Investigation of PRDM10 and PRDM13 Expression in Developing Mouse Embryos by an Optimized PACT-Based Embryo Clearing Method
Abstract
1. Introduction
2. Results
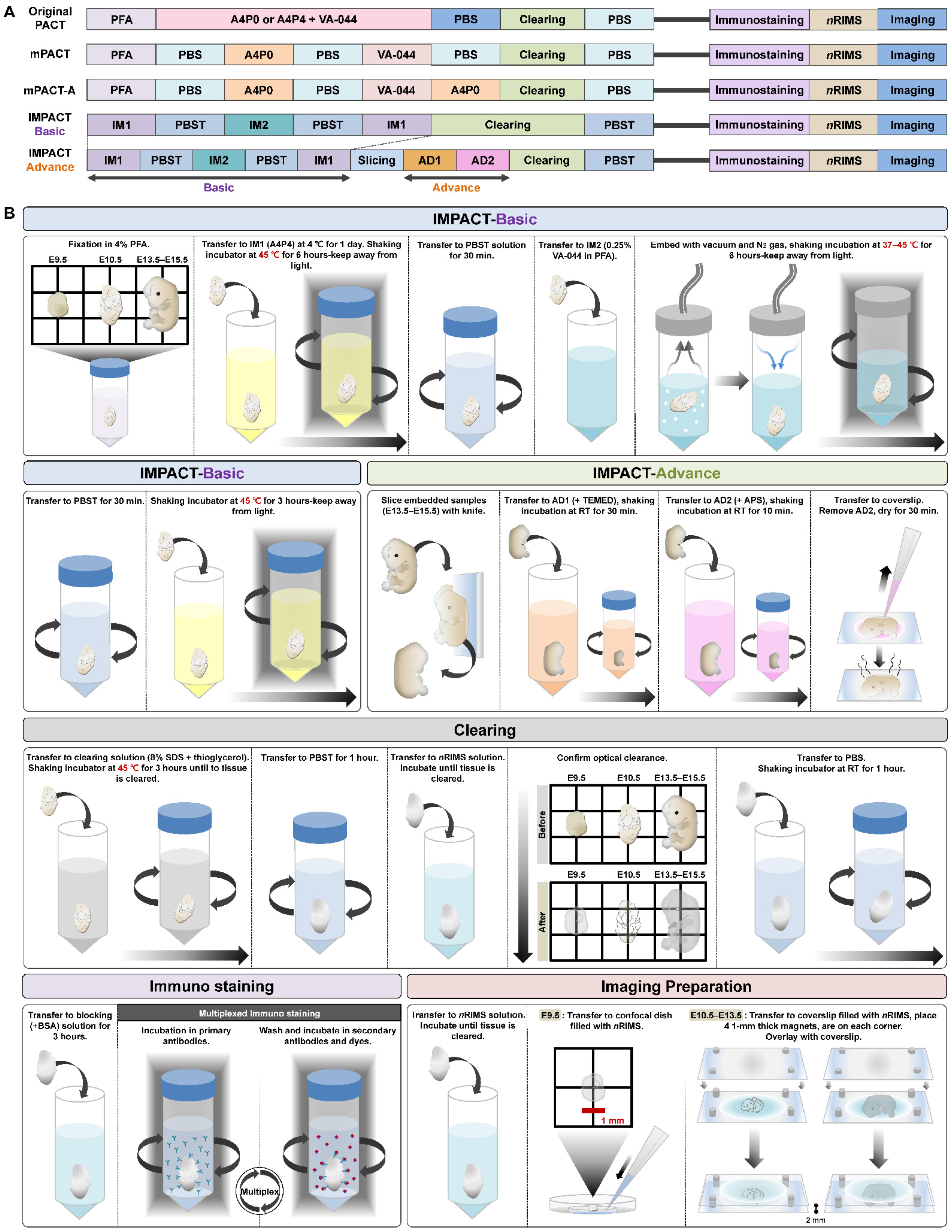
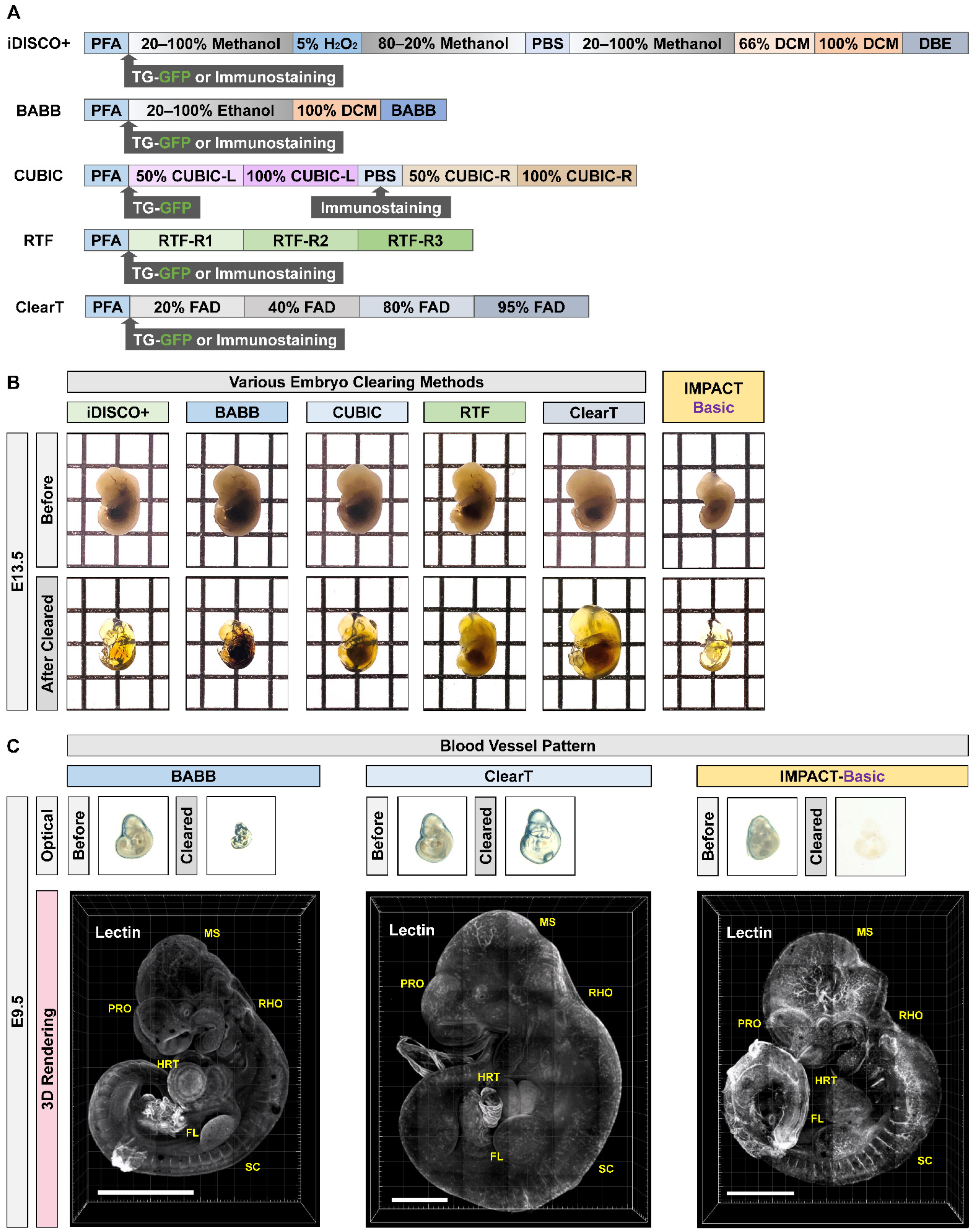
2.1. Generation of Transparent Mouse Embryos Using a PACT-Based Modified Tissue Clearing Method
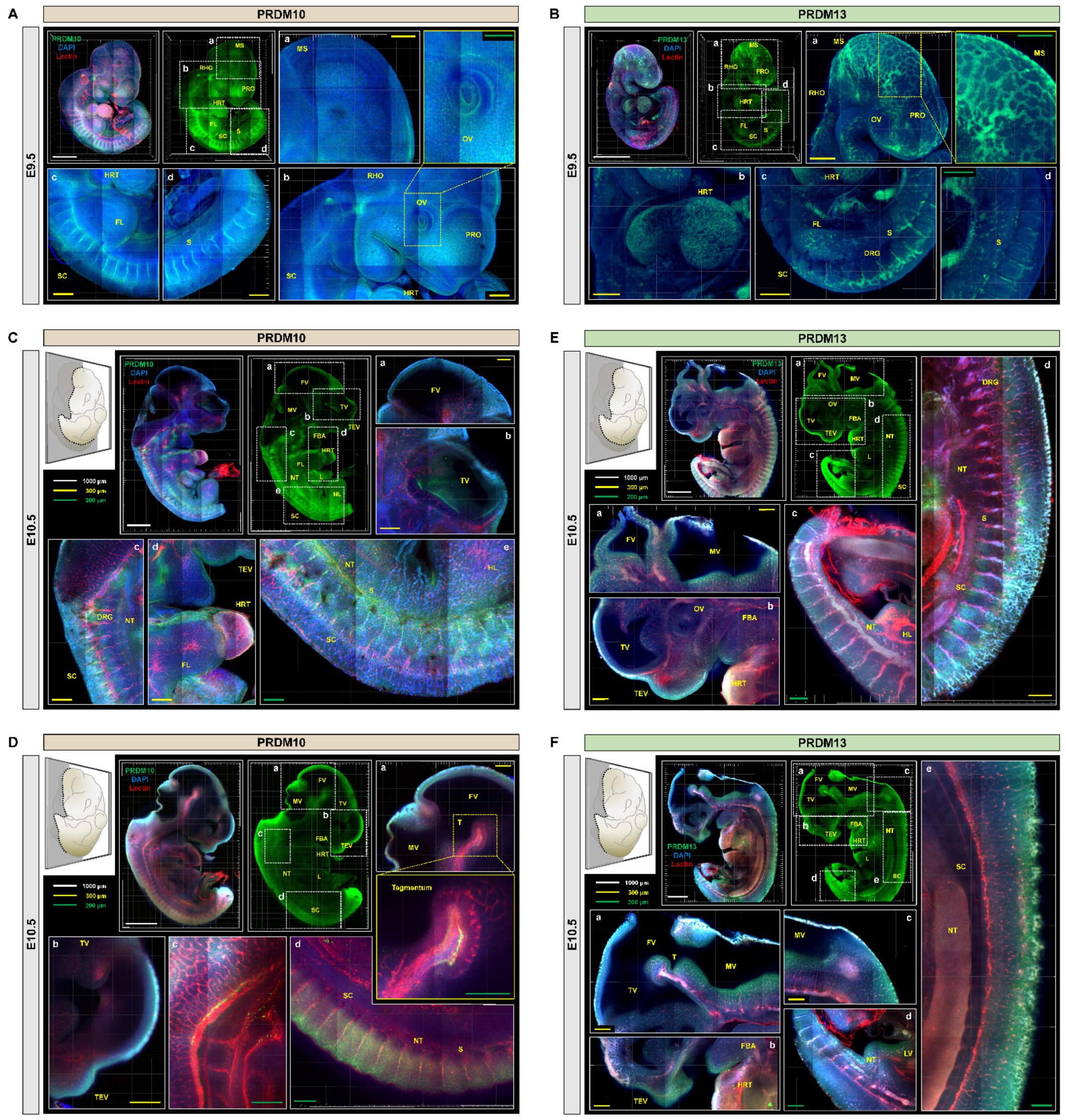
2.2. Profiling of PRDM10 and PRDM13 Expression in Early-Stage Mouse Embryos via IMPACT-Basic
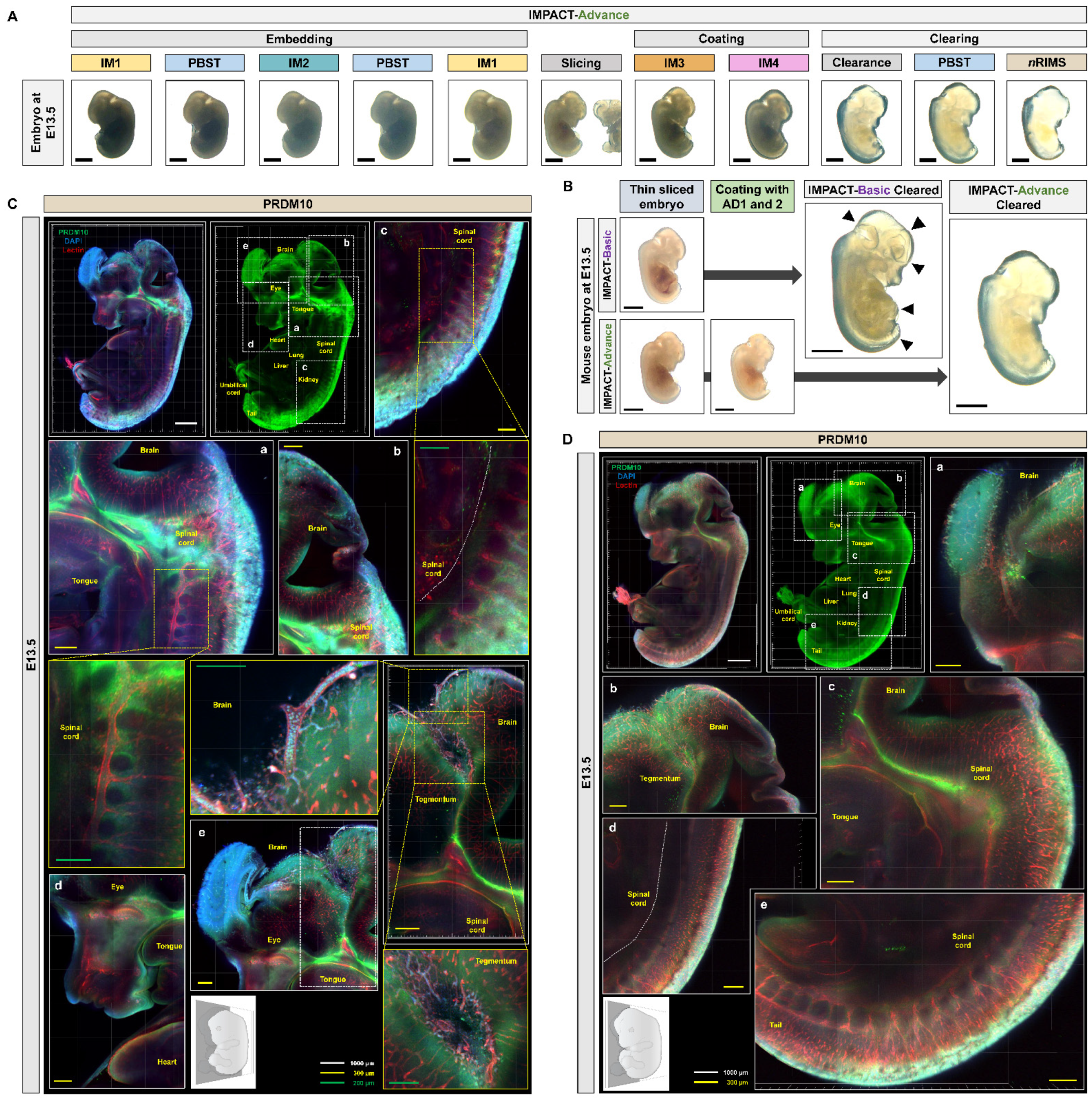
2.3. Development of Embryo-Specific IMPACT-Advance to Achieve Tissue Clarity and Retain Intact Organs in Large Embryo Section
2.4. Profiling of PRDM10 and PRDM13 Expression in E13.5 Mouse Embryos via IMPACT-Advance
3. Discussion
4. Materials and Methods
4.1. Animal
4.2. Isolation of Mouse Embryo and Tissue
4.3. Original PACT and mPACT
4.4. IMPACT-Basic and IMPACT-Advance
4.5. RTF
4.6. CUBIC
4.7. BABB
4.8. iDISCO+
4.9. ClearT
4.10. Immunostaining
4.11. Image Processing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PACT | Passive clearing technique |
| mPACT | Modified passive clearing technique |
| IMPACT | Initial embedding passive clearing technique |
| CNS | Central nervous system |
| PRDM | PR domain |
| CLARITY | Clear lipid-exchanged acrylamide-hybridized rigid imaging/immunostaining/in situ-hybridization-compatible tissue-hydrogel |
References
