Electron-Induced Decomposition of Uracil-5-yl O-(N,N-dimethylsulfamate): Role of Methylation in Molecular Stability
Abstract
1. Introduction
2. Results
3. Methods
3.1. CEMB Experiments
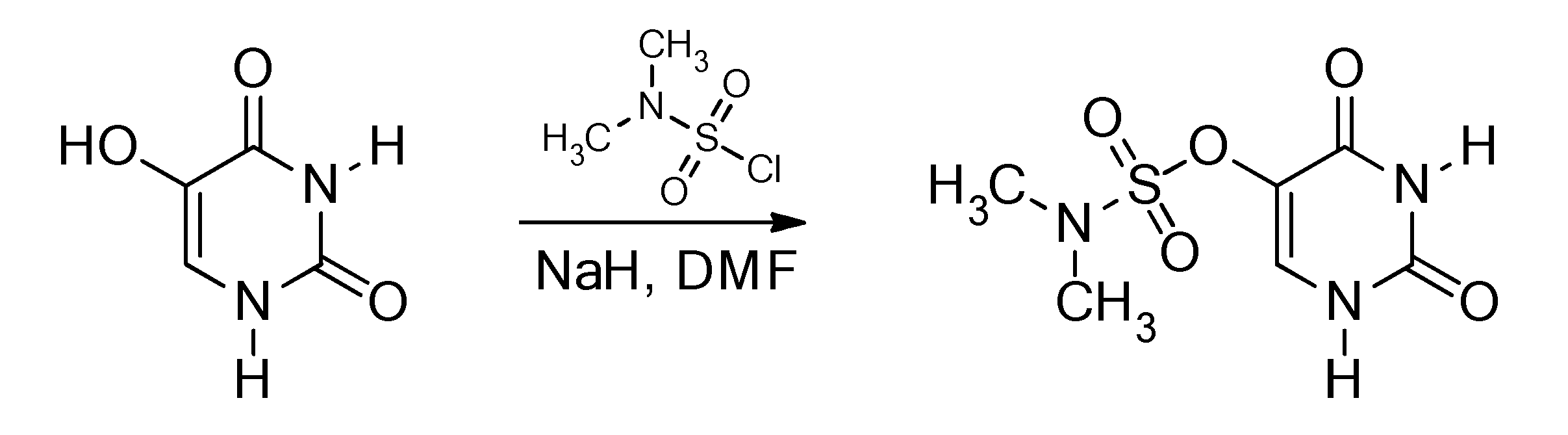
3.2. Synthesis
3.3. Computational Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Ruysscher, D.; Niedermann, G.; Burnet, N.G.; Siva, S.; Lee, A.W.M.; Hegi-Johnson, F. Radiotherapy toxicity. Nat. Rev. Dis. Prim. 2019, 5, 1–20. [Google Scholar] [CrossRef]
- Rami, M.; Dubois, L.; Parvathaneni, N.K.; Alterio, V.; Van Kuijk, S.J.A.; Monti, S.M.; Lambin, P.; De Simone, G.; Supuran, C.T.; Winum, J.Y. Hypoxia-targeting carbonic anhydrase IX inhibitors by a new series of nitroimidazole-sulfonamides/sulfamides/sulfamates. J. Med. Chem. 2013, 56, 8512–8520. [Google Scholar] [CrossRef]
- Wang, H.; Mu, X.; He, H.; Zhang, X.D. Cancer Radiosensitizers. Trends Pharmacol. Sci. 2018, 39, 24–48. [Google Scholar] [CrossRef] [PubMed]
- Pandeti, S.; Feketeová, L.; Reddy, T.J.; Abdoul-Carime, H.; Farizon, B.; Farizon, M.; Märk, T.D. Nitroimidazolic radiosensitizers investigated by electrospray ionization time-of-flight mass spectrometry and density functional theory. RSC Adv. 2017, 7, 45211–45221. [Google Scholar] [CrossRef]
- Overgaard, J.; Hansen, H.S.; Overgaard, M.; Bastholt, L.; Berthelsen, A.; Specht, L.; Lindeløv, B.; Jørgensen, K. A randomized double-blind phase III study of nimorazole as a hypoxic radiosensitizer of primary radiotherapy in supraglottic larynx and pharynx carcinoma. Results of the Danish Head and Neck Cancer Study (DAHANCA) Protocol 5-85. Radiother. Oncol. 1998, 46, 135–146. [Google Scholar] [CrossRef]
- Shen, X.; Gates, K.S. Enzyme-activated generation of reactive oxygen species from heterocyclic N-oxides under aerobic and anaerobic conditions and its relevance to hypoxia-selective prodrugs. Chem. Res. Toxicol. 2019, 32, 348–361. [Google Scholar] [CrossRef]
- Arthur-Baidoo, E.; Ameixa, J.; Ziegler, P.; Ferreira da Silva, F.; Ončák, M.; Denifl, S. Reactions in Tirapazamine Induced by the Attachment of Low-Energy Electrons: Dissociation Versus Roaming of OH. Angew. Chem. Int. Ed. 2020, 59, 17177–17181. [Google Scholar] [CrossRef]
- Kopyra, J.; Keller, A.; Bald, I. On the role of fluoro-substituted nucleosides in DNA radiosensitization for tumor radiation therapy. RSC Adv. 2014, 4, 6825–6829. [Google Scholar] [CrossRef]
- Saqib, M.; Arthur-Baidoo, E.; Ončák, M.; Denifl, S. Electron attachment studies with the potential radiosensitizer 2-nitrofuran. Int. J. Mol. Sci. 2020, 21, 8906. [Google Scholar] [CrossRef] [PubMed]
