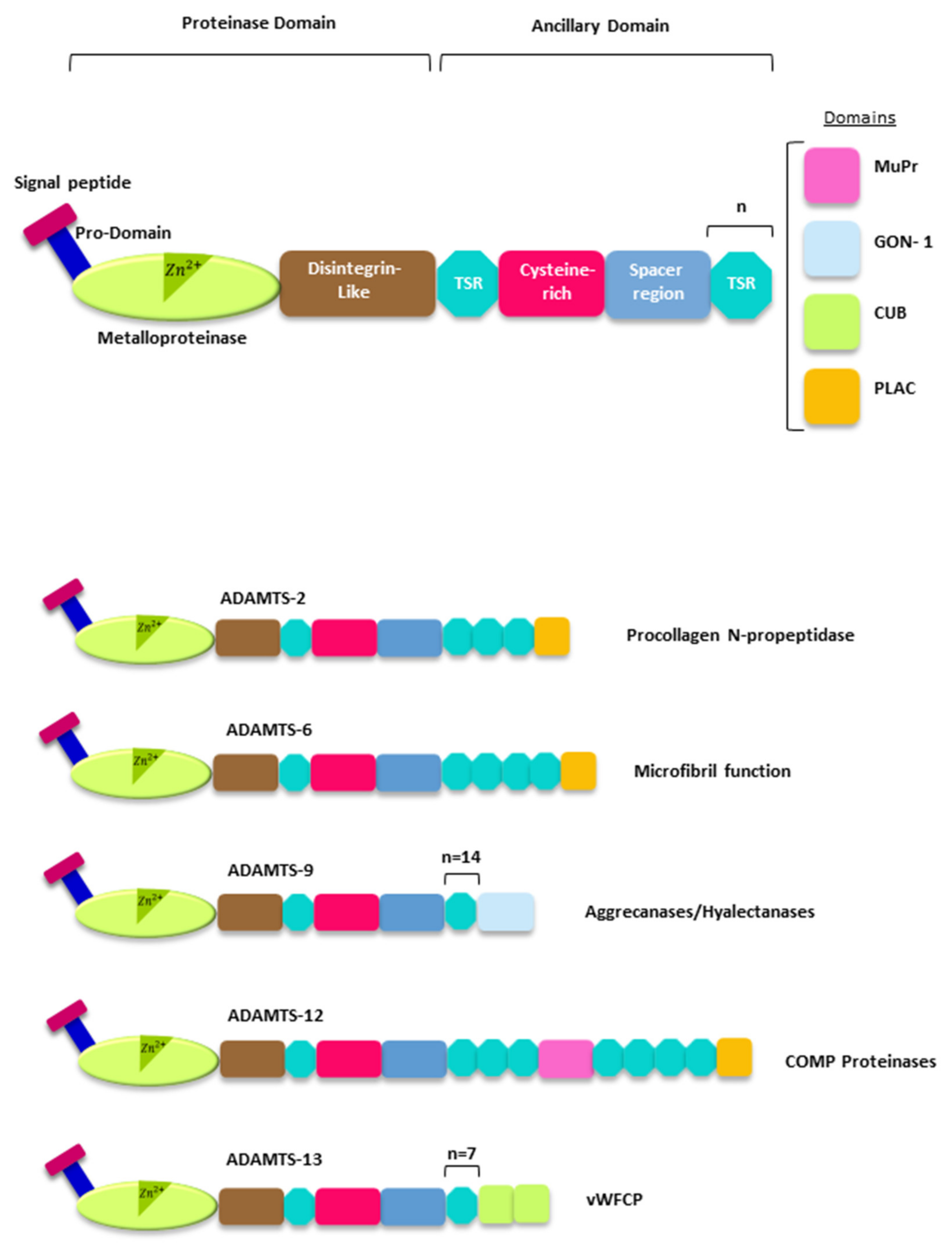
2. The ADAMTSs Metalloproteases
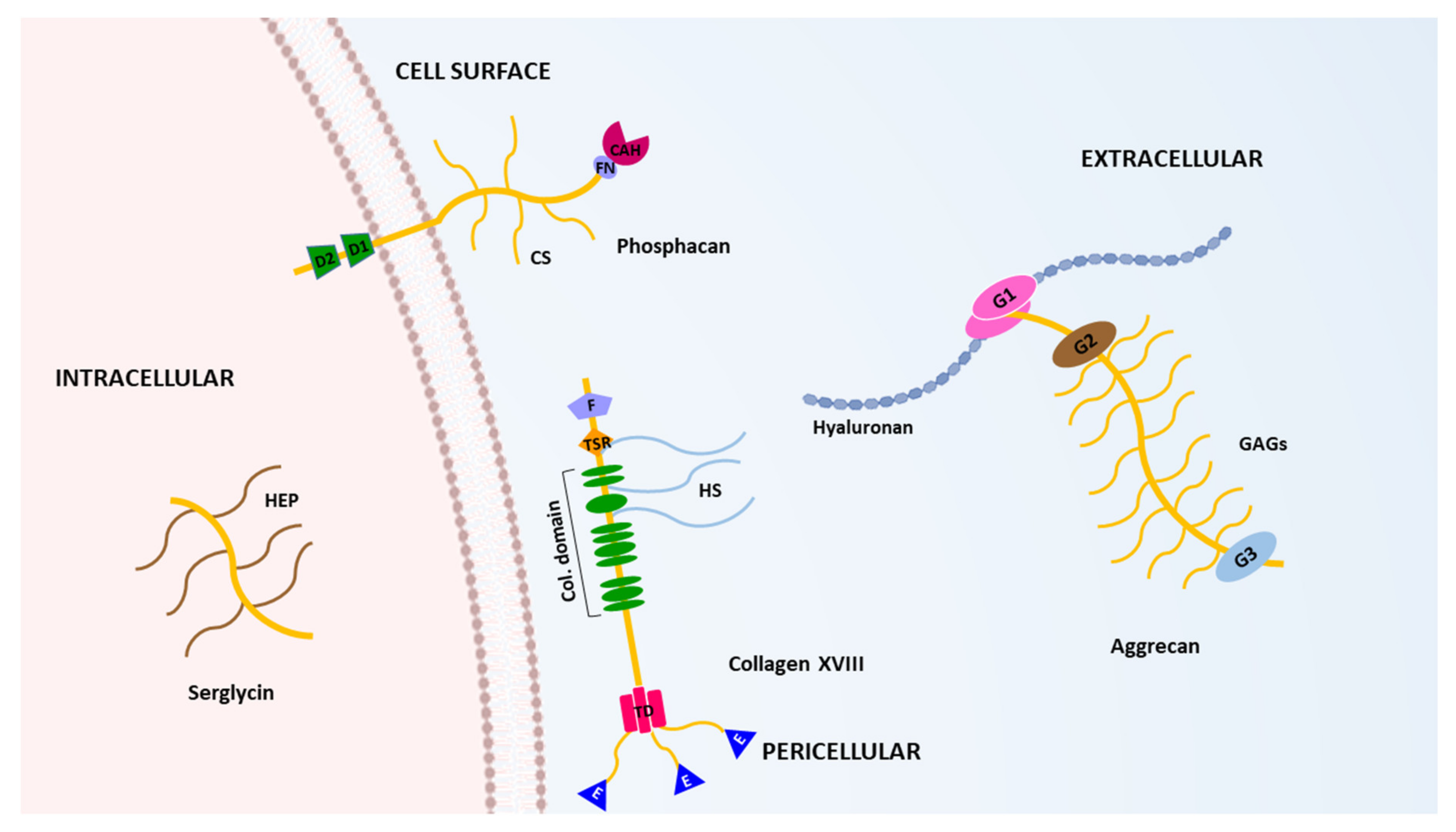
3. The Hyalectans
4. Hyalectans Cleavage by ADAMTSs
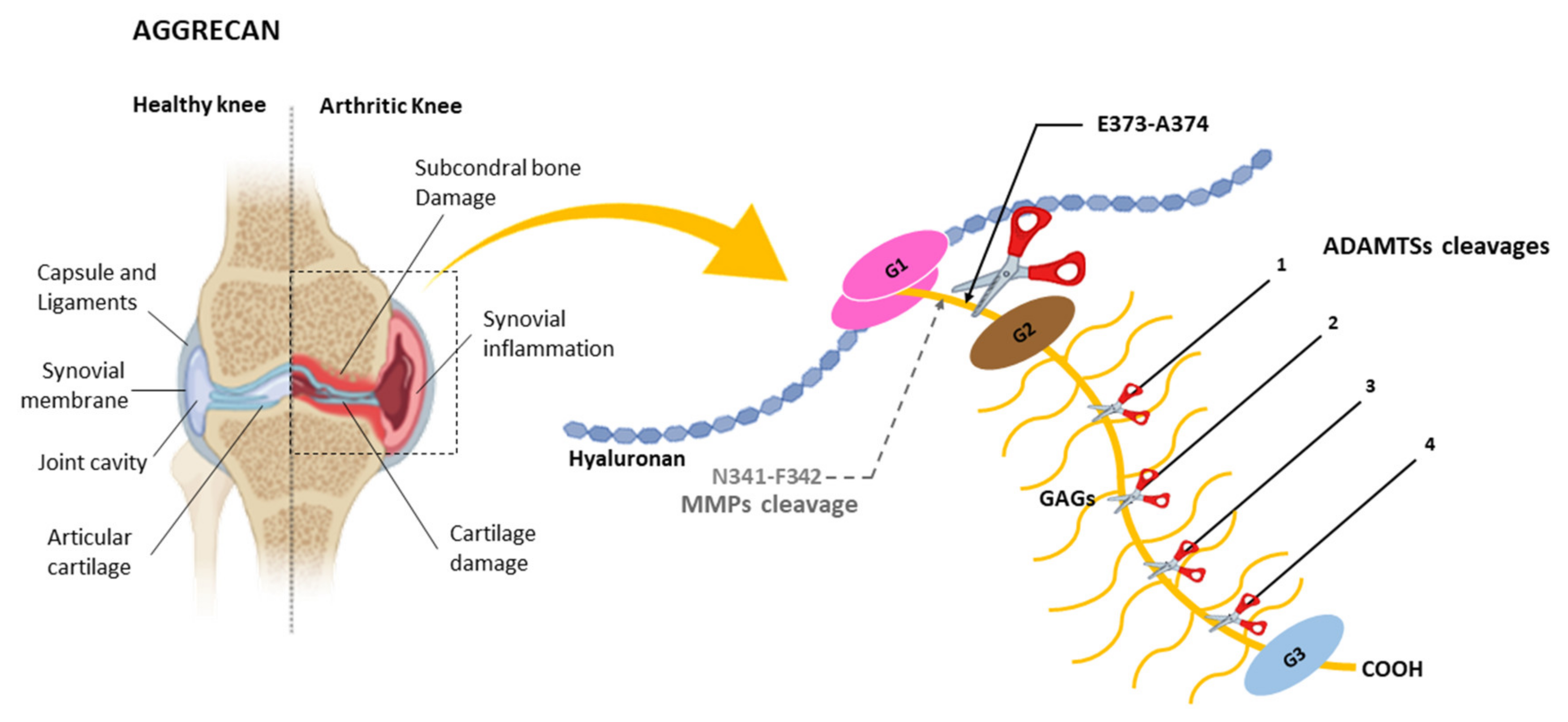
4.1. Cleavage of Aggrecan by ADAMTSs
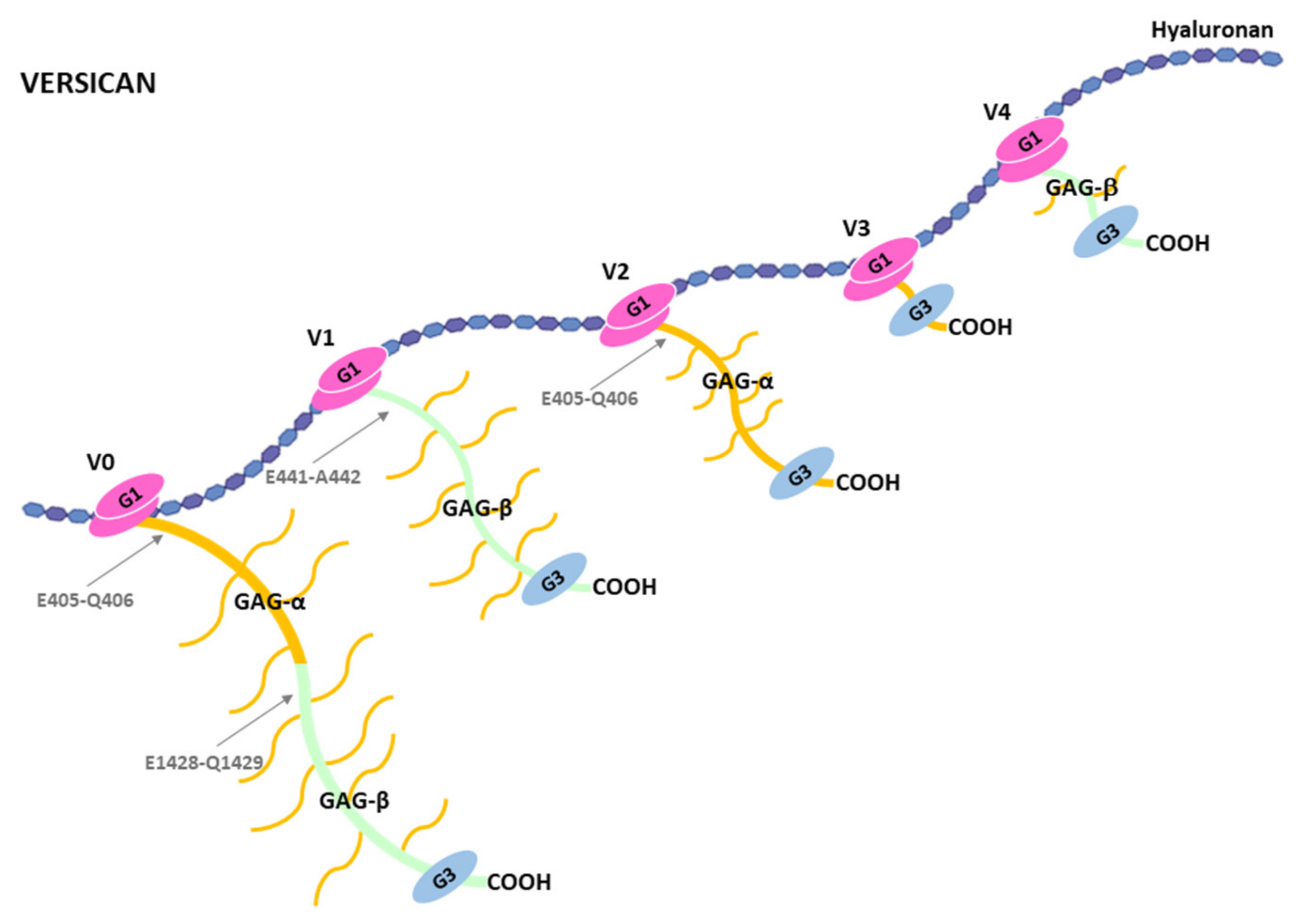
4.2. Cleavage of Versican by ADAMTSs
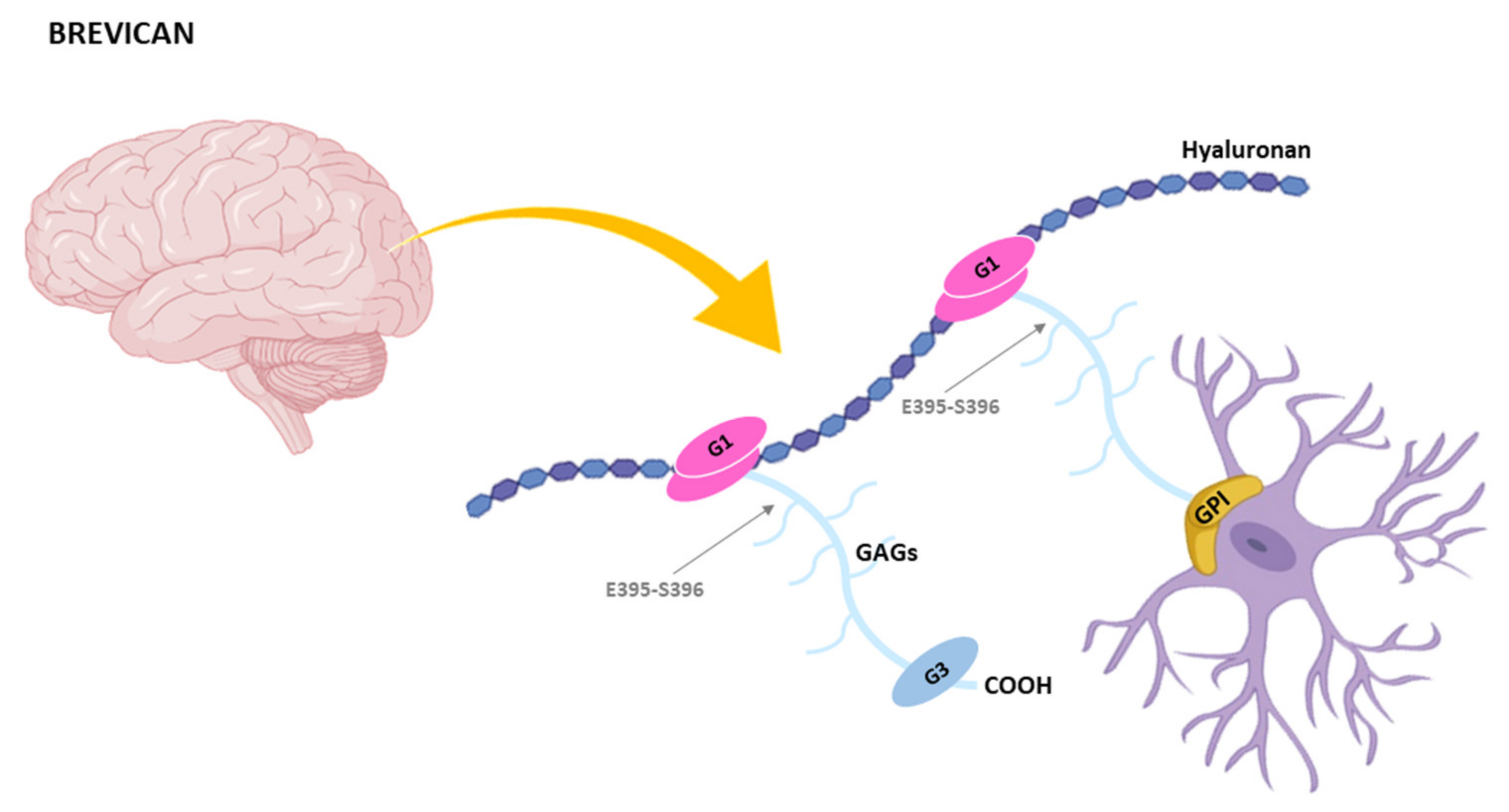
4.3. Cleavage of Brevican by ADAMTSs
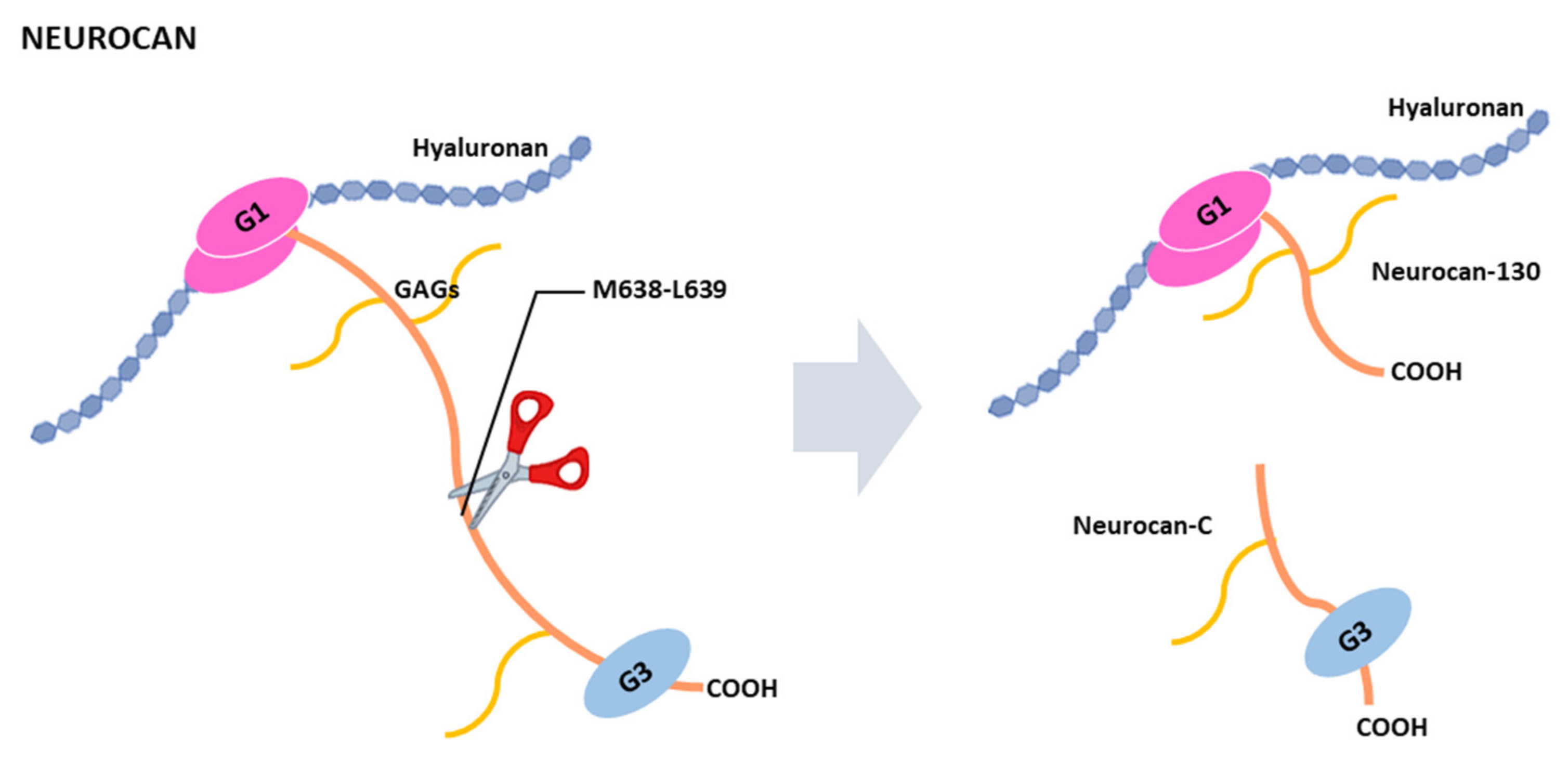
4.4. Cleavage of Neurocan by ADAMTSs
5. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Karamanos, N.K.; Theocharis, A.D.; Neill, T.; Iozzo, R.V. Matrix modeling and remodeling: A biological interplay regulating tissue homeostasis and diseases. Matrix Biol. 2019, 75–76, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.D.; Manou, D.; Karamanos, N.K. The extracellular matrix as a multitasking player in disease. FEBS J. 2019, 286, 2830–2869. [Google Scholar] [CrossRef] [PubMed]
- Chelyshev, Y.A.; Kabdesh, I.M.; Mukhamedshina, Y.O. Extracellular Matrix in Neural Plasticity and Regeneration. Cell. Mol. Neurobiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Grigorieva, E.V. Radiation Effects on Brain Extracellular Matrix. Front Oncol. 2020, 10, 576701. [Google Scholar] [CrossRef]
- Mencio, C.P.; Hussein, R.K.; Yu, P.; Geller, H.M. The Role of Chondroitin Sulfate Proteoglycans in Nervous System Development. J. Histochem. Cytochem. 2021, 69, 61–80. [Google Scholar] [CrossRef]
- Soleman, S.; Filippov, M.A.; Dityatev, A.; Fawcett, J.W. Targeting the neural extracellular matrix in neurological disorders. Neuroscience 2013, 253, 194–213. [Google Scholar] [CrossRef]
- Ma, J.; Ma, C.; Li, J.; Sun, Y.; Ye, F.; Liu, K.; Zhang, H. Extracellular Matrix Proteins Involved in Alzheimer’s Disease. Chemistry 2020, 26, 12101–12110. [Google Scholar] [CrossRef]
- Walker, C.; Mojares, E.; Del Rio Hernandez, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef]
- Iozzo, R.V.; Sanderson, R.D. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J. Cell. Mol. Med. 2011, 15, 1013–1031. [Google Scholar] [CrossRef] [PubMed]
- Santi, A.; Kugeratski, F.G.; Zanivan, S. Cancer Associated Fibroblasts: The Architects of Stroma Remodeling. Proteomics 2018, 18, e1700167. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.; Greve, B.; Espinoza-Sanchez, N.A.; Gotte, M. Cell-surface heparan sulfate proteoglycans as multifunctional integrators of signaling in cancer. Cell. Signal. 2021, 77, 109822. [Google Scholar] [CrossRef]
- Louault, K.; Li, R.R.; DeClerck, Y.A. Cancer-Associated Fibroblasts: Understanding Their Heterogeneity. Cancers 2020, 12, 3108. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Binder, M.J.; McCoombe, S.; Williams, E.D.; McCulloch, D.R.; Ward, A.C. The extracellular matrix in cancer progression: Role of hyalectan proteoglycans and ADAMTS enzymes. Cancer Lett. 2017, 385, 55–64. [Google Scholar] [CrossRef]
- Krishnan, Y.; Grodzinsky, A.J. Cartilage diseases. Matrix Biol. 2018, 71–72, 51–69. [Google Scholar] [CrossRef]
- Masutani, T.; Yamada, S.; Hara, A.; Takahashi, T.; Green, P.G.; Niwa, M. Exogenous Application of Proteoglycan to the Cell Surface Microenvironment Facilitates to Chondrogenic Differentiation and Maintenance. Int. J. Mol. Sci. 2020, 21, 7744. [Google Scholar] [CrossRef]
- Vincent, T.L.; Wann, A.K.T. Mechanoadaptation: Articular cartilage through thick and thin. J. Physiol. 2019, 597, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Karsdal, M.A.; Nielsen, S.H.; Leeming, D.J.; Langholm, L.L.; Nielsen, M.J.; Manon-Jensen, T.; Siebuhr, A.; Gudmann, N.S.; Ronnow, S.; Sand, J.M.; et al. The good and the bad collagens of fibrosis—Their role in signaling and organ function. Adv. Drug. Deliv. Rev. 2017, 121, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, A.; Li, X.; Kao, W.Y.; Viker, K.; Butters, K.; Masuoka, H.; Knudsen, B.; Gores, G.; Charlton, M. Lumican, an extracellular matrix proteoglycan, is a novel requisite for hepatic fibrosis. Lab. Investig. 2012, 92, 1712–1725. [Google Scholar] [CrossRef] [PubMed]
- Bukong, T.N.; Maurice, S.B.; Chahal, B.; Schaeffer, D.F.; Winwood, P.J. Versican: A novel modulator of hepatic fibrosis. Lab. Investig. 2016, 96, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Smith, L.R.; Khandekar, G.; Patel, P.; Yu, C.K.; Zhang, K.; Chen, C.S.; Han, L.; Wells, R.G. Distinct effects of different matrix proteoglycans on collagen fibrillogenesis and cell-mediated collagen reorganization. Sci. Rep. 2020, 10, 19065. [Google Scholar] [CrossRef] [PubMed]
- Lamande, S.R.; Bateman, J.F. Genetic Disorders of the Extracellular Matrix. Anat. Rec. 2020, 303, 1527–1542. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, A.; Nikopoulos, K.; Maugeri, A.; de Brouwer, A.P.; van Nouhuys, C.E.; Boon, C.J.; Perveen, R.; Zegers, H.A.; Wittebol-Post, D.; van den Biesen, P.R.; et al. Erosive vitreoretinopathy and wagner disease are caused by intronic mutations in CSPG2/Versican that result in an imbalance of splice variants. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3565–3572. [Google Scholar] [CrossRef]
