Subunits of the PBAP Chromatin Remodeler Are Capable of Mediating Enhancer-Driven Transcription in Drosophila
Abstract
1. Introduction
2. Results
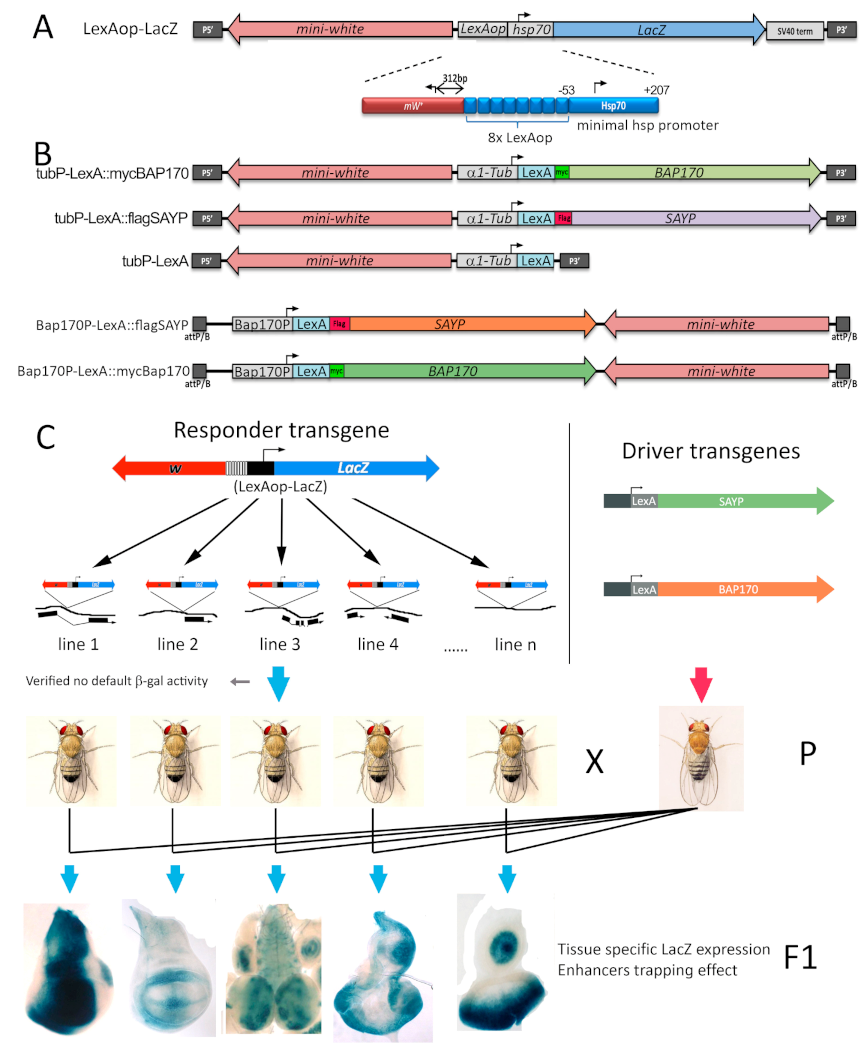
2.1. In Vivo Targeting of SAYP or BAP170 to a Minimal Promoter Mediates Enhancer-Dependent Transcriptional Activation
2.2. Promoter-Bound PBAP Subunit Functions as a Tethering Factor for the hsp70 Minimal Promoter
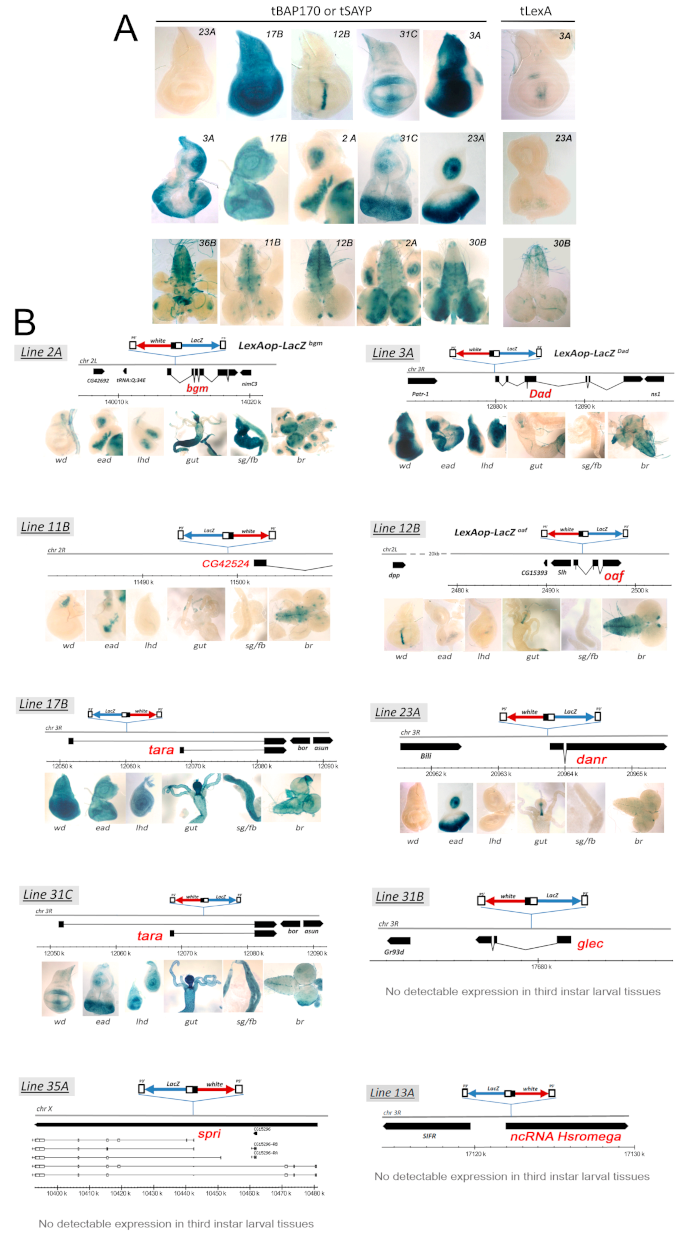
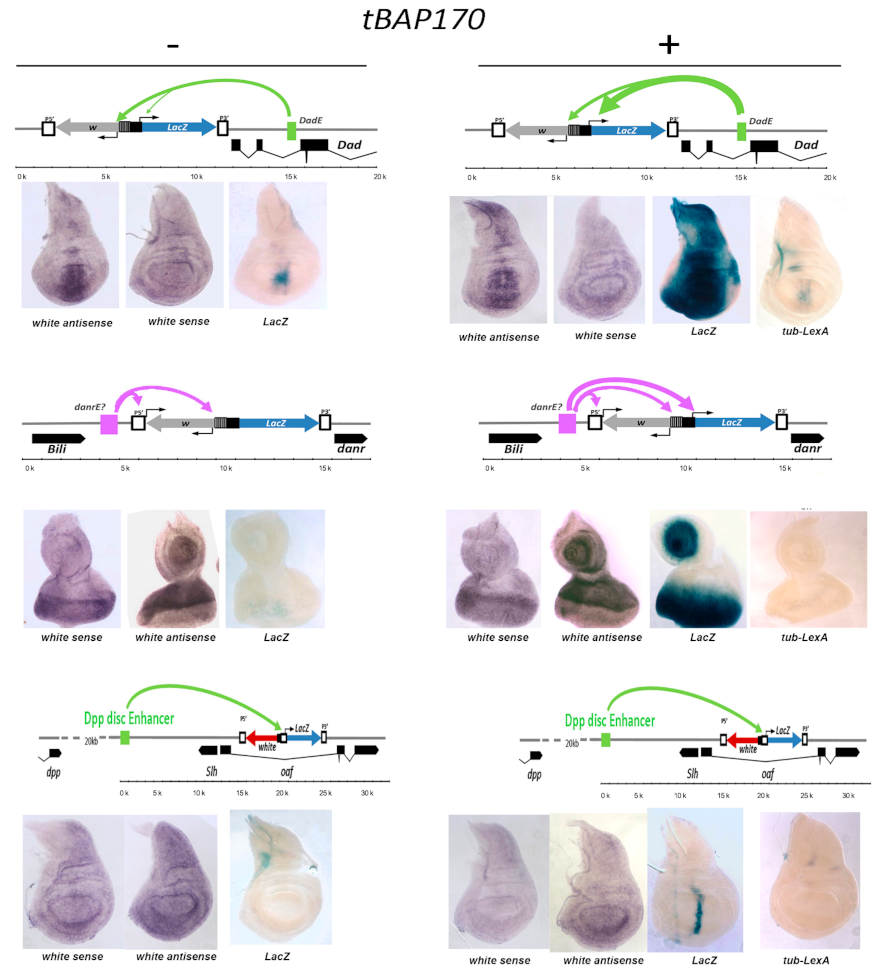
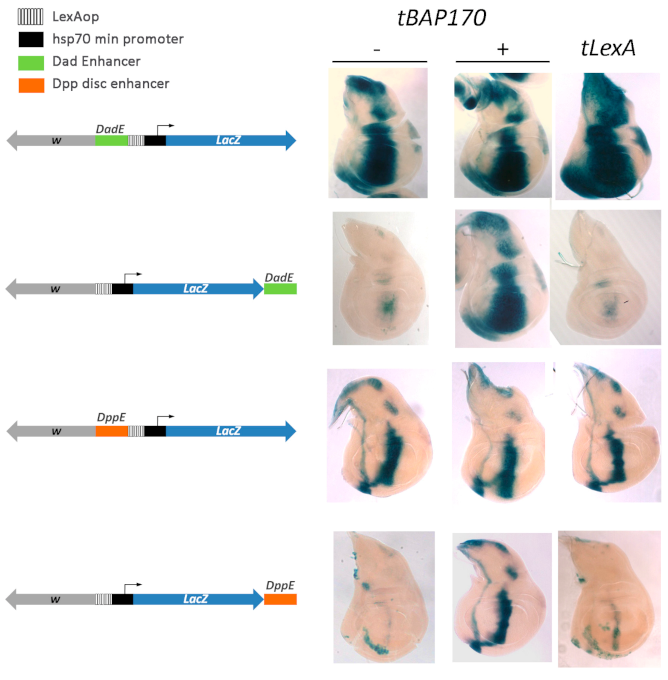
2.3. Tethered BAP170/SAYP Facilitates Capture of Distant Enhancers
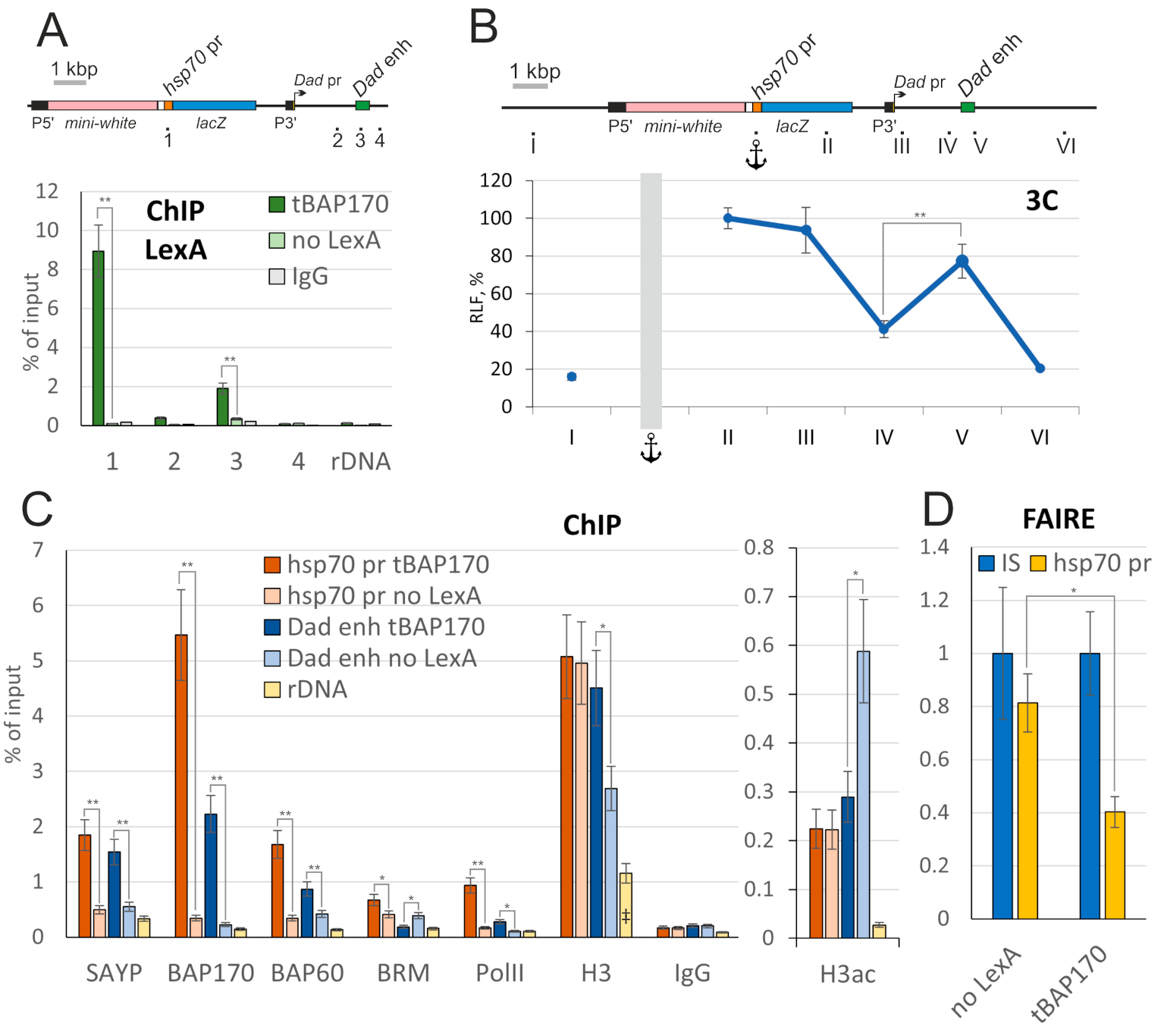
2.4. Tethered PBAP Subunit Recruits the Whole Complex onto the hsp70 Promoter
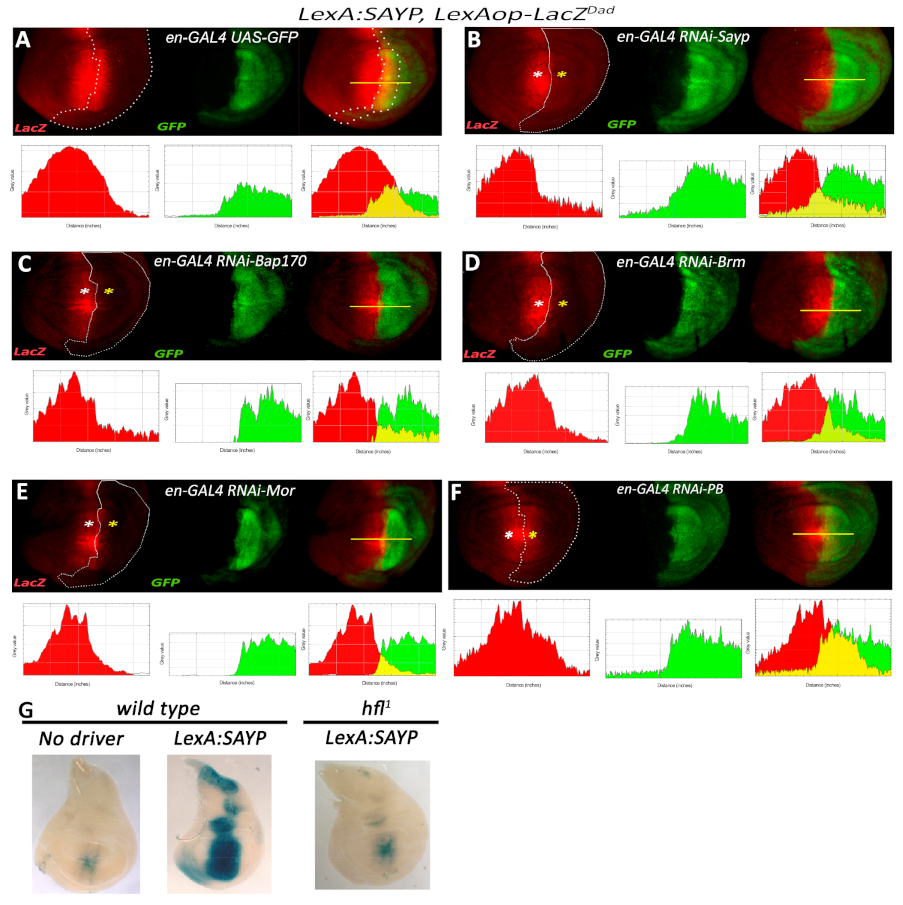
2.5. tSAYP-Dependent Reporter Transcription Crucially Depends on BAP170 and the Core PBAP Subunits
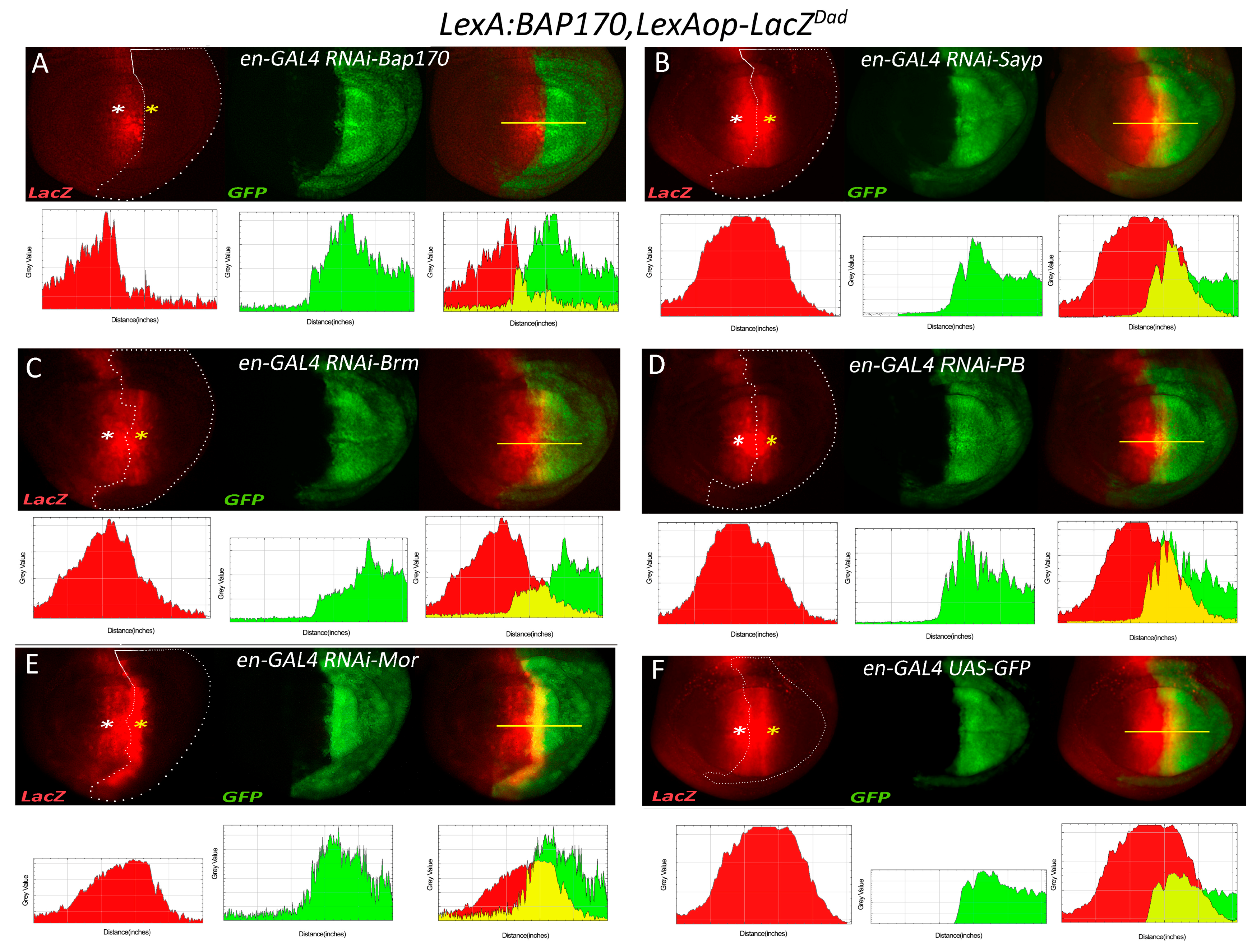
2.6. tBAP170-Driven Transcriptional Activation Is Resistant to Depletion of SAYP and PBAP Core Subunits
3. Discussion
4. Materials and Methods
4.1. Plasmid Preparation
4.2. Beta-Gal Staining, In Situ Hybridization, and Immunofluorescence
4.3. Transgenic Line Preparation
4.4. Drosophila Strains
4.5. Antibodies
4.6. Polytene Chromosome Analysis
