Putative Circulating MicroRNAs Are Able to Identify Patients with Mitral Valve Prolapse and Severe Regurgitation
Abstract
1. Introduction
2. Results
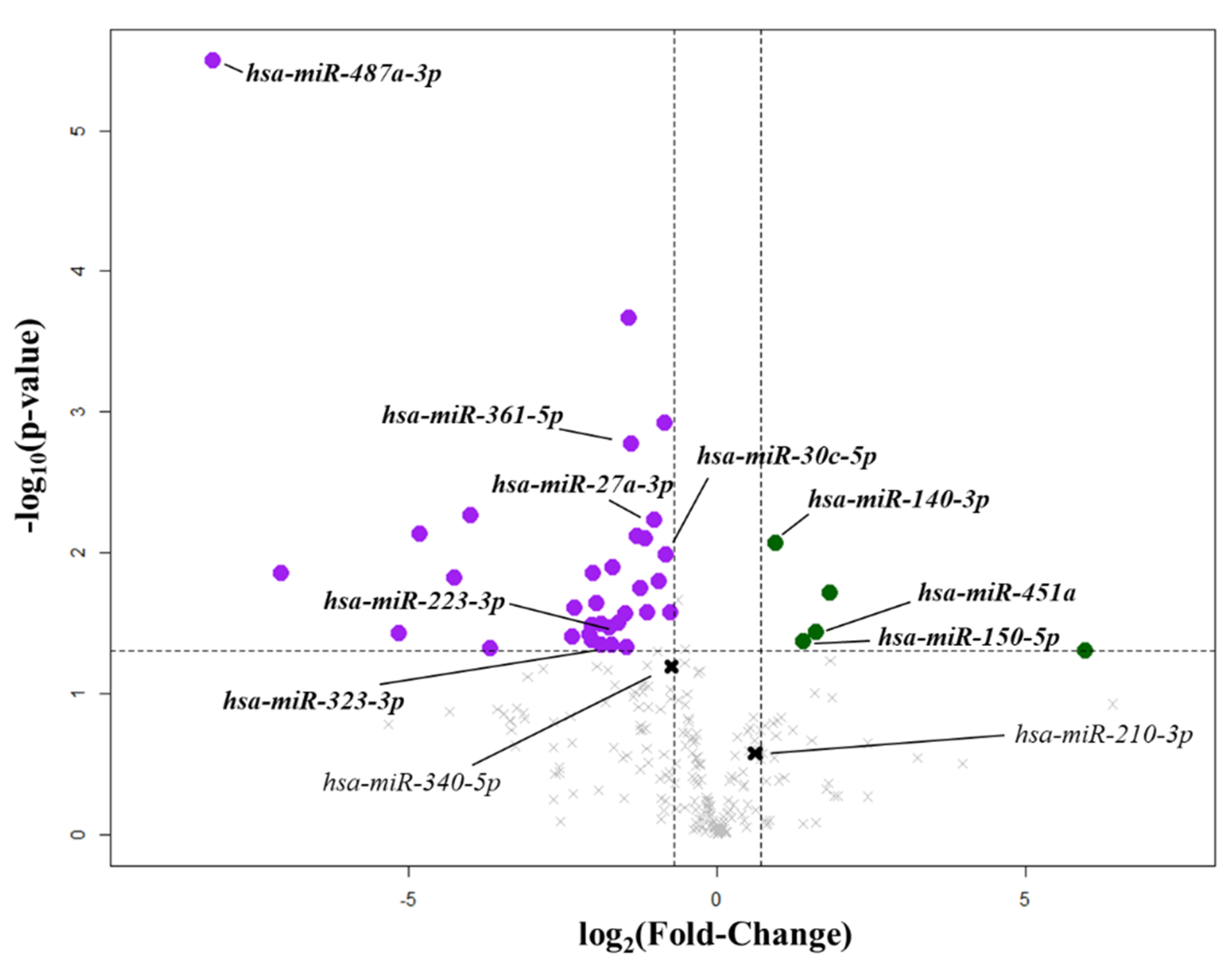
2.1. Circulating miRNA Linked to Mitral Valve Prolapse
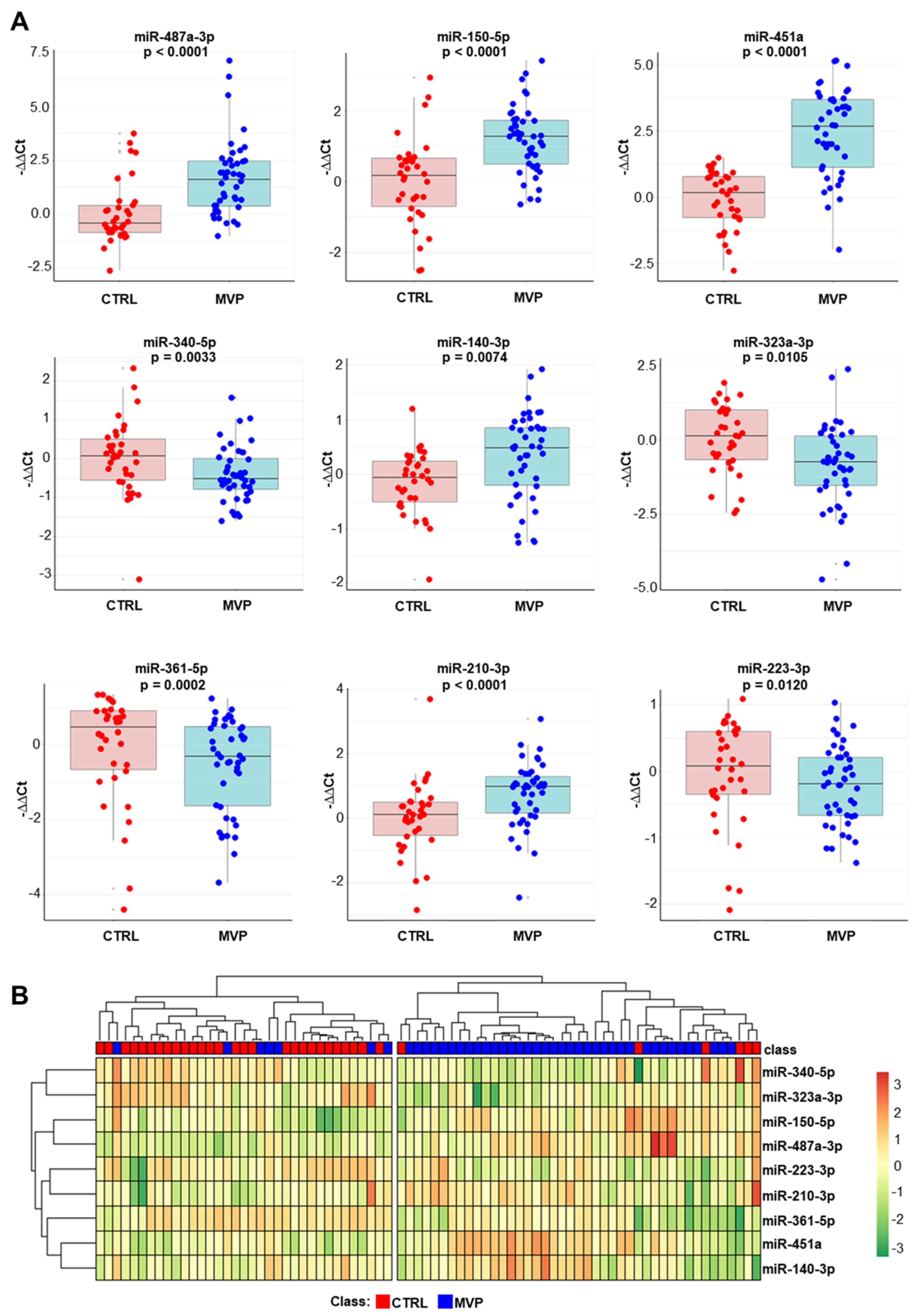
2.2. Validation Phase
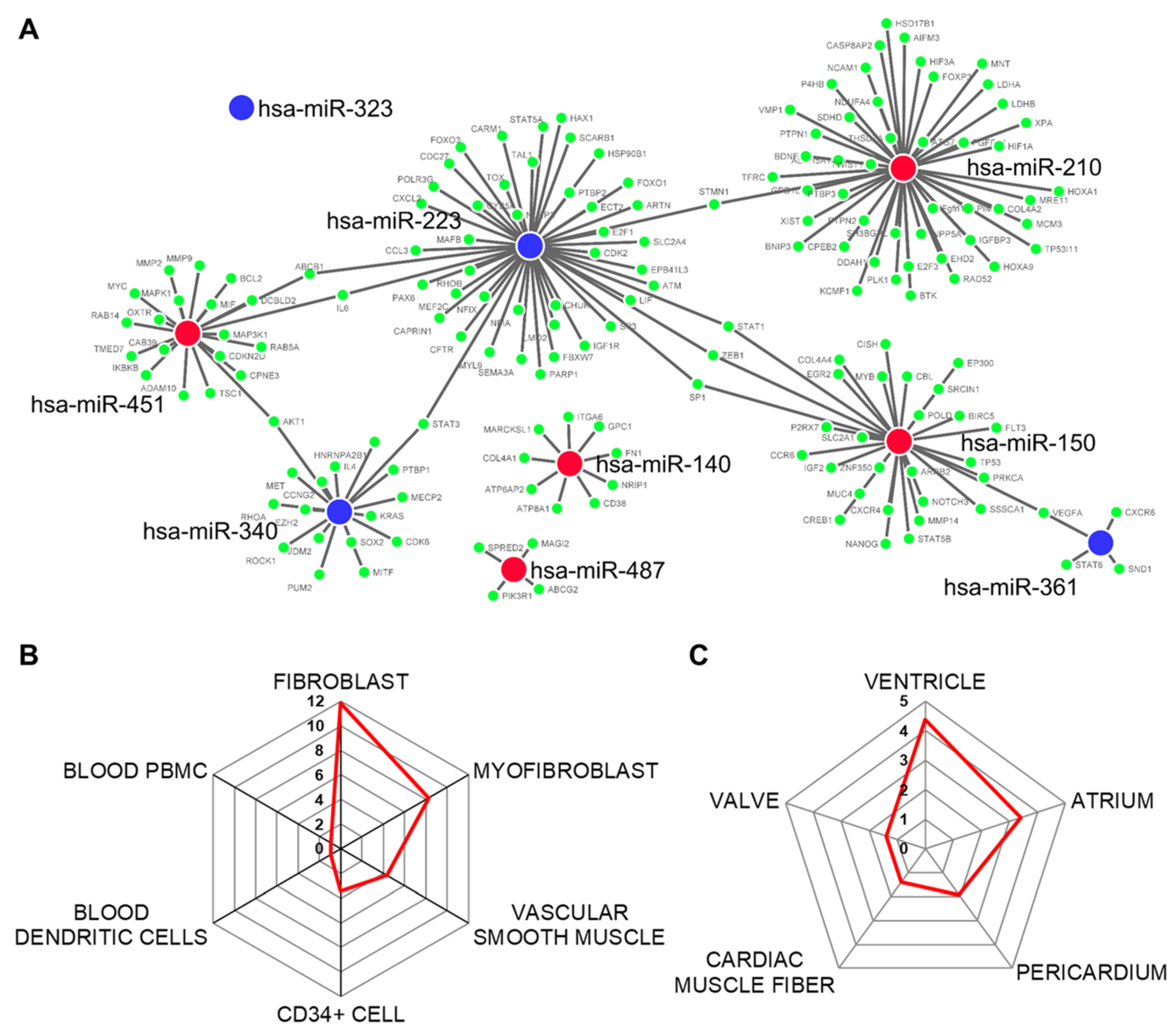
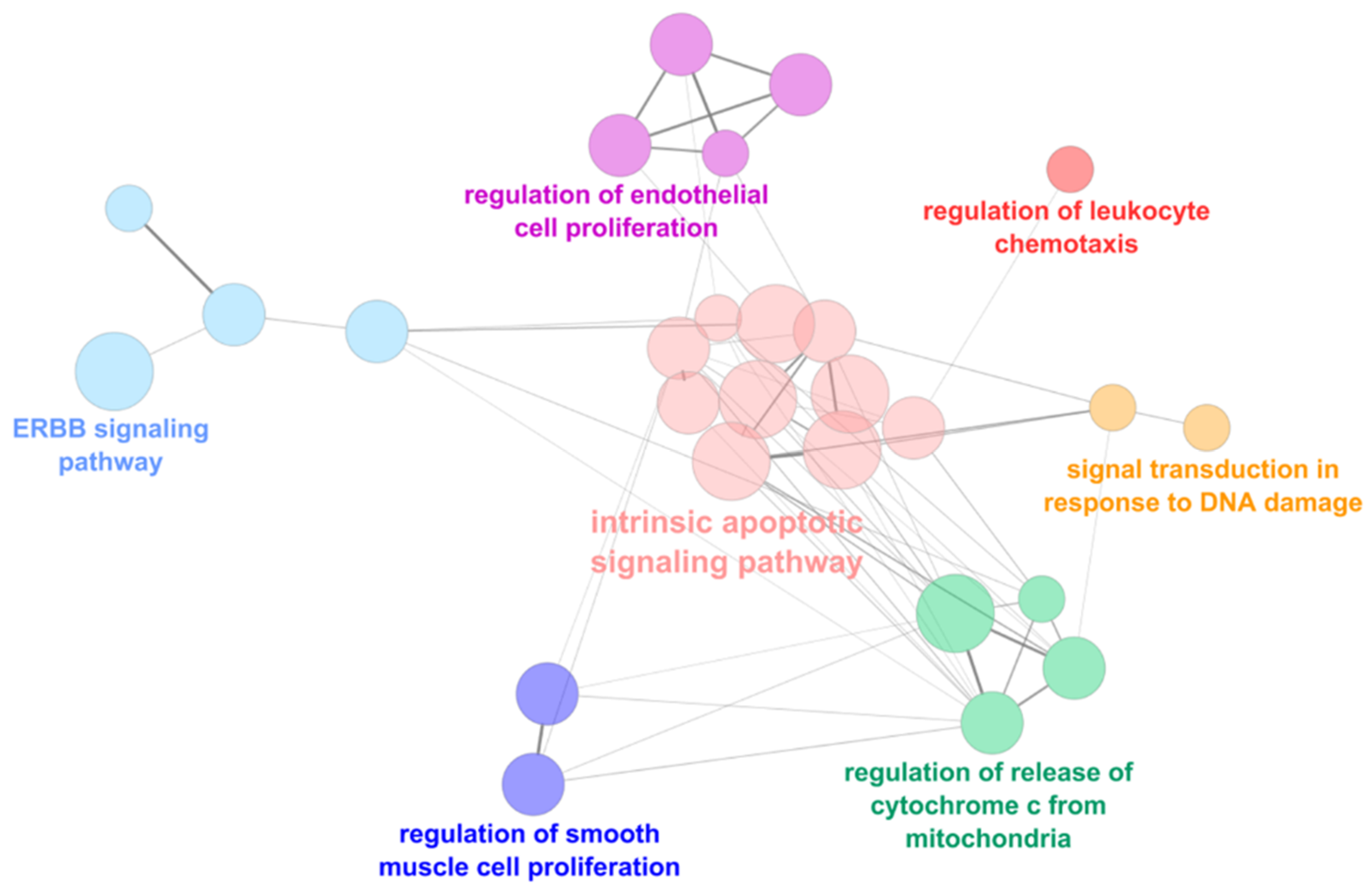
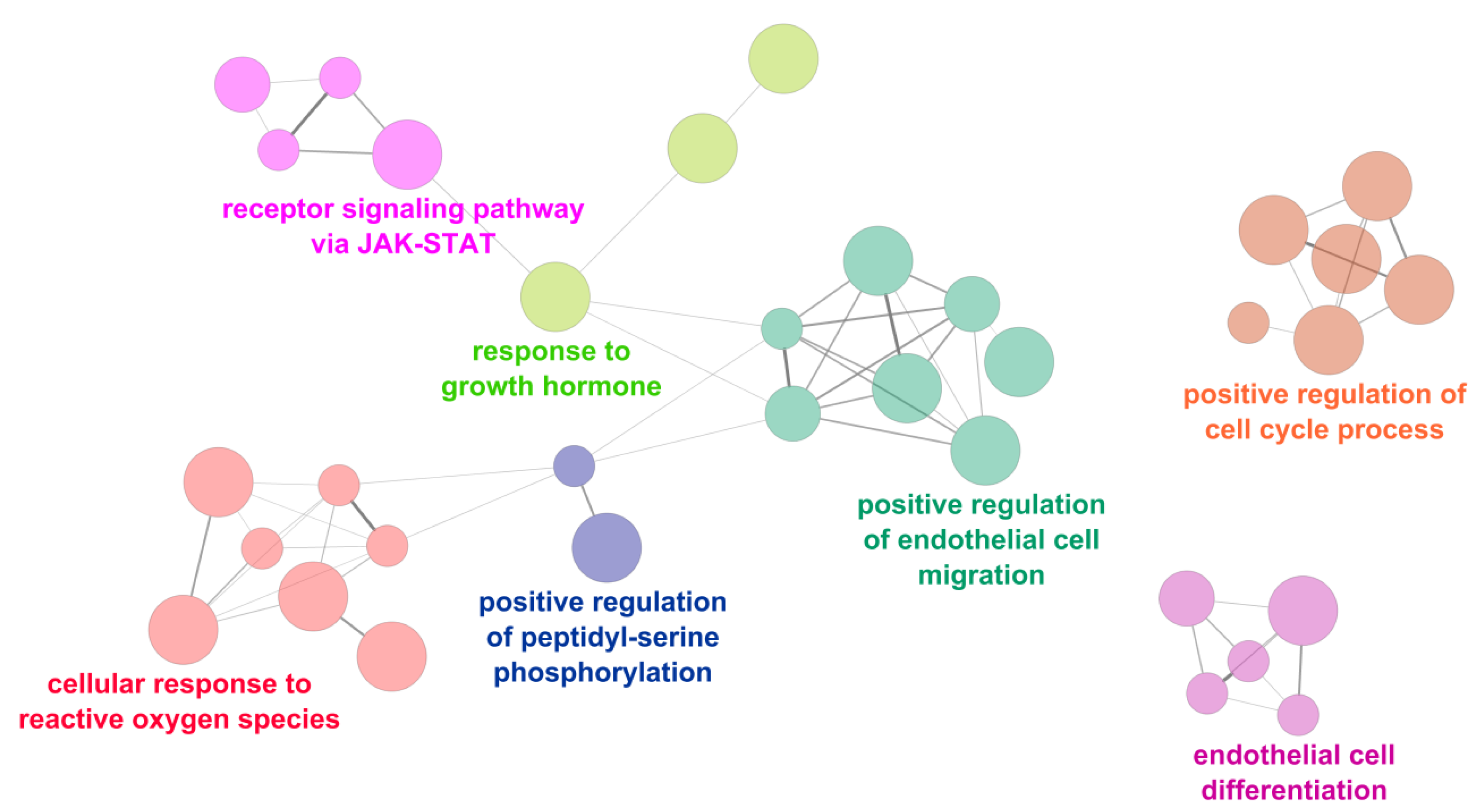
2.3. Functional Analysis
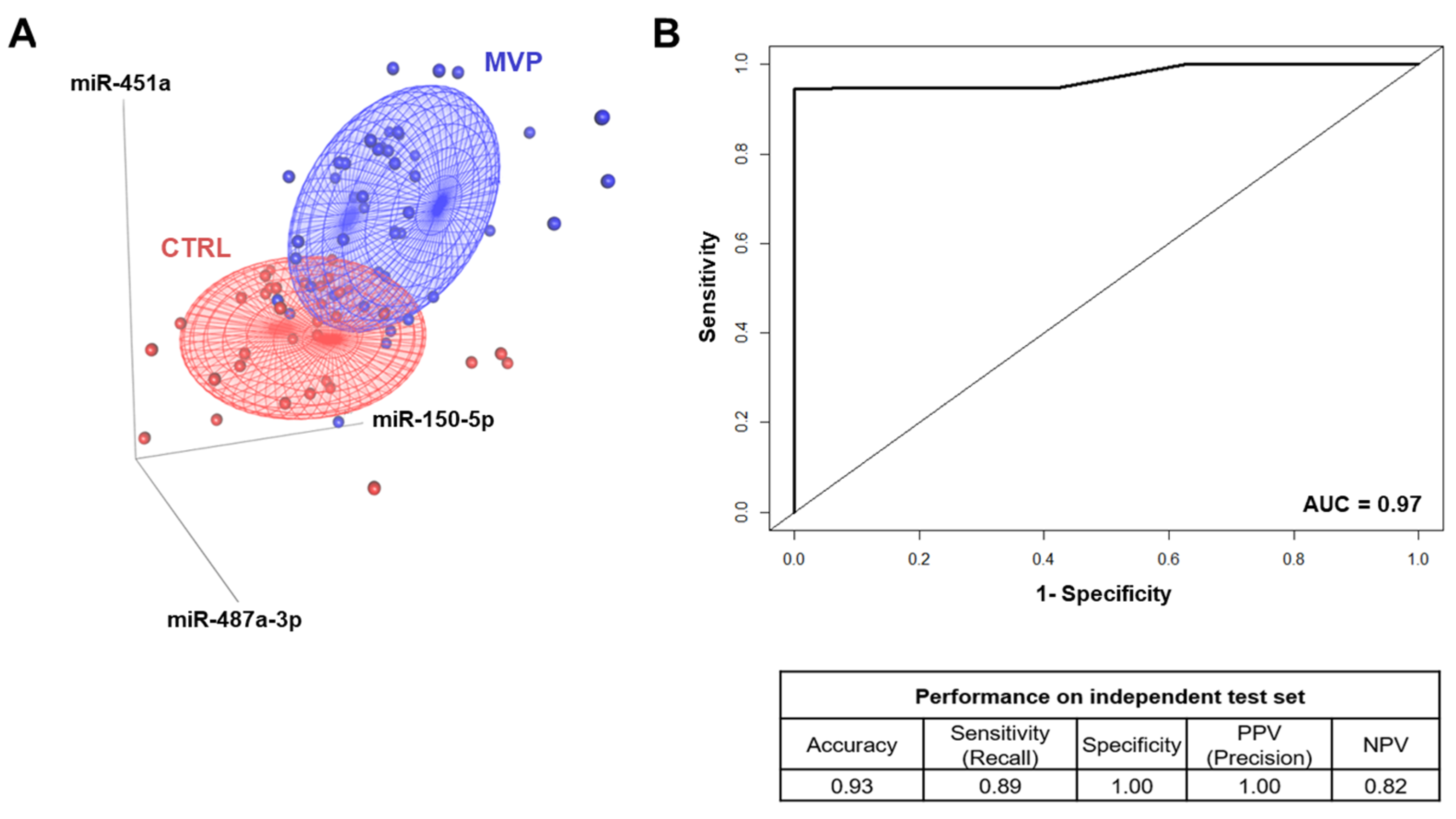
2.4. Mitral Valve Prolapse Circulating miRNA Signature Strength
3. Discussion
4. Materials and Methods
4.1. Patient Population
4.2. Blood Sampling
4.3. TaqMan Human miRNA Card A Arrays
4.4. Reverse Transcription and Real-Time PCR
4.5. MiRNA–mRNA Target Prediction
4.6. Functional and Cell Type Enrichment Analyses