- Meester, J.A.; Vandeweyer, G.; Pintelon, I.; Lammens, M.; Van Hoorick, L.; De Belder, S.; Waitzman, K.; Young, L.; Markham, L.W.; Vogt, J.; et al. Loss-of-function mutations in the X-linked biglycan gene cause a severe syndromic form of thoracic aortic aneurysms and dissections. Genet. Med. 2017, 19, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Nicole, S.; Davoine, C.S.; Topaloglu, H.; Cattolico, L.; Barral, D.; Beighton, P.; Hamida, C.B.; Hammouda, H.; Cruaud, C.; White, P.S.; et al. Perlecan, the major proteoglycan of basement membranes, is altered in patients with Schwartz-Jampel syndrome (chondrodystrophic myotonia). Nat. Genet. 2000, 26, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Bech-Hansen, N.T.; Naylor, M.J.; Maybaum, T.A.; Sparkes, R.L.; Koop, B.; Birch, D.G.; Bergen, A.A.; Prinsen, C.F.; Polomeno, R.C.; Gal, A.; et al. Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nat. Genet. 2000, 26, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Stanton, H.; Melrose, J.; Little, C.B.; Fosang, A.J. Proteoglycan degradation by the ADAMTS family of proteinases. Biochim. Biophys. Acta 2011, 1812, 1616–1629. [Google Scholar] [CrossRef]
- Kelwick, R.; Desanlis, I.; Wheeler, G.N.; Edwards, D.R. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015, 16, 113. [Google Scholar] [CrossRef] [PubMed]
- Colige, A.; Vandenberghe, I.; Thiry, M.; Lambert, C.A.; Van Beeumen, J.; Li, S.W.; Prockop, D.J.; Lapiere, C.M.; Nusgens, B.V. Cloning and characterization of ADAMTS-14, a novel ADAMTS displaying high homology with ADAMTS-2 and ADAMTS-3. J. Biol. Chem. 2002, 277, 5756–5766. [Google Scholar] [CrossRef]
- Liu, C.J.; Kong, W.; Ilalov, K.; Yu, S.; Xu, K.; Prazak, L.; Fajardo, M.; Sehgal, B.; Di Cesare, P.E. ADAMTS-7: A metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB J. 2006, 20, 988–990. [Google Scholar] [CrossRef]
- Courtneidge, S.A.; Kypta, R.M.; Cooper, J.A.; Kazlauskas, A. Platelet-derived growth factor receptor sequences important for binding of src family tyrosine kinases. Cell Growth Differ 1991, 2, 483–486. [Google Scholar] [PubMed]
- Fujikawa, K.; Suzuki, H.; McMullen, B.; Chung, D. Purification of human von Willebrand factor-cleaving protease and its identification as a new member of the metalloproteinase family. Blood 2001, 98, 1662–1666. [Google Scholar] [CrossRef] [PubMed]
- Karoulias, S.Z.; Taye, N.; Stanley, S.; Hubmacher, D. The ADAMTS/Fibrillin Connection: Insights into the Biological Functions of ADAMTS10 and ADAMTS17 and Their Respective Sister Proteases. Biomolecules 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.W.; Kutz, W.E.; Mead, T.J.; Beene, L.C.; Singh, S.; Jenkins, M.W.; Reinhardt, D.P.; Apte, S.S. Adamts10 inactivation in mice leads to persistence of ocular microfibrils subsequent to reduced fibrillin-2 cleavage. Matrix Biol. 2019, 77, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Cain, S.A.; Mularczyk, E.J.; Singh, M.; Massam-Wu, T.; Kielty, C.M. ADAMTS-10 and -6 differentially regulate cell-cell junctions and focal adhesions. Sci. Rep. 2016, 6, 35956. [Google Scholar] [CrossRef] [PubMed]
- Karoulias, S.Z.; Beyens, A.; Balic, Z.; Symoens, S.; Vandersteen, A.; Rideout, A.L.; Dickinson, J.; Callewaert, B.; Hubmacher, D. A novel ADAMTS17 variant that causes Weill-Marchesani syndrome 4 alters fibrillin-1 and collagen type I deposition in the extracellular matrix. Matrix Biol. 2020, 88, 1–18. [Google Scholar] [CrossRef]
- Morales, J.; Al-Sharif, L.; Khalil, D.S.; Shinwari, J.M.; Bavi, P.; Al-Mahrouqi, R.A.; Al-Rajhi, A.; Alkuraya, F.S.; Meyer, B.F.; Al Tassan, N. Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am. J. Hum. Genet. 2009, 85, 558–568. [Google Scholar] [CrossRef] [PubMed]
- Peluso, I.; Conte, I.; Testa, F.; Dharmalingam, G.; Pizzo, M.; Collin, R.W.; Meola, N.; Barbato, S.; Mutarelli, M.; Ziviello, C.; et al. The ADAMTS18 gene is responsible for autosomal recessive early onset severe retinal dystrophy. Orphanet J. Rare Dis. 2013, 8, 16. [Google Scholar] [CrossRef]
- Aldahmesh, M.A.; Khan, A.O.; Mohamed, J.Y.; Alkuraya, H.; Ahmed, H.; Bobis, S.; Al-Mesfer, S.; Alkuraya, F.S. Identification of ADAMTS18 as a gene mutated in Knobloch syndrome. J. Med. Genet. 2011, 48, 597–601. [Google Scholar] [CrossRef]
- Abdul-Majeed, S.; Mell, B.; Nauli, S.M.; Joe, B. Cryptorchidism and infertility in rats with targeted disruption of the Adamts16 locus. PLoS ONE 2014, 9, e100967. [Google Scholar] [CrossRef]
- Jacobi, C.L.; Rudigier, L.J.; Scholz, H.; Kirschner, K.M. Transcriptional regulation by the Wilms tumor protein, Wt1, suggests a role of the metalloproteinase Adamts16 in murine genitourinary development. J. Biol. Chem. 2013, 288, 18811–18824. [Google Scholar] [CrossRef] [PubMed]
- Pyun, J.A.; Kim, S.; Kwack, K. Interaction between thyroglobulin and ADAMTS16 in premature ovarian failure. Clin. Exp. Reprod. Med. 2014, 41, 120–124. [Google Scholar] [CrossRef]
- Mead, T.J.; Apte, S.S. ADAMTS proteins in human disorders. Matrix Biol. 2018, 71–72, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y. Lecticans: Organizers of the brain extracellular matrix. Cell. Mol. Life Sci. 2000, 57, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Mohamedi, Y.; Fontanil, T.; Cobo, T.; Cal, S.; Obaya, A.J. New Insights into ADAMTS Metalloproteases in the Central Nervous System. Biomolecules 2020, 10, 403. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V. Matrix proteoglycans: From molecular design to cellular function. Annu. Rev. Biochem. 1998, 67, 609–652. [Google Scholar] [CrossRef] [PubMed]