- Pimblott, S.M.; LaVerne, J.A. Production of low-energy electrons by ionizing radiation. Radiat. Phys. Chem. 2007, 76, 1244–1247. [Google Scholar] [CrossRef]
- Sulzer, P.; Ptasinska, S.; Zappa, F.; Mielewska, B.; Milosavljevic, A.R.; Scheier, P.; Märk, T.D.; Bald, I.; Gohlke, S.; Huels, M.A.; et al. Dissociative electron attachment to furan, tetrahydrofuran, and fructose. J. Chem. Phys. 2006, 125, 044304. [Google Scholar] [CrossRef] [PubMed]
- Ptasińska, S.; Alizadeh, E.; Sulzer, P.; Abouaf, R.; Mason, N.J.; Märk, T.D.; Scheier, P. Negative ion formation by low energy electron attachment to gas-phase 5-nitrouracil. Int. J. Mass Spectrom. 2008, 277, 291–295. [Google Scholar] [CrossRef]
- Sanche, L. Role of secondary low energy electrons in radiobiology and chemoradiation therapy of cancer. Chem. Phys. Lett. 2009, 474, 1–6. [Google Scholar] [CrossRef]
- Ptasinska, S.; Denifl, S.; Scheier, P.; Illenberger, E.; Märk, T.D. Bond-and Site-Selective Loss of H Atoms from Nucleobases by Very-Low-Energy Electrons (<3 eV). Angew. Chem. Int. Ed. 2005, 44, 6941–6943. [Google Scholar] [CrossRef] [PubMed]
- Abdoul-Carime, H.; Huels, M.A.; Brüning, F.; Illenberger, E.; Sanche, L. Dissociative electron attachment to gas-phase 5-bromouracil. J. Chem. Phys. 2000, 113, 2517–2521. [Google Scholar] [CrossRef]
- Denifl, S.; Matejcik, S.; Gstir, B.; Hanel, G.; Probst, M.; Scheier, P.; Märk, T.D. Electron attachment to 5-chloro uracil. J. Chem. Phys. 2003, 118, 4107–4114. [Google Scholar] [CrossRef]
- Denifl, S.; Matejcik, S.; Ptasinska, S.; Gstir, B.; Probst, M.; Scheier, P.; Illenberger, E.; Mark, T.D. Electron attachment to chlorouracil: A comparison between 6-ClU and 5-ClU. J. Chem. Phys. 2004, 120, 704–709. [Google Scholar] [CrossRef]
- Abdoul-Carime, H.; Huels, M.A.; Illenberger, E.; Sanche, L. Formation of negative ions from gas phase halo-uracils by low-energy (0–18 eV) electron impact. Int. J. Mass Spectrom. 2003, 228, 703–716. [Google Scholar] [CrossRef]
- Kossoski, F.; Bettega, M.H.F.; Varella, M.D.N. Shape resonance spectra of uracil, 5-fluorouracil, and 5-chlorouracil. J. Chem. Phys. 2014, 140, 24317. [Google Scholar] [CrossRef] [PubMed]
- Chomicz, L.; Rak, J.; Storoniak, P. Electron-induced elimination of the bromide anion from brominated nucleobases. A computational study. J. Phys. Chem. B 2012, 116, 5612–5619. [Google Scholar] [CrossRef]
- Ameixa, J.; Arthur-Baidoo, E.; Meißner, R.; Makurat, S.; Kozak, W.; Butowska, K.; da Silva, F.; Rak, J.; Denifl, S. Low-energy electron-induced decomposition of 5-trifluoromethanesulfonyl-uracil: A potential radiosensitizer. J. Chem. Phys. 2018, 149, 164307. [Google Scholar] [CrossRef] [PubMed]