4.7. Chromatin Immunoprecipitation, 3C and FAIRE Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bouazoune, K.; Brehm, A. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 2006, 14, 433–449. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef] [PubMed]
- Mohrmann, L.; Verrijzer, C.P. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 2005, 1681, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Cosma, M.P. Ordered recruitment: Gene-specific mechanism of transcription activation. Mol. Cell. 2002, 10, 227–236. [Google Scholar] [CrossRef]
- Hirschhorn, J.N.; Brown, S.A.; Clark, C.D.; Winston, F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992, 6, 2288–2298. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.L.; Workman, J.L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000, 10, 187–192. [Google Scholar] [CrossRef]
- Lorch, Y.; Maier-Davis, B.; Kornberg, R.D. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. USA 2006, 103, 3090–3093. [Google Scholar] [CrossRef]
- Lorch, Y.; Zhang, M.; Kornberg, R.D. Histone octamer transfer by a chromatin-remodeling complex. Cell 1999, 96, 389–392. [Google Scholar] [CrossRef]
- Whitehouse, I.; Flaus, A.; Cairns, B.R.; White, M.F.; Workman, J.L.; Owen-Hughes, T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 1999, 400, 784–787. [Google Scholar] [CrossRef]
- Mani, U.; Sankareswaran, A.; Goutham, R.N.A.; Mohan, S.S. SWI/SNF Infobase-An exclusive information portal for SWI/SNF remodeling complex subunits. PLoS ONE 2017, 12, e0184445. [Google Scholar] [CrossRef]
- Mohrmann, L.; Langenberg, K.; Krijgsveld, J.; Kal, A.J.; Heck, A.J.; Verrijzer, C.P. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 2004, 24, 3077–3088. [Google Scholar] [CrossRef]
- Moshkin, Y.M.; Mohrmann, L.; van Ijcken, W.F.; Verrijzer, C.P. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol. Cell. Biol. 2007, 27, 651–661. [Google Scholar] [CrossRef]
- Carrera, I.; Zavadil, J.; Treisman, J.E. Two subunits specific to the PBAP chromatin remodeling complex have distinct and redundant functions during drosophila development. Mol. Cell. Biol. 2008, 28, 5238–5250. [Google Scholar] [CrossRef]
- He, J.; Xuan, T.; Xin, T.; An, H.; Wang, J.; Zhao, G.; Li, M. Evidence for chromatin-remodeling complex PBAP-controlled maintenance of the Drosophila ovarian germline stem cells. PLoS ONE 2014, 9, e103473. [Google Scholar] [CrossRef]