- Howell, M.D.; Gottschall, P.E. Lectican proteoglycans, their cleaving metalloproteinases, and plasticity in the central nervous system extracellular microenvironment. Neuroscience 2012, 217, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Sandy, J.D. A contentious issue finds some clarity: On the independent and complementary roles of aggrecanase activity and MMP activity in human joint aggrecanolysis. Osteoarthr. Cartil. 2006, 14, 95–100. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kuno, K.; Kanada, N.; Nakashima, E.; Fujiki, F.; Ichimura, F.; Matsushima, K. Molecular cloning of a gene encoding a new type of metalloproteinase-disintegrin family protein with thrombospondin motifs as an inflammation associated gene. J. Biol. Chem. 1997, 272, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Sandy, J.D.; Flannery, C.R.; Neame, P.J.; Lohmander, L.S. The structure of aggrecan fragments in human synovial fluid. Evidence for the involvement in osteoarthritis of a novel proteinase which cleaves the Glu 373-Ala 374 bond of the interglobular domain. J. Clin. Investig. 1992, 89, 1512–1516. [Google Scholar] [CrossRef]
- Tortorella, M.D.; Burn, T.C.; Pratta, M.A.; Abbaszade, I.; Hollis, J.M.; Liu, R.; Rosenfeld, S.A.; Copeland, R.A.; Decicco, C.P.; Wynn, R.; et al. Purification and cloning of aggrecanase-1: A member of the ADAMTS family of proteins. Science 1999, 284, 1664–1666. [Google Scholar] [CrossRef]
- Abbaszade, I.; Liu, R.Q.; Yang, F.; Rosenfeld, S.A.; Ross, O.H.; Link, J.R.; Ellis, D.M.; Tortorella, M.D.; Pratta, M.A.; Hollis, J.M.; et al. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. J. Biol. Chem. 1999, 274, 23443–23450. [Google Scholar] [CrossRef]
- Fosang, A.J.; Little, C.B. Drug insight: Aggrecanases as therapeutic targets for osteoarthritis. Nat. Clin. Pract. Rheumatol. 2008, 4, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Little, C.B.; Meeker, C.T.; Golub, S.B.; Lawlor, K.E.; Farmer, P.J.; Smith, S.M.; Fosang, A.J. Blocking aggrecanase cleavage in the aggrecan interglobular domain abrogates cartilage erosion and promotes cartilage repair. J. Clin. Investig. 2007, 117, 1627–1636. [Google Scholar] [CrossRef] [PubMed]
- Azadi, M.; Nia, H.T.; Gauci, S.J.; Ortiz, C.; Fosang, A.J.; Grodzinsky, A.J. Wide bandwidth nanomechanical assessment of murine cartilage reveals protection of aggrecan knock-in mice from joint-overuse. J. Biomech. 2016, 49, 1634–1640. [Google Scholar] [CrossRef] [PubMed]
- Collins-Racie, L.A.; Flannery, C.R.; Zeng, W.; Corcoran, C.; Annis-Freeman, B.; Agostino, M.J.; Arai, M.; DiBlasio-Smith, E.; Dorner, A.J.; Georgiadis, K.E.; et al. ADAMTS-8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biol. 2004, 23, 219–230. [Google Scholar] [CrossRef]
- Santamaria, S. ADAMTS-5: A difficult teenager turning 20. Int. J. Exp. Pathol. 2020, 101, 4–20. [Google Scholar] [CrossRef] [PubMed]
- El Bakali, J.; Gras-Masse, H.; Maingot, L.; Deprez, B.; Dumont, J.; Leroux, F.; Deprez-Poulain, R. Inhibition of aggrecanases as a therapeutic strategy in osteoarthritis. Future Med. Chem. 2014, 6, 1399–1412. [Google Scholar] [CrossRef]
- Santamaria, S.; Yamamoto, K.; Teraz-Orosz, A.; Koch, C.; Apte, S.S.; de Groot, R.; Lane, D.A.; Ahnstrom, J. Exosites in Hypervariable Loops of ADAMTS Spacer Domains control Substrate Recognition and Proteolysis. Sci. Rep. 2019, 9, 10914. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, S.; de Groot, R. Monoclonal antibodies against metzincin targets. Br. J. Pharmacol. 2019, 176, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Lohr, T.A.; Elefante, L.; Shearin, J.; Matico, R.; Su, J.L.; Xue, Y.; Liu, F.; Genell, C.; Miller, R.E.; et al. Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification. Osteoarthr. Cartil. 2015, 23, 1254–1266. [Google Scholar] [CrossRef]
- Malemud, C.J. Inhibition of MMPs and ADAM/ADAMTS. Biochem. Pharmacol. 2019, 165, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Siebuhr, A.S.; Werkmann, D.; Bay-Jensen, A.C.; Thudium, C.S.; Karsdal, M.A.; Serruys, B.; Ladel, C.; Michaelis, M.; Lindemann, S. The Anti-ADAMTS-5 Nanobody((R)) M6495 Protects Cartilage Degradation Ex Vivo. Int. J. Mol. Sci. 2020, 21, 5992. [Google Scholar] [CrossRef]
- Glasson, S.S.; Askew, R.; Sheppard, B.; Carito, B.; Blanchet, T.; Ma, H.L.; Flannery, C.R.; Peluso, D.; Kanki, K.; Yang, Z.; et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature 2005, 434, 644–648. [Google Scholar] [CrossRef]
- Stanton, H.; Rogerson, F.M.; East, C.J.; Golub, S.B.; Lawlor, K.E.; Meeker, C.T.; Little, C.B.; Last, K.; Farmer, P.J.; Campbell, I.K.; et al. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature 2005, 434, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, M.K.; Askew, R.; Schelling, S.; Stedman, N.; Blanchet, T.; Hopkins, B.; Morris, E.A.; Glasson, S.S. Double-knockout of ADAMTS-4 and ADAMTS-5 in mice results in physiologically normal animals and prevents the progression of osteoarthritis. Arthritis Rheum. 2007, 56, 3670–3674. [Google Scholar] [CrossRef]
- Rogerson, F.M.; Stanton, H.; East, C.J.; Golub, S.B.; Tutolo, L.; Farmer, P.J.; Fosang, A.J. Evidence of a novel aggrecan-degrading activity in cartilage: Studies of mice deficient in both ADAMTS-4 and ADAMTS-5. Arthritis Rheum. 2008, 58, 1664–1673. [Google Scholar] [CrossRef]