- Chung, K.; Wallace, J.; Kim, S.Y.; Kalyanasundaram, S.; Andalman, A.S.; Davidson, T.J.; Mirzabekov, J.J.; Zalocusky, K.A.; Mattis, J.; Denisin, A.K.; et al. Structural and molecular interrogation of intact biological systems. Nature 2013, 497, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Treweek, J.B.; Kulkarni, R.P.; Deverman, B.E.; Chen, C.K.; Lubeck, E.; Shah, S.; Cai, L.; Gradinaru, V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 2014, 158, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.; Lee, M.; Seo, J.M.; Park, H.S.; Cho, Y.E. Optimization of the optical transparency of rodent tissues by modified PACT-based passive clearing. Exp. Mol. Med. 2016, 48, e274. [Google Scholar] [CrossRef]
- Woo, J.; Lee, E.Y.; Park, H.S.; Park, J.Y.; Cho, Y.E. Novel Passive Clearing Methods for the Rapid Production of Optical Transparency in Whole CNS Tissue. J. Vis. Exp. 2018. [Google Scholar] [CrossRef]
- Woo, J.; Kang, H.; Lee, E.Y.; Park, S.; Cho, Y.E. Investigation of PRDM7 and PRDM12 expression pattern during mouse embryonic development by using a modified passive clearing technique. Biochem. Biophys. Res. Commun. 2020, 524, 346–353. [Google Scholar] [CrossRef]