4.7. Machine Learning Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Delling, F.N.; Vasan, R.S. Epidemiology and Pathophysiology of Mitral Valve Prolapse. Circulation 2014, 129, 2158–2170. [Google Scholar] [CrossRef]
- Freed, L.A.; Levy, D.; Levine, R.A.; Larson, M.G.; Evans, J.C.; Fuller, D.L.; Lehman, B.; Benjamin, E.J. Prevalence and Clinical Outcome of Mitral-Valve Prolapse. N. Engl. J. Med. 1999, 341, 1–7. [Google Scholar] [CrossRef]
- Olson, L.J.; Subramanian, R.; Ackermann, D.M.; Orszulak, T.A.; Edwards, W.D. Surgical Pathology of the Mitral Valve: A Study of 712 Cases Spanning 21 Years. Mayo Clin. Proc. 1987, 62, 22–34. [Google Scholar] [CrossRef]
- Anyanwu, A.C.; Adams, D.H. Etiologic Classification of Degenerative Mitral Valve Disease: Barlow’s Disease and Fibroelastic Deficiency. Semin. Thorac. Cardiovasc. Surg. 2007, 19, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Barlow, J.B.; Pocock, W.A. Billowing, floppy, prolapsed or flail mitral valves? Am. J. Cardiol. 1985, 55, 501–502. [Google Scholar] [CrossRef]
- Carpentier, A.; Chauvaud, S.; Fabiani, J.N.; Deloche, A.; Relland, J.; Lessana, A.; D’Allaines, C.; Blondeau, P.; Piwnica, A.; Dubost, C. Reconstructive surgery of mitral valve incompetence: Ten-year appraisal. J. Thorac. Cardiovasc. Surg. 1980, 79, 338–348. [Google Scholar] [CrossRef]
- Hjortnaes, J.; Keegan, J.; Bruneval, P.; Schwartz, E.; Schoen, F.J.; Carpentier, A.; Levine, R.A.; Hagège, A.; Aikawa, E. Comparative Histopathological Analysis of Mitral Valves in Barlow Disease and Fibroelastic Deficiency. Semin. Thorac. Cardiovasc. Surg. 2016, 28, 757–767. [Google Scholar] [CrossRef]