- Nakayama, T.; Shimojima, T.; Hirose, S. The PBAP remodeling complex is required for histone H3.3 replacement at chromatin boundaries and for boundary functions. Development 2012, 139, 4582–4590. [Google Scholar] [CrossRef] [PubMed]
- Rendina, R.; Strangi, A.; Avallone, B.; Giordano, E. Bap170, a subunit of the Drosophila PBAP chromatin remodeling complex, negatively regulates the EGFR signaling. Genetics 2010, 186, 167–181. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Terriente-Felix, A.; de Celis, J.F. Osa, a subunit of the BAP chromatin-remodelling complex, participates in the regulation of gene expression in response to EGFR signalling in the Drosophila wing. Dev. Biol. 2009, 329, 350–361. [Google Scholar] [CrossRef]
- Chalkley, G.E.; Moshkin, Y.M.; Langenberg, K.; Bezstarosti, K.; Blastyak, A.; Gyurkovics, H.; Demmers, J.A.; Verrijzer, C.P. The transcriptional coactivator SAYP is a trithorax group signature subunit of the PBAP chromatin remodeling complex. Mol. Cell. Biol. 2008, 28, 2920–2929. [Google Scholar] [CrossRef][Green Version]
- Laurent, B.C.; Treitel, M.A.; Carlson, M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of Genes. Mol. Cell. Biol. 1990, 10, 5616–5625. [Google Scholar] [CrossRef]