- Spisz, P.; Zdrowowicz, M.; Kozak, W.; Chomicz-Mańka, L.; Falkiewicz, K.; Makurat, S.; Sikorski, A.; Wyrzykowski, D.; Rak, J.; Arthur-Baidoo, E.; et al. Uracil-5-yl O-Sulfamate: An Illusive Radiosensitizer. Pitfalls in Modeling the Radiosensitizing Derivatives of Nucleobases. J. Phys. Chem. B 2020, 124, 5600–5613. [Google Scholar] [CrossRef] [PubMed]
- Meißner, R.; Makurat, S.; Kozak, W.; Limão-Vieira, P.; Rak, J.; Denifl, S. Electron-Induced Dissociation of the Potential Radiosensitizer 5-Selenocyanato-2′-deoxyuridine. J. Phys. Chem. B 2019, 123, 1274–1282. [Google Scholar] [CrossRef] [PubMed]
- Spisz, P.; Kozak, W.; Chomicz-Mańka, L.; Makurat, S.; Falkiewicz, K.; Sikorski, A.; Czaja, A.; Rak, J.; Zdrowowicz, M. 5-(N-Trifluoromethylcarboxy) aminouracil as a Potential DNA Radiosensitizer and Its Radiochemical Conversion into N-Uracil-5-yloxamic Acid. Int. J. Mol. Sci. 2020, 21, 6352. [Google Scholar] [CrossRef]
- Chomicz, L.; Zdrowowicz, M.; Kasprzykowski, F.; Rak, J.; Buonaugurio, A.; Wang, Y.; Bowen, K.H. How to find out whether a 5-substituted uracil could be a potential DNA radiosensitizer. J. Phys. Chem. Lett. 2013, 4, 2853–2857. [Google Scholar] [CrossRef]
- Pogozelski, W.K.; Tullius, T.D. Oxidative strand scission of nucleic acids: Routes initiated by hydrogen abstraction from the sugar moiety. Chem. Rev. 1998, 98, 1089–1107. [Google Scholar] [CrossRef] [PubMed]
- Curtiss, L.A.; Redfern, P.C.; Raghavachari, K.; Rassolov, V.; Pople, J.A. Gaussian-3 theory using reduced Møller-Plesset order. J. Chem. Phys. 1999, 110, 4703–4709. [Google Scholar] [CrossRef]
- Denifl, S.; Ptasińska, S.; Hanel, G.; Gstir, B.; Probst, M.; Scheier, P.; Märk, T.D. Electron attachment to gas-phase uracil. J. Chem. Phys. 2004, 120, 6557–6565. [Google Scholar] [CrossRef]
- Rienstra-Kiracofe, J.C.; Tschumper, G.S.; Schaefer, H.F.; Nandi, S.; Ellison, G.B. Atomic and molecular electron affinities: Photoelectron experiments and theoretical computations. Chem. Rev. 2002, 102, 231–282. [Google Scholar] [CrossRef]
- Limão-Vieira, P.; da Silva, F.F.; Gómez-Tejedor, G.G. Electron transfer-induced fragmentation in (bio) molecules by atom-molecule collisions. In Radiation Damage in Biomolecular Systems; Springer: Dordrecht, The Netherlands, 2012; pp. 59–70. [Google Scholar]
- Manura, J.J.; Manura, D.J. Isotope Distribution Calculator and Mass Spec Plotter. Sci. Instrum. Serv. 2009, 1996–2009. Available online: http://www.sisWeb.com/mstools/isotope.htm (accessed on 21 February 2021).