- Sharpe, J.; Ahlgren, U.; Perry, P.; Hill, B.; Ross, A.; Hecksher-Sorensen, J.; Baldock, R.; Davidson, D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science 2002, 296, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, J. Optical projection tomography as a new tool for studying embryo anatomy. J. Anat. 2003, 202, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Hohenauer, T.; Moore, A.W. The Prdm family: Expanding roles in stem cells and development. Development 2012, 139, 2267–2282. [Google Scholar] [CrossRef] [PubMed]
- Fog, C.K.; Galli, G.G.; Lund, A.H. PRDM proteins: Important players in differentiation and disease. Bioessays 2012, 34, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Hofvander, J.; Puls, F.; Pillay, N.; Steele, C.D.; Flanagan, A.M.; Magnusson, L.; Nilsson, J.; Mertens, F. Undifferentiated pleomorphic sarcomas with PRDM10 fusions have a distinct gene expression profile. J. Pathol. 2019, 249, 425–434. [Google Scholar] [CrossRef]
- Hofvander, J.; Tayebwa, J.; Nilsson, J.; Magnusson, L.; Brosjo, O.; Larsson, O.; Vult von Steyern, F.; Mandahl, N.; Fletcher, C.D.; Mertens, F. Recurrent PRDM10 gene fusions in undifferentiated pleomorphic sarcoma. Clin. Cancer Res. 2015, 21, 864–869. [Google Scholar] [CrossRef] [PubMed]
- Zamanian Azodi, M.; Rezaei Tavirani, M.; Rezaei Tavirani, M.; Vafaee, R.; Rostami-Nejad, M. Nasopharyngeal Carcinoma Protein Interaction Mapping Analysis via Proteomic Approaches. Asian Pac. J. Cancer Prev. 2018, 19, 845–851. [Google Scholar] [CrossRef]