- Laurent, B.C.; Treitel, M.A.; Carlson, M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc. Natl. Acad. Sci. USA 1991, 88, 2687–2691. [Google Scholar] [CrossRef]
- Vorobyeva, N.E.; Soshnikova, N.V.; Nikolenko, J.V.; Kuzmina, J.L.; Nabirochkina, E.N.; Georgieva, S.G.; Shidlovskii, Y.V. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc. Natl. Acad. Sci. USA 2009, 106, 11049–11054. [Google Scholar] [CrossRef] [PubMed]
- Tsuneizumi, K.; Nakayama, T.; Kamoshida, Y.; Kornberg, T.B.; Christian, J.L.; Tabata, T. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature 1997, 389, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Masucci, J.D.; Miltenberger, R.J.; Hoffmann, F.M. Pattern-specific expression of the Drosophila decapentaplegic gene in imaginal disks is regulated by 3’ cis-regulatory elements. Genes Dev. 1990, 4, 2011–2023. [Google Scholar] [CrossRef] [PubMed]
- Emerald, B.S.; Curtiss, J.; Mlodzik, M.; Cohen, S.M. Distal antenna and distal antenna related encode nuclear proteins containing pipsqueak motifs involved in antenna development in Drosophila. Development 2003, 130, 1171–1180. [Google Scholar] [CrossRef]
- FlyBase. Available online: https://flybase.org/reports/FBrf0083714.html (accessed on 10 March 2021).