- Ameixa, J.; Arthur-Baidoo, E.; da Silva, J.; Ryszka, M.; Carmichael, I.; Cornetta, L.M.; Varella, M.T.; da Silva, F.; Ptasinska, S.; Denifl, S. Formation of resonances and anionic fragments upon electron attachment to benzaldehyde. Phys. Chem. Chem. Phys. 2020, 22, 8171–8181. [Google Scholar] [CrossRef] [PubMed]
- Ribar, A.; Huber, S.E.; Śmiałek, M.A.; Tanzer, K.; Neustetter, M.; Schürmann, R.; Bald, I.; Denifl, S. Hydroperoxyl radical and formic acid formation from common DNA stabilizers upon low energy electron attachment. Phys. Chem. Chem. Phys. 2018, 20, 5578–5585. [Google Scholar] [CrossRef]
- Alizadeh, E.; Gschliesser, D.; Bartl, P.; Hager, M.; Edtbauer, A.; Vizcaino, V.; Mauracher, A.; Probst, M.; Märk, T.D.; Ptasińska, S.; et al. Bond dissociation of the dipeptide dialanine and its derivative alanine anhydride induced by low energy electrons. J. Chem. Phys. 2011, 134, 54305. [Google Scholar] [CrossRef]
- Huber, S.E.; Śmiałek, M.A.; Tanzer, K.; Denifl, S. Dissociative electron attachment to the radiosensitizing chemotherapeutic agent hydroxyurea. J. Chem. Phys. 2016, 144, 224309. [Google Scholar] [CrossRef]