- East, C.J.; Stanton, H.; Golub, S.B.; Rogerson, F.M.; Fosang, A.J. ADAMTS-5 deficiency does not block aggrecanolysis at preferred cleavage sites in the chondroitin sulfate-rich region of aggrecan. J. Biol. Chem. 2007, 282, 8632–8640. [Google Scholar] [CrossRef] [PubMed]
- Stanton, H.; Golub, S.B.; Rogerson, F.M.; Last, K.; Little, C.B.; Fosang, A.J. Investigating ADAMTS-mediated aggrecanolysis in mouse cartilage. Nat. Protoc. 2011, 6, 388–404. [Google Scholar] [CrossRef]
- Santamaria, S.; Yamamoto, K. Analysis of Aggrecanase Activity Using Neoepitope Antibodies. Methods Mol. Biol. 2020, 2043, 125–136. [Google Scholar] [CrossRef]
- Rodriguez-Manzaneque, J.C.; Westling, J.; Thai, S.N.; Luque, A.; Knauper, V.; Murphy, G.; Sandy, J.D.; Iruela-Arispe, M.L. ADAMTS1 cleaves aggrecan at multiple sites and is differentially inhibited by metalloproteinase inhibitors. Biochem. Biophys. Res. Commun. 2002, 293, 501–508. [Google Scholar] [CrossRef]
- Rogerson, F.M.; Last, K.; Golub, S.B.; Gauci, S.J.; Stanton, H.; Bell, K.M.; Fosang, A.J. ADAMTS-9 in Mouse Cartilage Has Aggrecanase Activity That Is Distinct from ADAMTS-4 and ADAMTS-5. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Koch, C.D.; Lee, C.M.; Apte, S.S. Aggrecan in Cardiovascular Development and Disease. J. Histochem. Cytochem. 2020, 68, 777–795. [Google Scholar] [CrossRef]
- Suna, G.; Wojakowski, W.; Lynch, M.; Barallobre-Barreiro, J.; Yin, X.; Mayr, U.; Baig, F.; Lu, R.; Fava, M.; Hayward, R.; et al. Extracellular Matrix Proteomics Reveals Interplay of Aggrecan and Aggrecanases in Vascular Remodeling of Stented Coronary Arteries. Circulation 2018, 137, 166–183. [Google Scholar] [CrossRef]
- Cikach, F.S.; Koch, C.D.; Mead, T.J.; Galatioto, J.; Willard, B.B.; Emerton, K.B.; Eagleton, M.J.; Blackstone, E.H.; Ramirez, F.; Roselli, E.E.; et al. Massive aggrecan and versican accumulation in thoracic aortic aneurysm and dissection. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, L.E.; Nelson, E.L.; Hozik, B.; Porto, S.C.; Rogers-DeCotes, A.; Fosang, A.; Kern, C.B. Adamts5(-/-) Mice Exhibit Altered Aggrecan Proteolytic Profiles That Correlate With Ascending Aortic Anomalies. Arterioscler Thromb. Vasc. Biol. 2019, 39, 2067–2081. [Google Scholar] [CrossRef] [PubMed]
- Tola, E.N.; Koroglu, N.D.; Yalcin, S.E.; Oral, H.B. The role of serum ADAMTS-1 and aggrecan on polycystic ovary syndrome in adolescents and younger-aged females. Arch. Gynecol. Obstet. 2018, 297, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.A.; Dos Santos, L.; Turri, J.A.; Nonogaki, S.; Buim, M.; Lima, J.F.; de Jesus Viana Pinheiro, J.; Bueno de Toledo Osorio, C.A.; Soares, F.A.; Freitas, V.M. Prognostic Value of ADAMTS Proteases and Their Substrates in Epithelial Ovarian Cancer. Pathobiology 2016, 83, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Morawski, M.; Bruckner, G.; Arendt, T.; Matthews, R.T. Aggrecan: Beyond cartilage and into the brain. Int. J. Biochem. Cell Biol. 2012, 44, 690–693. [Google Scholar] [CrossRef]
- Lemarchant, S.; Pruvost, M.; Montaner, J.; Emery, E.; Vivien, D.; Kanninen, K.; Koistinaho, J. ADAMTS proteoglycanases in the physiological and pathological central nervous system. J. Neuroinflammation 2013, 10, 133. [Google Scholar] [CrossRef]
- Islam, S.; Watanabe, H. Versican: A Dynamic Regulator of the Extracellular Matrix. J. Histochem. Cytochem. 2020, 68, 763–775. [Google Scholar] [CrossRef]
- Henderson, D.J.; Copp, A.J. Versican expression is associated with chamber specification, septation, and valvulogenesis in the developing mouse heart. Circ. Res. 1998, 83, 523–532. [Google Scholar] [CrossRef]
- Zanin, M.K.; Bundy, J.; Ernst, H.; Wessels, A.; Conway, S.J.; Hoffman, S. Distinct spatial and temporal distributions of aggrecan and versican in the embryonic chick heart. Anat. Rec. 1999, 256, 366–380. [Google Scholar] [CrossRef]
- Nandadasa, S.; Foulcer, S.; Apte, S.S. The multiple, complex roles of versican and its proteolytic turnover by ADAMTS proteases during embryogenesis. Matrix Biol. 2014, 35, 34–41. [Google Scholar] [CrossRef]
- Bode-Lesniewska, B.; Dours-Zimmermann, M.T.; Odermatt, B.F.; Briner, J.; Heitz, P.U.; Zimmermann, D.R. Distribution of the large aggregating proteoglycan versican in adult human tissues. J. Histochem. Cytochem. 1996, 44, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Schmalfeldt, M.; Dours-Zimmermann, M.T.; Winterhalter, K.H.; Zimmermann, D.R. Versican V2 is a major extracellular matrix component of the mature bovine brain. J. Biol. Chem. 1998, 273, 15758–15764. [Google Scholar] [CrossRef] [PubMed]
- Dours-Zimmermann, M.T.; Maurer, K.; Rauch, U.; Stoffel, W.; Fassler, R.; Zimmermann, D.R. Versican V2 assembles the extracellular matrix surrounding the nodes of ranvier in the CNS. J. NeuroSci. 2009, 29, 7731–7742. [Google Scholar] [CrossRef] [PubMed]
- Cattaruzza, S.; Schiappacassi, M.; Ljungberg-Rose, A.; Spessotto, P.; Perissinotto, D.; Morgelin, M.; Mucignat, M.T.; Colombatti, A.; Perris, R. Distribution of PG-M/versican variants in human tissues and de novo expression of isoform V3 upon endothelial cell activation, migration, and neoangiogenesis in vitro. J. Biol. Chem. 2002, 277, 47626–47635. [Google Scholar] [CrossRef] [PubMed]