- Chen, N.; Hu, T.; Gui, Y.; Gao, J.; Li, Z.; Huang, S. Transcriptional regulation of Bcl-2 gene by the PR/SET domain family member PRDM10. PeerJ 2019, 7, e6941. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, A.; Federico, A.; Rienzo, M.; Gazzerro, P.; Bifulco, M.; Ciccodicola, A.; Casamassimi, A.; Abbondanza, C. PR/SET Domain Family and Cancer: Novel Insights from the Cancer Genome Atlas. Int. J. Mol. Sci. 2018, 19, 3250. [Google Scholar] [CrossRef] [PubMed]
- Behrends, U.; Schneider, I.; Rossler, S.; Frauenknecht, H.; Golbeck, A.; Lechner, B.; Eigenstetter, G.; Zobywalski, C.; Muller-Weihrich, S.; Graubner, U.; et al. Novel tumor antigens identified by autologous antibody screening of childhood medulloblastoma cDNA libraries. Int. J. Cancer 2003, 106, 244–251. [Google Scholar] [CrossRef]
- Bessodes, N.; Parain, K.; Bronchain, O.; Bellefroid, E.J.; Perron, M. Prdm13 forms a feedback loop with Ptf1a and is required for glycinergic amacrine cell genesis in the Xenopus Retina. Neural. Dev. 2017, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Goodson, N.B.; Nahreini, J.; Randazzo, G.; Uruena, A.; Johnson, J.E.; Brzezinski, J.A.T. Prdm13 is required for Ebf3+ amacrine cell formation in the retina. Dev. Biol. 2018, 434, 149–163. [Google Scholar] [CrossRef]