- Mauracher, A.; Denifl, S.; Aleem, A.; Wendt, N.; Zappa, F.; Cicman, P.; Probst, M.; Märk, T.D.; Scheier, P.; Flosadóttir, H.D.; et al. Dissociative electron attachment to gas phase glycine: Exploring the decomposition pathways by mass separation of isobaric fragment anions. Phys. Chem. Chem. Phys. 2007, 9, 5680. [Google Scholar] [CrossRef] [PubMed]
- Meißner, R.; Kočišek, J.; Feketeová, L.; Fedor, J.; Fárník, M.; Limão-Vieira, P.; Illenberger, E.; Denifl, S. Low-energy electrons transform the nimorazole molecule into a radiosensitiser. Nat. Commun. 2019, 10, 2388. [Google Scholar] [CrossRef]
- Woo, L.W.L.; Howarth, N.M.; Purohit, A.; Hejaz, H.A.M.; Reed, M.J.; Potter, B.V.L. Steroidal and nonsteroidal sulfamates as potent inhibitors of steroid sulfatase. J. Med. Chem. 1998, 41, 1068–1083. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other function. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Kendall, R.A.; Dunning, T.H., Jr.; Harrison, R.J. Electron affinities of the first-row atoms revisited. Systematic basis sets and wave functions. J. Chem. Phys. 1992, 96, 6796–6806. [Google Scholar] [CrossRef]
- Ochterski, J.W. Thermochemistry in Gaussian; Gaussian Inc.: Wallingford, CT, USA, 2000; Volume 1, pp. 1–19. [Google Scholar]
- Simons, J. Resonances in Electron-Molecule Scattering, Van der Waals Complexes, and Reactive Chemical Dynamics; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1984; pp. 3–16. [Google Scholar]