- Baumgartner, H.; Falk, V.; Bax, J.J.; De Bonis, M.; Hamm, C.; Holm, P.J.; Iung, B.; Lancellotti, P.; Lansac, E.; Muñoz, D.R.; et al. 2017 ESC/EACTS Guidelines for the management of valvular heart disease. Eur. Heart J. 2017, 38, 2739–2791. [Google Scholar] [CrossRef]
- Nishimura, R.A.; Otto, C.M.; Bonow, R.O.; Carabello, B.A.; Erwin, J.P.; Guyton, R.A.; O’Gara, P.T.; Ruiz, C.E.; Skubas, N.J.; Sorajja, P.; et al. 2014 AHA/ACC Guideline for the Management of Patients with Valvular Heart Disease. J. Am. Coll. Cardiol. 2014, 63, e57–e185. [Google Scholar] [CrossRef]
- Lang, R.M.; Adams, D.H. 3D Echocardiographic Quantification in Functional Mitral Regurgitation⁎⁎Editorials published in JACC: Cardiovascular Imaging reflect the views of the authors and do not necessarily represent the views of JACC: Cardiovascular Imaging or the American College of Cardiology. JACC Cardiovasc. Imaging 2012, 5, 346–347. [Google Scholar] [CrossRef]
- Lang, R.M.; Badano, L.P.; Tsang, W.; Adams, D.H.; Agricola, E.; Buck, T.; Faletra, F.F.; Franke, A.; Hung, J.; De Isla, L.P.; et al. EAE/ASE recommendations for image acquisition and display using three-dimensional echocardiography. Eur. Heart J. Cardiovasc. Imaging 2012, 13, 1–46. [Google Scholar] [CrossRef]
- Nishimura, R.A.; Otto, C. 2014 ACC/AHA valve guidelines: Earlier intervention for chronic mitral regurgitation. Heart 2014, 100, 905–907. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, R.A.; Otto, C.M.; Bonow, R.O.; Carabello, B.A.; Erwin, J.P.; Fleisher, L.A.; Jneid, H.; Mack, M.J.; McLeod, C.J.; O’Gara, P.T.; et al. 2017 AHA/ACC Focused Update of the 2014 AHA/ACC Guideline for the Management of