- Lemire, J.M.; Merrilees, M.J.; Braun, K.R.; Wight, T.N. Overexpression of the V3 variant of versican alters arterial smooth muscle cell adhesion, migration, and proliferation in vitro. J. Cell Physiol. 2002, 190, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Kischel, P.; Waltregny, D.; Dumont, B.; Turtoi, A.; Greffe, Y.; Kirsch, S.; De Pauw, E.; Castronovo, V. Versican overexpression in human breast cancer lesions: Known and new isoforms for stromal tumor targeting. Int. J. Cancer 2010, 126, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, S.; de Groot, R. ADAMTS proteases in cardiovascular physiology and disease. Open Biol. 2020, 10, 200333. [Google Scholar] [CrossRef]
- Sandy, J.D.; Westling, J.; Kenagy, R.D.; Iruela-Arispe, M.L.; Verscharen, C.; Rodriguez-Mazaneque, J.C.; Zimmermann, D.R.; Lemire, J.M.; Fischer, J.W.; Wight, T.N.; et al. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441-Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS-4. J. Biol. Chem. 2001, 276, 13372–13378. [Google Scholar] [CrossRef] [PubMed]
- Kern, C.B.; Twal, W.O.; Mjaatvedt, C.H.; Fairey, S.E.; Toole, B.P.; Iruela-Arispe, M.L.; Argraves, W.S. Proteolytic cleavage of versican during cardiac cushion morphogenesis. Dev. Dyn. 2006, 235, 2238–2247. [Google Scholar] [CrossRef] [PubMed]
- Stankunas, K.; Hang, C.T.; Tsun, Z.Y.; Chen, H.; Lee, N.V.; Wu, J.I.; Shang, C.; Bayle, J.H.; Shou, W.; Iruela-Arispe, M.L.; et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev. Cell 2008, 14, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Jonsson-Rylander, A.C.; Nilsson, T.; Fritsche-Danielson, R.; Hammarstrom, A.; Behrendt, M.; Andersson, J.O.; Lindgren, K.; Andersson, A.K.; Wallbrandt, P.; Rosengren, B.; et al. Role of ADAMTS-1 in atherosclerosis: Remodeling of carotid artery, immunohistochemistry, and proteolysis of versican. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Hirohata, S.; Murakami, T.; Miyoshi, T.; Demircan, K.; Oohashi, T.; Ogawa, H.; Koten, K.; Toeda, K.; Kusachi, S.; et al. Dynamic induction of ADAMTS1 gene in the early phase of acute myocardial infarction. J. Biochem. 2004, 136, 439–446. [Google Scholar] [CrossRef]
- Kumar, S.; Chen, M.; Li, Y.; Wong, F.H.; Thiam, C.W.; Hossain, M.Z.; Poh, K.K.; Hirohata, S.; Ogawa, H.; Angeli, V.; et al. Loss of ADAMTS4 reduces high fat diet-induced atherosclerosis and enhances plaque stability in ApoE(-/-) mice. Sci. Rep. 2016, 6, 31130. [Google Scholar] [CrossRef]
- Dancevic, C.M.; Fraser, F.W.; Smith, A.D.; Stupka, N.; Ward, A.C.; McCulloch, D.R. Biosynthesis and expression of a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats-15: A novel versican-cleaving proteoglycanase. J. Biol. Chem. 2013, 288, 37267–37276. [Google Scholar] [CrossRef]
- Dupuis, L.E.; Osinska, H.; Weinstein, M.B.; Hinton, R.B.; Kern, C.B. Insufficient versican cleavage and Smad2 phosphorylation results in bicuspid aortic and pulmonary valves. J. Mol. Cell. Cardiol. 2013, 60, 50–59. [Google Scholar] [CrossRef]
- Dupuis, L.E.; McCulloch, D.R.; McGarity, J.D.; Bahan, A.; Wessels, A.; Weber, D.; Diminich, A.M.; Nelson, C.M.; Apte, S.S.; Kern, C.B. Altered versican cleavage in ADAMTS5 deficient mice; a novel etiology of myxomatous valve disease. Dev. Biol. 2011, 357, 152–164. [Google Scholar] [CrossRef]
- Islam, S.; Chuensirikulchai, K.; Khummuang, S.; Keratibumrungpong, T.; Kongtawelert, P.; Kasinrerk, W.; Hatano, S.; Nagamachi, A.; Honda, H.; Watanabe, H. Accumulation of versican facilitates wound healing: Implication of its initial ADAMTS-cleavage site. Matrix Biol. 2020, 87, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Nagyova, E.; Salustri, A.; Nemcova, L.; Scsukova, S.; Kalous, J.; Camaioni, A. Versican G1 Fragment Establishes a Strongly Stabilized Interaction with Hyaluronan-Rich Expanding Matrix during Oocyte Maturation. Int. J. Mol. Sci. 2020, 21, 2267. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, H.; Nelson, C.M.; Somerville, R.P.; Mielke, K.; Dixon, L.J.; Powell, K.; Apte, S.S. Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development 2010, 137, 4029–4038. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, D.R.; Nelson, C.M.; Dixon, L.J.; Silver, D.L.; Wylie, J.D.; Lindner, V.; Sasaki, T.; Cooley, M.A.; Argraves, W.S.; Apte, S.S. ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev. Cell 2009, 17, 687–698. [Google Scholar] [CrossRef]
- Hong, C.C.; Tang, A.T.; Detter, M.R.; Choi, J.P.; Wang, R.; Yang, X.; Guerrero, A.A.; Wittig, C.F.; Hobson, N.; Girard, R.; et al. Cerebral cavernous malformations are driven by ADAMTS5 proteolysis of versican. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Hope, C.; Emmerich, P.B.; Papadas, A.; Pagenkopf, A.; Matkowskyj, K.A.; Van De Hey, D.R.; Payne, S.N.; Clipson, L.; Callander, N.S.; Hematti, P.; et al. Versican-Derived Matrikines Regulate Batf3-Dendritic Cell Differentiation and Promote T Cell Infiltration in Colorectal Cancer. J. Immunol. 2017, 199, 1933–1941. [Google Scholar] [CrossRef]
- Asano, K.; Nelson, C.M.; Nandadasa, S.; Aramaki-Hattori, N.; Lindner, D.J.; Alban, T.; Inagaki, J.; Ohtsuki, T.; Oohashi, T.; Apte, S.S.; et al. Stromal Versican Regulates Tumor Growth by Promoting Angiogenesis. Sci. Rep. 2017, 7, 17225. [Google Scholar] [CrossRef]