- Weiss, A.; Charbonnier, E.; Ellertsdottir, E.; Tsirigos, A.; Wolf, C.; Schuh, R.; Pyrowolakis, G.; Affolter, M. A conserved activation element in BMP signaling during Drosophila development. Nat. Struct Mol. Biol. 2010, 17, 69–76. [Google Scholar] [CrossRef]
- Muller, B.; Basler, K. The repressor and activator forms of Cubitus interruptus control Hedgehog target genes through common generic gli-binding sites. Development 2000, 127, 2999–3007. [Google Scholar]
- Blackman, R.K.; Sanicola, M.; Raftery, L.A.; Gillevet, T.; Gelbart, W.M. An extensive 3’ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development 1991, 111, 657–666. [Google Scholar]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar]
- Corbin, V.; Maniatis, T. The role of specific enhancer-promoter interactions in the Drosophila Adh promoter switch. Genes Dev. 1989, 3, 2091–2120. [Google Scholar] [CrossRef]
- Kwon, D.; Mucci, D.; Langlais, K.K.; Americo, J.L.; DeVido, S.K.; Cheng, Y.; Kassis, J.A. Enhancer-promoter communication at the Drosophila engrailed locus. Development 2009, 136, 3067–3075. [Google Scholar] [CrossRef]
- O’Kane, C.J.; Gehring, W.J. Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. USA 1987, 84, 9123–9127. [Google Scholar] [CrossRef]
- Qian, S.; Varjavand, B.; Pirrotta, V. Molecular analysis of the zeste-white interaction reveals a promoter-proximal element essential for distant enhancer-promoter communication. Genetics 1992, 131, 79–90. [Google Scholar] [CrossRef]
- Attrill, H.; Falls, K.; Goodman, J.L.; Millburn, G.H.; Antonazzo, G.; Rey, A.J.; Marygold, S.J.; FlyBase, C. FlyBase: Establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 2016, 44, D786–D792. [Google Scholar] [CrossRef] [PubMed]
- Bittencourt, D.; Wu, D.Y.; Jeong, K.W.; Gerke, D.S.; Herviou, L.; Ianculescu, I.; Chodankar, R.; Siegmund, K.D.; Stallcup, M.R. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target Genes. Proc. Natl. Acad. Sci. USA 2012, 109, 19673–19678. [Google Scholar] [CrossRef]
- Shapira, S.N.; Lim, H.W.; Rajakumari, S.; Sakers, A.P.; Ishibashi, J.; Harms, M.J.; Won, K.J.; Seale, P. EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev. 2017, 31, 660–673. [Google Scholar] [CrossRef] [PubMed]
- Baum, S.; Reimer-Michalski, E.M.; Jaskiewicz, M.R.; Conrath, U. Formaldehyde-assisted isolation of regulatory DNA elements from Arabidopsis leaves. Nat. Protoc. 2020, 15, 713–733. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, B.D.; Jenett, A.; Hammonds, A.S.; Ngo, T.T.; Misra, S.; Murphy, C.; Scully, A.; Carlson, J.W.; Wan, K.H.; Laverty, T.R.; et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 2008, 105, 9715–9720. [Google Scholar] [CrossRef]
- Pfeiffer, B.D.; Ngo, T.T.; Hibbard, K.L.; Murphy, C.; Jenett, A.; Truman, J.W.; Rubin, G.M. Refinement of tools for targeted gene expression in Drosophila. Genetics 2010, 186, 735–755. [Google Scholar] [CrossRef]
- Nakayama, R.T.; Pulice, J.L.; Valencia, A.M.; McBride, M.J.; McKenzie, Z.M.; Gillespie, M.A.; Ku, W.L.; Teng, M.; Cui, K.; Williams, R.T.; et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat. Genet. 2017, 49, 1613–1623. [Google Scholar] [CrossRef]
- Wang, X.; Lee, R.S.; Alver, B.H.; Haswell, J.R.; Wang, S.; Mieczkowski, J.; Drier, Y.; Gillespie, S.M.; Archer, T.C.; Wu, J.N.; et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 2017, 49, 289–295. [Google Scholar] [CrossRef]
- Mathur, R.; Alver, B.H.; San Roman, A.K.; Wilson, B.G.; Wang, X.; Agoston, A.T.; Park, P.J.; Shivdasani, R.A.; Roberts, C.W. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 2017, 49, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Moshkin, Y.M.; Chalkley, G.E.; Kan, T.W.; Reddy, B.A.; Ozgur, Z.; van Ijcken, W.F.; Dekkers, D.H.; Demmers, J.A.; Travers, A.A.; Verrijzer, C.P. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol. Cell. Biol. 2012, 32, 675–688. [Google Scholar] [CrossRef]
- Tolstorukov, M.Y.; Sansam, C.G.; Lu, P.; Koellhoffer, E.C.; Helming, K.C.; Alver, B.H.; Tillman, E.J.; Evans, J.A.; Wilson, B.G.; Park, P.J.; et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc. Natl. Acad. Sci. USA 2013, 110, 10165–10170. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.M.; Hota, S.K.; He, D.; Thomas, S.; Ho, L.; Pennacchio, L.A.; Bruneau, B.G. Brg1 modulates enhancer activation in mesoderm lineage commitment. Development 2015, 142, 1418–1430. [Google Scholar] [CrossRef]
- Alver, B.H.; Kim, K.H.; Lu, P.; Wang, X.; Manchester, H.E.; Wang, W.; Haswell, J.R.; Park, P.J.; Roberts, C.W. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat. Commun. 2017, 8, 14648. [Google Scholar] [CrossRef] [PubMed]
- Bossen, C.; Murre, C.S.; Chang, A.N.; Mansson, R.; Rodewald, H.R.; Murre, C. The chromatin remodeler Brg1 activates enhancer repertoires to establish B cell identity and modulate cell growth. Nat. Immunol. 2015, 16, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Schones, D.E.; Cui, K.; Ybarra, R.; Northrup, D.; Tang, Q.; Gattinoni, L.; Restifo, N.P.; Huang, S.; Zhao, K. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 2011, 21, 1650–1658. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Abou El Hassan, M.; Xu, Z.; Yu, T.; Bremner, R. The chromatin-remodeling enzyme BRG1 coordinates CIITA induction through many interdependent distal enhancers. Nat. Immunol. 2008, 9, 785–793. [Google Scholar] [CrossRef]