- Simons, J. How Do Low-Energy (0.1–2 eV) Electrons Cause DNA-Strand Breaks? Acc. Chem. Res. 2006, 39, 772–779. [Google Scholar] [CrossRef]
- Berdys, J.; Anusiewicz, I.; Skurski, P.; Simons, J. Damage to model DNA fragments from very low-energy (<1 eV) electrons. J. Am. Chem. Soc. 2004, 126, 6441–6447. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision A. 02; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016; p. 29. [Google Scholar]
- Tanzer, K.; Feketeová, L.; Puschnigg, B.; Scheier, P.; Illenberger, E.; Denifl, S. Reactions in Nitroimidazole Triggered by Low-Energy (0-2 eV) Electrons: Methylation at N1-H Completely Blocks Reactivity. Angew. Chemie Int. Ed. 2014, 53, 12240–12243. [Google Scholar] [CrossRef]
- Tanzer, K.; Feketeová, L.; Puschnigg, B.; Scheier, P.; Illenberger, E.; Denifl, S. Reactions in Nitroimidazole and Methylnitroimidazole Triggered by Low-Energy (0-8 eV) Electrons. J. Phys. Chem. A 2015, 119, 6668–6675. [Google Scholar] [CrossRef]
- Kossoski, F.; Varella, M.T.D.N. How does methylation suppress the electron-induced decomposition of 1-methyl-nitroimidazoles? J. Chem. Phys. 2017, 147, 164310. [Google Scholar] [CrossRef]