- Hope, C.; Foulcer, S.; Jagodinsky, J.; Chen, S.X.; Jensen, J.L.; Patel, S.; Leith, C.; Maroulakou, I.; Callander, N.; Miyamoto, S.; et al. Immunoregulatory roles of versican proteolysis in the myeloma microenvironment. Blood 2016, 128, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Jensen, B.M.; Kruse-Andersen, S.; Andersen, K. Thoracic actinomycosis. Scand. J. Thorac. Cardiovasc. Surg. 1989, 23, 181–184. [Google Scholar] [CrossRef]
- Papadas, A.; Arauz, G.; Cicala, A.; Wiesner, J.; Asimakopoulos, F. Versican and Versican-matrikines in Cancer Progression, Inflammation, and Immunity. J. Histochem. Cytochem. 2020, 68, 871–885. [Google Scholar] [CrossRef] [PubMed]
- Papadas, A.; Asimakopoulos, F. Versican in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1272, 55–72. [Google Scholar] [CrossRef]
- Gueye, N.A.; Mead, T.J.; Koch, C.D.; Biscotti, C.V.; Falcone, T.; Apte, S.S. Versican Proteolysis by ADAMTS Proteases and Its Influence on Sex Steroid Receptor Expression in Uterine Leiomyoma. J. Clin. Endocrinol. Metab. 2017, 102, 1631–1641. [Google Scholar] [CrossRef]
- Reymond, M.A.; de Gottrau, P.; Fournier, P.E.; Arnold, T.; Jacomet, H.; Rigo, M. Traumatology in hang-gliding accidents. Studies based on 100 cases. Chirurg 1988, 59, 777–781. [Google Scholar] [PubMed]
- Bignami, A.; Hosley, M.; Dahl, D. Hyaluronic acid and hyaluronic acid-binding proteins in brain extracellular matrix. Anat. Embryol. 1993, 188, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Westling, J.; Gottschall, P.E.; Thompson, V.P.; Cockburn, A.; Perides, G.; Zimmermann, D.R.; Sandy, J.D. ADAMTS4 (aggrecanase-1) cleaves human brain versican V2 at Glu405-Gln406 to generate glial hyaluronate binding protein. Biochem. J. 2004, 377, 787–795. [Google Scholar] [CrossRef]
- Fischbarg, J.; Maurice, D.M. An update on corneal hydration control. Exp. Eye Res. 2004, 78, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Wight, T.N. Versican: A versatile extracellular matrix proteoglycan in cell biology. Curr. Opin. Cell Biol. 2002, 14, 617–623. [Google Scholar] [CrossRef]
- Yamada, H.; Watanabe, K.; Shimonaka, M.; Yamaguchi, Y. Molecular cloning of brevican, a novel brain proteoglycan of the aggrecan/versican family. J. Biol. Chem. 1994, 269, 10119–10126. [Google Scholar] [CrossRef]
- Frischknecht, R.; Seidenbecher, C.I. Brevican: A key proteoglycan in the perisynaptic extracellular matrix of the brain. Int. J. Biochem. Cell Biol. 2012, 44, 1051–1054. [Google Scholar] [CrossRef]
- John, N.; Krugel, H.; Frischknecht, R.; Smalla, K.H.; Schultz, C.; Kreutz, M.R.; Gundelfinger, E.D.; Seidenbecher, C.I. Brevican-containing perineuronal nets of extracellular matrix in dissociated hippocampal primary cultures. Mol. Cell NeuroSci. 2006, 31, 774–784. [Google Scholar] [CrossRef]
- Morawski, M.; Bruckner, G.; Jager, C.; Seeger, G.; Matthews, R.T.; Arendt, T. Involvement of perineuronal and perisynaptic extracellular matrix in Alzheimer’s disease neuropathology. Brain Pathol. 2012, 22, 547–561. [Google Scholar] [CrossRef]
- Del Bueno, D.J. The renal humoral system: A cause of hypertension? RN 1975, 38, 109–110, 113. [Google Scholar]
- Fernald, H.T. The Cotton Worm Moth Again. Science 1914, 40, 785. [Google Scholar] [CrossRef] [PubMed]
- Forehand, C.J. Morphology of sympathetic preganglionic neurons in the neonatal rat spinal cord: An intracellular horseradish peroxidase study. J. Comp. Neurol. 1990, 298, 334–342. [Google Scholar] [CrossRef]
- Jones, L.L.; Margolis, R.U.; Tuszynski, M.H. The chondroitin sulfate proteoglycans neurocan, brevican, phosphacan, and versican are differentially regulated following spinal cord injury. Exp. Neurol. 2003, 182, 399–411. [Google Scholar] [CrossRef]
- Quaglia, X.; Beggah, A.T.; Seidenbecher, C.; Zurn, A.D. Delayed priming promotes CNS regeneration post-rhizotomy in Neurocan and Brevican-deficient mice. Brain 2008, 131, 240–249. [Google Scholar] [CrossRef][Green Version]
- Beggah, A.T.; Dours-Zimmermann, M.T.; Barras, F.M.; Brosius, A.; Zimmermann, D.R.; Zurn, A.D. Lesion-induced differential expression and cell association of Neurocan, Brevican, Versican V1 and V2 in the mouse dorsal root entry zone. Neuroscience 2005, 133, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.L.; Sajed, D.; Tuszynski, M.H. Axonal regeneration through regions of chondroitin sulfate proteoglycan deposition after spinal cord injury: A balance of permissiveness and inhibition. J. NeuroSci. 2003, 23, 9276–9288. [Google Scholar] [CrossRef]
- Davies, J.E.; Tang, X.; Denning, J.W.; Archibald, S.J.; Davies, S.J. Decorin suppresses neurocan, brevican, phosphacan and NG2 expression and promotes axon growth across adult rat spinal cord injuries. Eur. J. NeuroSci. 2004, 19, 1226–1242. [Google Scholar] [CrossRef]
- Seidenbecher, C.I.; Richter, K.; Rauch, U.; Fassler, R.; Garner, C.C.; Gundelfinger, E.D. Brevican, a chondroitin sulfate proteoglycan of rat brain, occurs as secreted and cell surface glycosylphosphatidylinositol-anchored isoforms. J. Biol. Chem. 1995, 270, 27206–27212. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Fujii, Y.; Inoki, I.; Sugimoto, K.; Tanzawa, K.; Matsuki, H.; Miura, R.; Yamaguchi, Y.; Okada, Y. Brevican is degraded by matrix metalloproteinases and aggrecanase-1 (ADAMTS4) at different sites. J. Biol. Chem. 2000, 275, 38885–38890. [Google Scholar] [CrossRef]