- Shi, J.; Whyte, W.A.; Zepeda-Mendoza, C.J.; Milazzo, J.P.; Shen, C.; Roe, J.S.; Minder, J.L.; Mercan, F.; Wang, E.; Eckersley-Maslin, M.A.; et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013, 27, 2648–2662. [Google Scholar] [CrossRef]
- Wood, C.D.; Veenstra, H.; Khasnis, S.; Gunnell, A.; Webb, H.M.; Shannon-Lowe, C.; Andrews, S.; Osborne, C.S.; West, M.J. MYC activation and BCL2L11 silencing by a tumour virus through the large-scale reconfiguration of enhancer-promoter hubs. eLife 2016, 5. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, Y.; Kim, B.; Wang, H.; Zhao, C.; He, X.; Liu, L.; Liu, W.; Wu, L.M.; Mao, M.; et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 2013, 152, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Pillidge, Z.; Bray, S.J. SWI/SNF chromatin remodeling controls Notch-responsive enhancer accessibility. EMBO Rep. 2019, 20, e46944. [Google Scholar] [CrossRef] [PubMed]
- Vo Ngoc, L.; Wang, Y.L.; Kassavetis, G.A.; Kadonaga, J.T. The punctilious RNA polymerase II core promoter. Genes Dev. 2017, 31, 1289–1301. [Google Scholar] [CrossRef]
- Zabidi, M.A.; Stark, A. Regulatory Enhancer-Core-Promoter Communication via Transcription Factors and Cofactors. Trends Genet. 2016, 32, 801–814. [Google Scholar] [CrossRef]
- Cubenas-Potts, C.; Corces, V.G. Architectural proteins, transcription, and the three-dimensional organization of the genome. FEBS Lett. 2015, 589, 2923–2930. [Google Scholar] [CrossRef] [PubMed]
- Cubenas-Potts, C.; Rowley, M.J.; Lyu, X.; Li, G.; Lei, E.P.; Corces, V.G. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res. 2017, 45, 1714–1730. [Google Scholar] [CrossRef] [PubMed]
- Maksimenko, O.; Georgiev, P. Mechanisms and proteins involved in long-distance interactions. Front. Genet. 2014, 5, 28. [Google Scholar] [CrossRef]
- Bao, X.; Rubin, A.J.; Qu, K.; Zhang, J.; Giresi, P.G.; Chang, H.Y.; Khavari, P.A. A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 2015, 16, 284. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Yoshihara, E.; He, N.; Hah, N.; Fan, W.; Pinto, A.F.M.; Huddy, T.; Wang, Y.; Ross, B.; Estepa, G.; et al. Vitamin D Switches BAF Complexes to Protect beta Cells. Cell 2018, 173, 1135–1149.e1115. [Google Scholar] [CrossRef] [PubMed]
- Jordan-Pla, A.; Yu, S.; Waldholm, J.; Kallman, T.; Ostlund Farrants, A.K.; Visa, N. SWI/SNF regulates half of its targets without the need of ATP-driven nucleosome remodeling by Brahma. BMC Genom. 2018, 19, 367. [Google Scholar] [CrossRef]
- Panov, V.V.; Kuzmina, J.L.; Doronin, S.A.; Kopantseva, M.R.; Nabirochkina, E.N.; Georgieva, S.G.; Vorobyeva, N.E.; Shidlovskii, Y.V. Transcription co-activator SAYP mediates the action of STAT activator. Nucleic Acids Res. 2012, 40, 2445–2453. [Google Scholar] [CrossRef] [PubMed]
- Vorobyeva, N.E.; Nikolenko, J.V.; Krasnov, A.N.; Kuzmina, J.L.; Panov, V.V.; Nabirochkina, E.N.; Georgieva, S.G.; Shidlovskii, Y.V. SAYP interacts with DHR3 nuclear receptor and participates in ecdysone-dependent transcription regulation. Cell. Cycle 2011, 10, 1821–1827. [Google Scholar] [CrossRef]
- Bylino, O.V.; Ibragimov, A.N.; Shidlovskii, Y.V. Evolution of Regulated Transcription. Cells 2020, 9, 1675. [Google Scholar] [CrossRef]
- Furlong, E.E.M.; Levine, M. Developmental enhancers and chromosome topology. Science 2018, 361, 1341–1345. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Wittmeyer, J.; Cairns, B.R. Chromatin remodelling: The industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006, 7, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Dechassa, M.L.; Sabri, A.; Pondugula, S.; Kassabov, S.R.; Chatterjee, N.; Kladde, M.P.; Bartholomew, B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell 2010, 38, 590–602. [Google Scholar] [CrossRef]
- Kassabov, S.R.; Zhang, B.; Persinger, J.; Bartholomew, B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell 2003, 11, 391–403. [Google Scholar] [CrossRef]
- Zhang, Y.; Smith, C.L.; Saha, A.; Grill, S.W.; Mihardja, S.; Smith, S.B.; Cairns, B.R.; Peterson, C.L.; Bustamante, C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell 2006, 24, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Bazett-Jones, D.P.; Cote, J.; Landel, C.C.; Peterson, C.L.; Workman, J.L. The SWI/SNF complex creates loop domains in DNA and polynucleosome arrays and can disrupt DNA-histone contacts within these domains. Mol. Cell. Biol. 1999, 19, 1470–1478. [Google Scholar] [CrossRef]
- Kim, S.I.; Bresnick, E.H.; Bultman, S.J. BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Res. 2009, 37, 6019–6027. [Google Scholar] [CrossRef]
- Kim, S.I.; Bultman, S.J.; Kiefer, C.M.; Dean, A.; Bresnick, E.H. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc. Natl. Acad. Sci. USA 2009, 106, 2259–2264. [Google Scholar] [CrossRef] [PubMed]
- Laurent, B.C.; Carlson, M. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes Dev. 1992, 6, 1707–1715. [Google Scholar] [CrossRef][Green Version]
- Ibragimov, A.N.; Bylino, O.V.; Shidlovskii, Y.V. Molecular Basis of the Function of Transcriptional Enhancers. Cells 2020, 9, 1620. [Google Scholar] [CrossRef]