- Mona, B.; Uruena, A.; Kollipara, R.K.; Ma, Z.; Borromeo, M.D.; Chang, J.C.; Johnson, J.E. Repression by PRDM13 is critical for generating precision in neuronal identity. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Hanotel, J.; Bessodes, N.; Thelie, A.; Hedderich, M.; Parain, K.; Van Driessche, B.; Brandao Kde, O.; Kricha, S.; Jorgensen, M.C.; Grapin-Botton, A.; et al. The Prdm13 histone methyltransferase encoding gene is a Ptf1a-Rbpj downstream target that suppresses glutamatergic and promotes GABAergic neuronal fate in the dorsal neural tube. Dev. Biol. 2014, 386, 340–357. [Google Scholar] [CrossRef] [PubMed]
- Casamassimi, A.; Rienzo, M.; Di Zazzo, E.; Sorrentino, A.; Fiore, D.; Proto, M.C.; Moncharmont, B.; Gazzerro, P.; Bifulco, M.; Abbondanza, C. Multifaceted Role of PRDM Proteins in Human Cancer. Int. J. Mol. Sci. 2020, 21, 2648. [Google Scholar] [CrossRef]
- Gomez-Gaviro, M.V.; Balaban, E.; Bocancea, D.; Lorrio, M.T.; Pompeiano, M.; Desco, M.; Ripoll, J.; Vaquero, J.J. Optimized CUBIC protocol for three-dimensional imaging of chicken embryos at single-cell resolution. Development 2017, 144, 2092–2097. [Google Scholar] [CrossRef] [PubMed]
- Park, J.A.; Kim, K.C. Expression patterns of PRDM10 during mouse embryonic development. BMB Rep. 2010, 43, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Kolesova, H.; Capek, M.; Radochova, B.; Janacek, J.; Sedmera, D. Comparison of different tissue clearing methods and 3D imaging techniques for visualization of GFP-expressing mouse embryos and embryonic hearts. Histochem. Cell Biol. 2016, 146, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Hama, H.; Kurokawa, H.; Kawano, H.; Ando, R.; Shimogori, T.; Noda, H.; Fukami, K.; Sakaue-Sawano, A.; Miyawaki, A. Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 2011, 14, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Kuwajima, T.; Sitko, A.A.; Bhansali, P.; Jurgens, C.; Guido, W.; Mason, C. ClearT: A detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 2013, 140, 1364–1368. [Google Scholar] [CrossRef] [PubMed]
- Ke, M.T.; Fujimoto, S.; Imai, T. SeeDB: A simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 2013, 16, 1154–1161. [Google Scholar] [CrossRef] [PubMed]
- Renier, N.; Wu, Z.; Simon, D.J.; Yang, J.; Ariel, P.; Tessier-Lavigne, M. iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 2014, 159, 896–910. [Google Scholar] [CrossRef]
- Dodt, H.U.; Leischner, U.; Schierloh, A.; Jahrling, N.; Mauch, C.P.; Deininger, K.; Deussing, J.M.; Eder, M.; Zieglgansberger, W.; Becker, K. Ultramicroscopy: Three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 2007, 4, 331–336. [Google Scholar] [CrossRef]
- Yu, T.; Zhu, J.; Li, Y.; Ma, Y.; Wang, J.; Cheng, X.; Jin, S.; Sun, Q.; Li, X.; Gong, H.; et al. RTF: A rapid and versatile tissue optical clearing method. Sci. Rep. 2018, 8, 1964. [Google Scholar] [CrossRef]