- Viapiano, M.S.; Hockfield, S.; Matthews, R.T. BEHAB/brevican requires ADAMTS-mediated proteolytic cleavage to promote glioma invasion. J. Neurooncol. 2008, 88, 261–272. [Google Scholar] [CrossRef]
- Held-Feindt, J.; Paredes, E.B.; Blomer, U.; Seidenbecher, C.; Stark, A.M.; Mehdorn, H.M.; Mentlein, R. Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. Int. J. Cancer 2006, 118, 55–61. [Google Scholar] [CrossRef]
- Nakada, M.; Miyamori, H.; Kita, D.; Takahashi, T.; Yamashita, J.; Sato, H.; Miura, R.; Yamaguchi, Y.; Okada, Y. Human glioblastomas overexpress ADAMTS-5 that degrades brevican. Acta Neuropathol. 2005, 110, 239–246. [Google Scholar] [CrossRef]
- Matthews, R.T.; Gary, S.C.; Zerillo, C.; Pratta, M.; Solomon, K.; Arner, E.C.; Hockfield, S. Brain-enriched hyaluronan binding (BEHAB)/brevican cleavage in a glioma cell line is mediated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family member. J. Biol. Chem. 2000, 275, 22695–22703. [Google Scholar] [CrossRef]
- Guo, Y.; Yu, L.; Wen, Z.; Li, M. Using support vector machine combined with auto covariance to predict protein-protein interactions from protein sequences. Nucleic. Acids. Res. 2008, 36, 3025–3030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kelly, G.; Zerillo, C.; Jaworski, D.M.; Hockfield, S. Expression of a cleaved brain-specific extracellular matrix protein mediates glioma cell invasion In vivo. J. NeuroSci. 1998, 18, 2370–2376. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Wu, C.; Guo, L.; Liu, Y.; Mo, W.; Wang, H.; Ding, J.; Wong, E.T.; Yu, M. The role of brevican in glioma: Promoting tumor cell motility in vitro and in vivo. BMC Cancer 2012, 12, 607. [Google Scholar] [CrossRef] [PubMed]
- Mayer, J.; Hamel, M.G.; Gottschall, P.E. Evidence for proteolytic cleavage of brevican by the ADAMTSs in the dentate gyrus after excitotoxic lesion of the mouse entorhinal cortex. BMC NeuroSci. 2005, 6, 52. [Google Scholar] [CrossRef]
- Minta, K.; Brinkmalm, G.; Thelin, E.P.; Al Nimer, F.; Piehl, F.; Tullberg, M.; Jeppsson, A.; Portelius, E.; Zetterberg, H.; Blennow, K.; et al. Cerebrospinal fluid brevican and neurocan fragment patterns in human traumatic brain injury. Clin. Chim. Acta 2021, 512, 74–83. [Google Scholar] [CrossRef]
- Jonesco, D.S.; Karsdal, M.A.; Henriksen, K. The CNS-specific proteoglycan, brevican, and its ADAMTS4-cleaved fragment show differential serological levels in Alzheimer’s disease, other types of dementia and non-demented controls: A cross-sectional study. PLoS ONE 2020, 15, e0234632. [Google Scholar] [CrossRef]
- Tauchi, R.; Imagama, S.; Natori, T.; Ohgomori, T.; Muramoto, A.; Shinjo, R.; Matsuyama, Y.; Ishiguro, N.; Kadomatsu, K. The endogenous proteoglycan-degrading enzyme ADAMTS-4 promotes functional recovery after spinal cord injury. J. Neuroinflammation 2012, 9, 53. [Google Scholar] [CrossRef]
- Mitlohner, J.; Kaushik, R.; Niekisch, H.; Blondiaux, A.; Gee, C.E.; Happel, M.F.K.; Gundelfinger, E.; Dityatev, A.; Frischknecht, R.; Seidenbecher, C. Dopamine Receptor Activation Modulates the Integrity of the Perisynaptic Extracellular Matrix at Excitatory Synapses. Cells 2020, 9, 260. [Google Scholar] [CrossRef]
- Demircan, K.; Topcu, V.; Takigawa, T.; Akyol, S.; Yonezawa, T.; Ozturk, G.; Ugurcu, V.; Hasgul, R.; Yigitoglu, M.R.; Akyol, O.; et al. ADAMTS4 and ADAMTS5 knockout mice are protected from versican but not aggrecan or brevican proteolysis during spinal cord injury. Biomed. Res. Int. 2014, 2014, 693746. [Google Scholar] [CrossRef] [PubMed]
- Margolis, R.K.; Rauch, U.; Maurel, P.; Margolis, R.U. Neurocan and phosphacan: Two major nervous tissue-specific chondroitin sulfate proteoglycans. Perspect Dev. Neurobiol. 1996, 3, 273–290. [Google Scholar] [PubMed]
- Rauch, U.; Karthikeyan, L.; Maurel, P.; Margolis, R.U.; Margolis, R.K. Cloning and primary structure of neurocan, a developmentally regulated, aggregating chondroitin sulfate proteoglycan of brain. J. Biol. Chem. 1992, 267, 19536–19547. [Google Scholar] [CrossRef]
- Meyer-Puttlitz, B.; Junker, E.; Margolis, R.U.; Margolis, R.K. Chondroitin sulfate proteoglycans in the developing central nervous system. II. Immunocytochemical localization of neurocan and phosphacan. J. Comp. Neurol. 1996, 366, 44–54. [Google Scholar] [CrossRef]
- Milev, P.; Maurel, P.; Chiba, A.; Mevissen, M.; Popp, S.; Yamaguchi, Y.; Margolis, R.K.; Margolis, R.U. Differential regulation of expression of hyaluronan-binding proteoglycans in developing brain: Aggrecan, versican, neurocan, and brevican. Biochem. Biophys Res. Commun. 1998, 247, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.; Arendt, T.; Morawski, M.; Sonntag, M. Neurocan Contributes to Perineuronal Net Development. Neuroscience 2020, 442, 69–86. [Google Scholar] [CrossRef]
- Sullivan, C.S.; Gotthard, I.; Wyatt, E.V.; Bongu, S.; Mohan, V.; Weinberg, R.J.; Maness, P.F. Perineuronal Net Protein Neurocan Inhibits NCAM/EphA3 Repellent Signaling in GABAergic Interneurons. Sci. Rep. 2018, 8, 6143. [Google Scholar] [CrossRef]