- Kyrchanova, O.; Georgiev, P. Mechanisms of Enhancer-Promoter Interactions in Higher Eukaryotes. Int. J. Mol. Sci. 2021, 22, 671. [Google Scholar] [CrossRef] [PubMed]
- Barutcu, A.R.; Lajoie, B.R.; Fritz, A.J.; McCord, R.P.; Nickerson, J.A.; van Wijnen, A.J.; Lian, J.B.; Stein, J.L.; Dekker, J.; Stein, G.S.; et al. SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 2016, 26, 1188–1201. [Google Scholar] [CrossRef]
- Euskirchen, G.M.; Auerbach, R.K.; Davidov, E.; Gianoulis, T.A.; Zhong, G.; Rozowsky, J.; Bhardwaj, N.; Gerstein, M.B.; Snyder, M. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011, 7, e1002008. [Google Scholar] [CrossRef]
- Wu, S.; Fatkhutdinov, N.; Rosin, L.; Luppino, J.M.; Iwasaki, O.; Tanizawa, H.; Tang, H.Y.; Kossenkov, A.V.; Gardini, A.; Noma, K.I.; et al. ARID1A spatially partitions interphase chromosomes. Sci. Adv. 2019, 5, eaaw5294. [Google Scholar] [CrossRef] [PubMed]
- Thummel, C.S.; Boulet, A.M.; Lipshitz, H.D. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene 1988, 74, 445–456. [Google Scholar] [CrossRef]
- Giordano, E.; Peluso, I.; Senger, S.; Furia, M. minifly, a Drosophila gene required for ribosome biogenesis. J. Cell. Biol. 1999, 144, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Giordano, E.; Rendina, R.; Peluso, I.; Furia, M. RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics 2002, 160, 637–648. [Google Scholar] [PubMed]
- Robertson, H.M.; Preston, C.R.; Phillis, R.W.; Johnson-Schlitz, D.M.; Benz, W.K.; Engels, W.R. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 1988, 118, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Shidlovskii, Y.V.; Krasnov, A.N.; Nikolenko, J.V.; Lebedeva, L.A.; Kopantseva, M.; Ermolaeva, M.A.; Ilyin, Y.V.; Nabirochkina, E.N.; Georgiev, P.G.; Georgieva, S.G. A novel multidomain transcription coactivator SAYP can also repress transcription in heterochromatin. EMBO J. 2005, 24, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Mazina, M.Y.; Kocheryzhkina, E.V.; Derevyanko, P.K.; Vorobyeva, N.E. The Composition of Swi/Snf Chromatin Remodeling Complex Is Stable during Gene Transcription. Tsitologiia 2016, 58, 285–289. [Google Scholar] [PubMed]
- Giordano, E. Immunofluorescence on polytene chromosomes of Drosophila larvae imginal discs. Protoc. Io 2020. [Google Scholar] [CrossRef]
- Comet, I.; Schuettengruber, B.; Sexton, T.; Cavalli, G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc. Natl. Acad. Sci. USA 2011, 108, 2294–2299. [Google Scholar] [CrossRef]
- Aoki, T.; Wolle, D.; Preger-Ben Noon, E.; Dai, Q.; Lai, E.C.; Schedl, P. Bi-functional cross-linking reagents efficiently capture protein-DNA complexes in Drosophila embryos. Fly Austin 2014, 8, 43–51. [Google Scholar] [CrossRef][Green Version]
- Simon, J.M.; Giresi, P.G.; Davis, I.J.; Lieb, J.D. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 2012, 7, 256–267. [Google Scholar] [CrossRef]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef]
- Golov, A.K.; Ulianov, S.V.; Luzhin, A.V.; Kalabusheva, E.P.; Kantidze, O.L.; Flyamer, I.M.; Razin, S.V.; Gavrilov, A.A. C-TALE, a new cost-effective method for targeted enrichment of Hi-C/3C-seq libraries. Methods 2020, 170, 48–60. [Google Scholar] [CrossRef]
- Nagano, T.; Lubling, Y.; Yaffe, E.; Wingett, S.W.; Dean, W.; Tanay, A.; Fraser, P. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat. Protoc. 2015, 10, 1986–2003. [Google Scholar] [CrossRef]
- Nagano, T.; Varnai, C.; Schoenfelder, S.; Javierre, B.M.; Wingett, S.W.; Fraser, P. Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome Biol. 2015, 16, 175. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shidlovskii, Y.V.; Bylino, O.V.; Shaposhnikov, A.V.; Kachaev, Z.M.; Lebedeva, L.A.; Kolesnik, V.V.; Amendola, D.; De Simone, G.; Formicola, N.; Schedl, P.; et al. Subunits of the PBAP Chromatin Remodeler Are Capable of Mediating Enhancer-Driven Transcription in Drosophila. Int. J. Mol. Sci. 2021, 22, 2856. https://doi.org/10.3390/ijms22062856
Shidlovskii YV, Bylino OV, Shaposhnikov AV, Kachaev ZM, Lebedeva LA, Kolesnik VV, Amendola D, De Simone G, Formicola N, Schedl P, et al. Subunits of the PBAP Chromatin Remodeler Are Capable of Mediating Enhancer-Driven Transcription in Drosophila. International Journal of Molecular Sciences. 2021; 22(6):2856. https://doi.org/10.3390/ijms22062856
Chicago/Turabian StyleShidlovskii, Yulii V., Oleg V. Bylino, Alexander V. Shaposhnikov, Zaur M. Kachaev, Lyubov A. Lebedeva, Valeria V. Kolesnik, Diego Amendola, Giovanna De Simone, Nadia Formicola, Paul Schedl, and et al. 2021. "Subunits of the PBAP Chromatin Remodeler Are Capable of Mediating Enhancer-Driven Transcription in Drosophila" International Journal of Molecular Sciences 22, no. 6: 2856. https://doi.org/10.3390/ijms22062856
APA StyleShidlovskii, Y. V., Bylino, O. V., Shaposhnikov, A. V., Kachaev, Z. M., Lebedeva, L. A., Kolesnik, V. V., Amendola, D., De Simone, G., Formicola, N., Schedl, P., Digilio, F. A., & Giordano, E. (2021). Subunits of the PBAP Chromatin Remodeler Are Capable of Mediating Enhancer-Driven Transcription in Drosophila. International Journal of Molecular Sciences, 22(6), 2856. https://doi.org/10.3390/ijms22062856