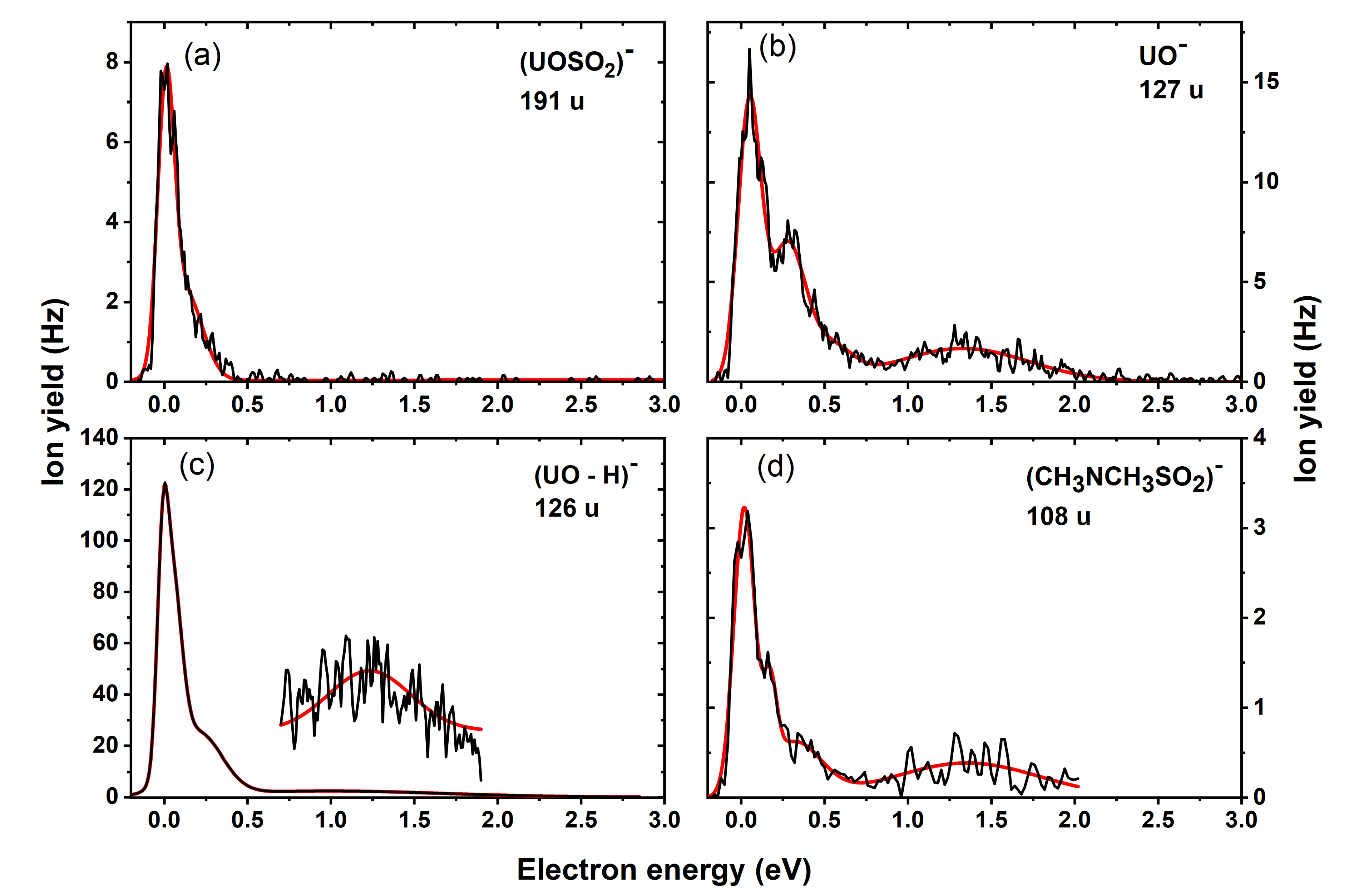
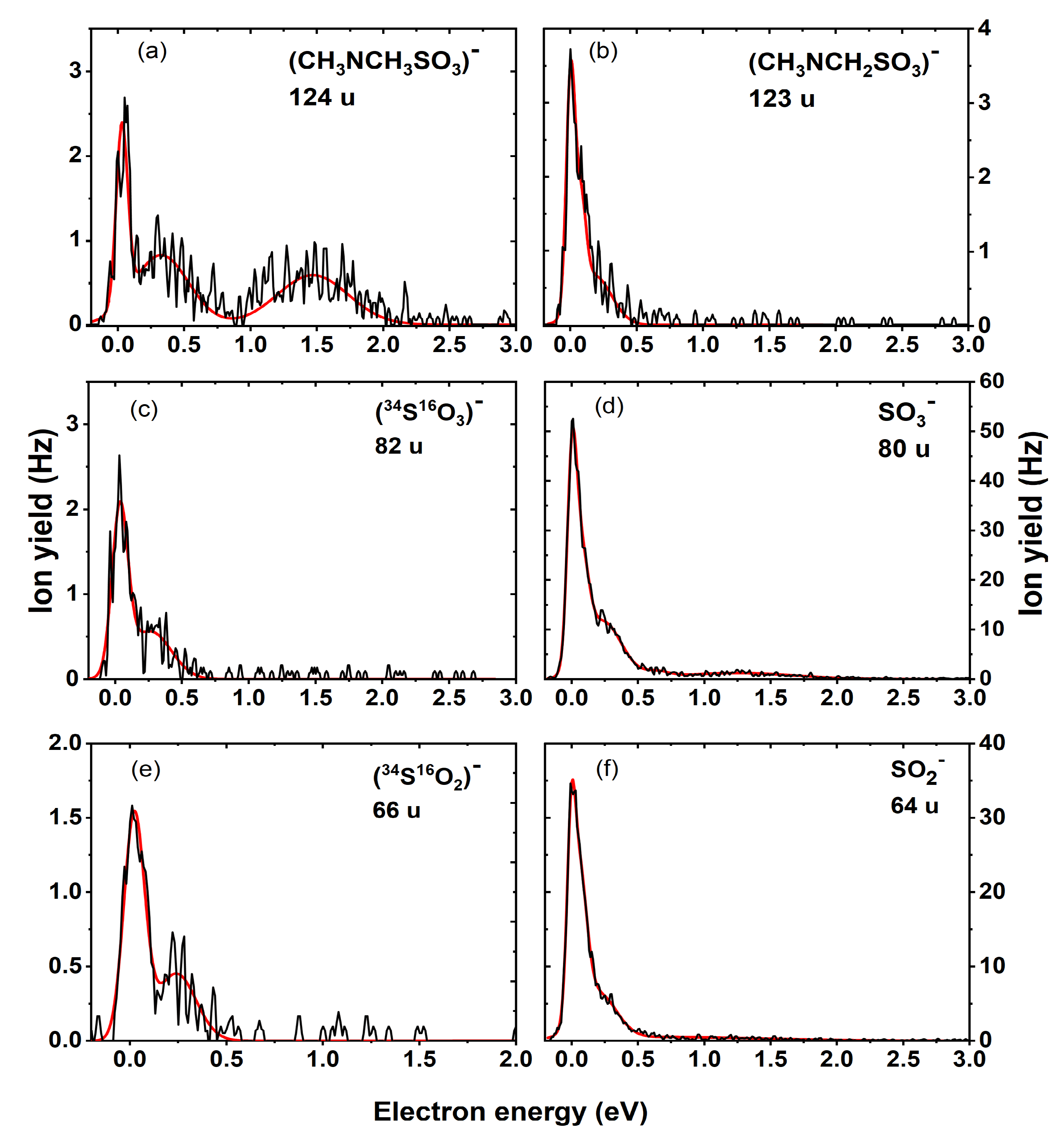
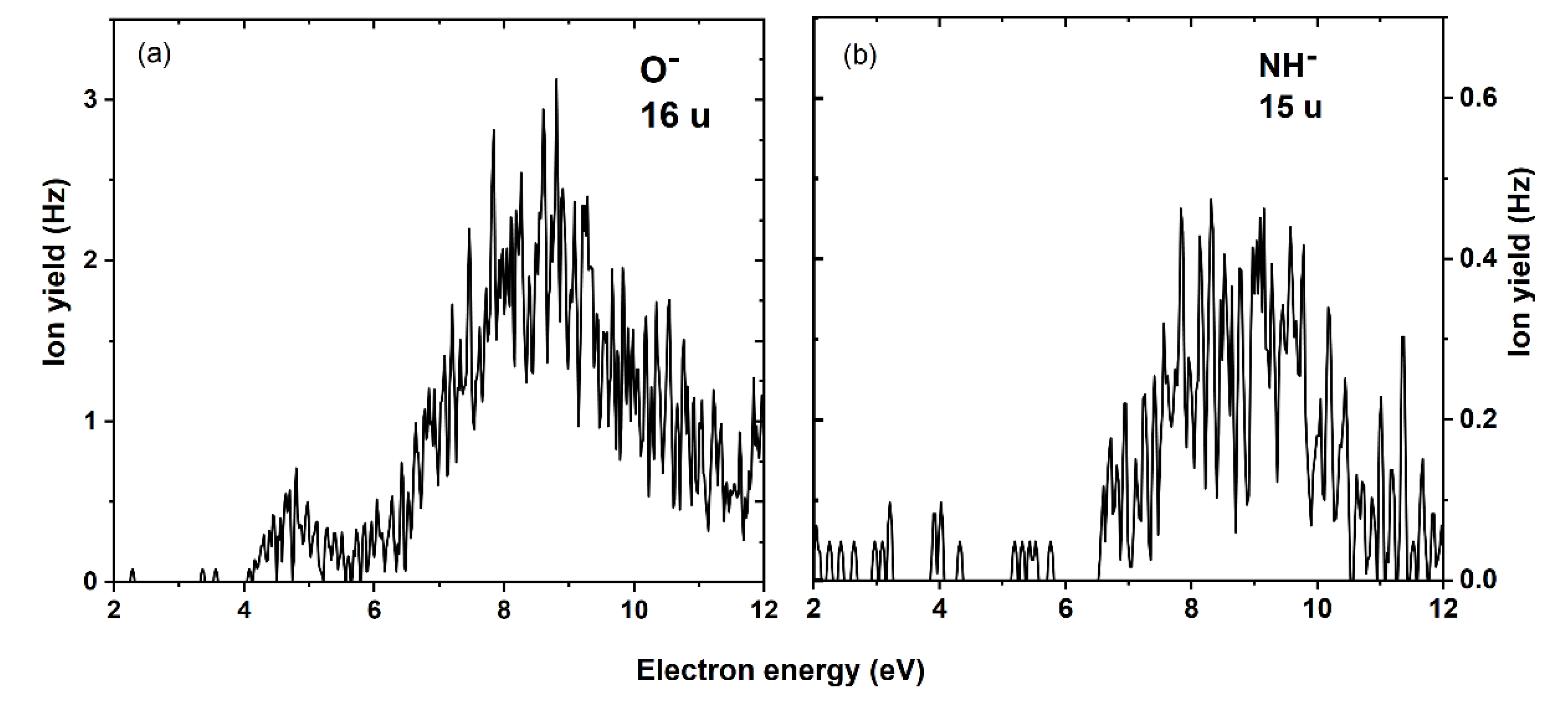

| Mass (u) | Anion | Maxima of Peak Positions (eV) | Thresholds (eV) 5.33 × 10−11 atm | AEAE (eV) | |||||
| 1. | 2. | 3. | 4. | Exp. | Calc. | Calc. | |||
| 386.15 K | 298.15 K | 386.15 K | |||||||
| 191 | UOSO2− | 0 | 0.1 | ~0 | −1.41 | −1.78 | 4.11 | ||
| 127 | UO− | 0 | 0.3 | 1.3 | ~0 | −0.60 | −0.95 | 2.40 | |
| 126 | (UO-H)− | 0 | 0.2 | 1.2 | ~0 | −1.07 a | −1.41 a | 4.84a | |
| 124 | (CH3)2NSO3− | 0 | 0.3 | 1.4 | ~0 | −0.30 | −0.64 | 4.66 | |
| 123 | ((CH3)2NSO3-H)− | 0 | 0.1 | ~0 | −1.45 | −1.79 | 4.88 | ||
| 108 | (CH3)2NSO2− | 0 | 0.2 | 0.3 | 1.4 | ~0 | −0.97 | −1.32 | 2.75 |
| 82 | H2SO3− | - * | - * | −0.48 | −1.18 | −0.45 (0.01 b) | |||
| 80 | SO3− | 0 | 0.3 | 1.3 | ~0 | −1.75 | −2.09 | 5.09 | |
| 66 | H2SO2− | - * | - * | 0.14 | −0.55 | −0.59 (0.08 b) | |||
| 64 | SO2− | 0 | 0.3 | ~0 | −0.31 | −0.66 | 1.43 | ||
| 16 | O− | 4.7 | 8.8 | ~4 | 2.61 c | 2.32 c | 1.41 | ||
| 15 | CH3− | 8.5 | ~6 | 3.46 | 3.12 | 0.001 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arthur-Baidoo, E.; Falkiewicz, K.; Chomicz-Mańka, L.; Czaja, A.; Demkowicz, S.; Biernacki, K.; Kozak, W.; Rak, J.; Denifl, S. Electron-Induced Decomposition of Uracil-5-yl O-(N,N-dimethylsulfamate): Role of Methylation in Molecular Stability. Int. J. Mol. Sci. 2021, 22, 2344. https://doi.org/10.3390/ijms22052344