- Susaki, E.A.; Tainaka, K.; Perrin, D.; Yukinaga, H.; Kuno, A.; Ueda, H.R. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 2015, 10, 1709–1727. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woo, J.; Jin, B.-H.; Lee, M.; Lee, E.Y.; Moon, H.-S.; Park, J.-Y.; Cho, Y.-E. Investigation of PRDM10 and PRDM13 Expression in Developing Mouse Embryos by an Optimized PACT-Based Embryo Clearing Method. Int. J. Mol. Sci. 2021, 22, 2892. https://doi.org/10.3390/ijms22062892
Woo J, Jin B-H, Lee M, Lee EY, Moon H-S, Park J-Y, Cho Y-E. Investigation of PRDM10 and PRDM13 Expression in Developing Mouse Embryos by an Optimized PACT-Based Embryo Clearing Method. International Journal of Molecular Sciences. 2021; 22(6):2892. https://doi.org/10.3390/ijms22062892
Chicago/Turabian StyleWoo, Jiwon, Byung-Ho Jin, Mirae Lee, Eunice Yoojin Lee, Hyung-Seok Moon, Jeong-Yoon Park, and Yong-Eun Cho. 2021. "Investigation of PRDM10 and PRDM13 Expression in Developing Mouse Embryos by an Optimized PACT-Based Embryo Clearing Method" International Journal of Molecular Sciences 22, no. 6: 2892. https://doi.org/10.3390/ijms22062892
APA StyleWoo, J., Jin, B.-H., Lee, M., Lee, E. Y., Moon, H.-S., Park, J.-Y., & Cho, Y.-E. (2021). Investigation of PRDM10 and PRDM13 Expression in Developing Mouse Embryos by an Optimized PACT-Based Embryo Clearing Method. International Journal of Molecular Sciences, 22(6), 2892. https://doi.org/10.3390/ijms22062892