- Mohan, V.; Wyatt, E.V.; Gotthard, I.; Phend, K.D.; Diestel, S.; Duncan, B.W.; Weinberg, R.J.; Tripathy, A.; Maness, P.F. Neurocan Inhibits Semaphorin 3F Induced Dendritic Spine Remodeling Through NrCAM in Cortical Neurons. Front. Cell. NeuroSci. 2018, 12, 346. [Google Scholar] [CrossRef]
- Zhou, X.H.; Brakebusch, C.; Matthies, H.; Oohashi, T.; Hirsch, E.; Moser, M.; Krug, M.; Seidenbecher, C.I.; Boeckers, T.M.; Rauch, U.; et al. Neurocan is dispensable for brain development. Mol. Cell Biol. 2001, 21, 5970–5978. [Google Scholar] [CrossRef]
- Gottschling, C.; Wegrzyn, D.; Denecke, B.; Faissner, A. Elimination of the four extracellular matrix molecules tenascin-C, tenascin-R, brevican and neurocan alters the ratio of excitatory and inhibitory synapses. Sci. Rep. 2019, 9, 13939. [Google Scholar] [CrossRef] [PubMed]
- Rauch, U.; Feng, K.; Zhou, X.H. Neurocan: A brain chondroitin sulfate proteoglycan. Cell Mol. Life Sci. 2001, 58, 1842–1856. [Google Scholar] [CrossRef] [PubMed]
- Asher, R.A.; Morgenstern, D.A.; Fidler, P.S.; Adcock, K.H.; Oohira, A.; Braistead, J.E.; Levine, J.M.; Margolis, R.U.; Rogers, J.H.; Fawcett, J.W. Neurocan is upregulated in injured brain and in cytokine-treated astrocytes. J. NeuroSci. 2000, 20, 2427–2438. [Google Scholar] [CrossRef] [PubMed]
- McKeon, R.J.; Jurynec, M.J.; Buck, C.R. The chondroitin sulfate proteoglycans neurocan and phosphacan are expressed by reactive astrocytes in the chronic CNS glial scar. J. NeuroSci. 1999, 19, 10778–10788. [Google Scholar] [CrossRef] [PubMed]
- Didangelos, A.; Puglia, M.; Iberl, M.; Sanchez-Bellot, C.; Roschitzki, B.; Bradbury, E.J. High-throughput proteomics reveal alarmins as amplifiers of tissue pathology and inflammation after spinal cord injury. Sci. Rep. 2016, 6, 21607. [Google Scholar] [CrossRef]
- Friedlander, D.R.; Milev, P.; Karthikeyan, L.; Margolis, R.K.; Margolis, R.U.; Grumet, M. The neuronal chondroitin sulfate proteoglycan neurocan binds to the neural cell adhesion molecules Ng-CAM/L1/NILE and N-CAM, and inhibits neuronal adhesion and neurite outgrowth. J. Cell Biol. 1994, 125, 669–680. [Google Scholar] [CrossRef]
- Retzler, C.; Gohring, W.; Rauch, U. Analysis of neurocan structures interacting with the neural cell adhesion molecule N-CAM. J. Biol. Chem. 1996, 271, 27304–27310. [Google Scholar] [CrossRef]
- Fontanil, T.; Mohamedi, Y.; Moncada-Pazos, A.; Cobo, T.; Vega, J.A.; Cobo, J.L.; Garcia-Suarez, O.; Cobo, J.; Obaya, A.J.; Cal, S. Neurocan is a New Substrate for the ADAMTS12 Metalloprotease: Potential Implications in Neuropathies. Cell. Physiol. Biochem. 2019, 52, 1003–1016. [Google Scholar] [CrossRef]
- Su, Z.; Kishida, S.; Tsubota, S.; Sakamoto, K.; Cao, D.; Kiyonari, S.; Ohira, M.; Kamijo, T.; Narita, A.; Xu, Y.; et al. Neurocan, an extracellular chondroitin sulfate proteoglycan, stimulates neuroblastoma cells to promote malignant phenotypes. Oncotarget 2017, 8, 106296–106310. [Google Scholar] [CrossRef]
- Meyer-Puttlitz, B.; Milev, P.; Junker, E.; Zimmer, I.; Margolis, R.U.; Margolis, R.K. Chondroitin sulfate and chondroitin/keratan sulfate proteoglycans of nervous tissue: Developmental changes of neurocan and phosphacan. J. Neurochem. 1995, 65, 2327–2337. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.P.; Siao, C.J.; Lu, W.; Sung, T.C.; Frohman, M.A.; Milev, P.; Bugge, T.H.; Degen, J.L.; Levine, J.M.; Margolis, R.U.; et al. The tissue plasminogen activator (tPA)/plasmin extracellular proteolytic system regulates seizure-induced hippocampal mossy fiber outgrowth through a proteoglycan substrate. J. Cell Biol. 2000, 148, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Turk, B.E.; Huang, L.L.; Piro, E.T.; Cantley, L.C. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat. Biotechnol. 2001, 19, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Bespalova, I.N.; Angelo, G.W.; Ritter, B.P.; Hunter, J.; Reyes-Rabanillo, M.L.; Siever, L.J.; Silverman, J.M. Genetic variations in the ADAMTS12 gene are associated with schizophrenia in Puerto Rican patients of Spanish descent. Neuromolecular Med. 2012, 14, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Cai, J.; Ni, J.; Zhang, J.; Tang, W.; Zhang, C. The NCAN gene: Schizophrenia susceptibility and cognitive dysfunction. Neuropsychiatr. Dis. Treat. 2016, 12, 2875–2883. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fontanil, T.; Mohamedi, Y.; Espina-Casado, J.; Obaya, Á.J.; Cobo, T.; Cal, S. Hyalectanase Activities by the ADAMTS Metalloproteases. Int. J. Mol. Sci. 2021, 22, 2988. https://doi.org/10.3390/ijms22062988
Fontanil T, Mohamedi Y, Espina-Casado J, Obaya ÁJ, Cobo T, Cal S. Hyalectanase Activities by the ADAMTS Metalloproteases. International Journal of Molecular Sciences. 2021; 22(6):2988. https://doi.org/10.3390/ijms22062988
Chicago/Turabian StyleFontanil, Tania, Yamina Mohamedi, Jorge Espina-Casado, Álvaro J. Obaya, Teresa Cobo, and Santiago Cal. 2021. "Hyalectanase Activities by the ADAMTS Metalloproteases" International Journal of Molecular Sciences 22, no. 6: 2988. https://doi.org/10.3390/ijms22062988
APA StyleFontanil, T., Mohamedi, Y., Espina-Casado, J., Obaya, Á. J., Cobo, T., & Cal, S. (2021). Hyalectanase Activities by the ADAMTS Metalloproteases. International Journal of Molecular Sciences, 22(6), 2988. https://doi.org/10.3390/ijms22062988