Arthur-Baidoo E, Falkiewicz K, Chomicz-Mańka L, Czaja A, Demkowicz S, Biernacki K, Kozak W, Rak J, Denifl S. Electron-Induced Decomposition of Uracil-5-yl O-(N,N-dimethylsulfamate): Role of Methylation in Molecular Stability. International Journal of Molecular Sciences. 2021; 22(5):2344. https://doi.org/10.3390/ijms22052344
Chicago/Turabian StyleArthur-Baidoo, Eugene, Karina Falkiewicz, Lidia Chomicz-Mańka, Anna Czaja, Sebastian Demkowicz, Karol Biernacki, Witold Kozak, Janusz Rak, and Stephan Denifl. 2021. "Electron-Induced Decomposition of Uracil-5-yl O-(N,N-dimethylsulfamate): Role of Methylation in Molecular Stability" International Journal of Molecular Sciences 22, no. 5: 2344. https://doi.org/10.3390/ijms22052344
APA StyleArthur-Baidoo, E., Falkiewicz, K., Chomicz-Mańka, L., Czaja, A., Demkowicz, S., Biernacki, K., Kozak, W., Rak, J., & Denifl, S. (2021). Electron-Induced Decomposition of Uracil-5-yl O-(N,N-dimethylsulfamate): Role of Methylation in Molecular Stability. International Journal of Molecular Sciences, 22(5), 2344. https://doi.org/10.3390/ijms22052344