Patients with Valvular Heart Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation 2017, 135, e1159–e1195. [Google Scholar] [CrossRef] [PubMed]
- Mayeux, R. Biomarkers: Potential uses and limitations. NeuroRX 2004, 1, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Songia, P.; Branchetti, E.; Parolari, A.; Myasoedova, V.; Ferrari, G.; Alamanni, F.; Tremoli, E.; Poggio, P. Mitral valve endothelial cells secrete osteoprotegerin during endothelial mesenchymal transition. J. Mol. Cell. Cardiol. 2016, 98, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Songia, P.; Porro, B.; Chiesa, M.; Myasoedova, V.; Alamanni, F.; Tremoli, E.; Poggio, P. Identification of Patients Affected by Mitral Valve Prolapse with Severe Regurgitation: A Multivariable Regression Model. Oxidative Med. Cell. Longev. 2017, 2017, 1–6. [Google Scholar] [CrossRef]
- Tan, H.T.; Ling, L.H.; Dolor-Torres, M.C.; Yip, J.W.-L.; Richards, A.M.; Chung, M.C. Proteomics discovery of biomarkers for mitral regurgitation caused by mitral valve prolapse. J. Proteom. 2013, 94, 337–345. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- Kumar, D.; Narang, R.; Sreenivas, V.; Rastogi, V.; Bhatia, J.; Saluja, D.; Srivastava, K. Circulatory miR-133b and miR-21 as Novel Biomarkers in Early Prediction and Diagnosis of Coronary Artery Disease. Genes 2020, 11, 164. [Google Scholar] [CrossRef]
- Ikeda, S.; Kong, S.W.; Lu, J.; Bisping, E.; Zhang, H.; Allen, P.D.; Golub, T.R.; Pieske, B.; Pu, W.T. Altered microRNA expression in human heart disease. Physiol. Genom. 2007, 31, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Chen, Y.; Lu, Y.; Li, P.; Yu, H.; Diao, F.-R.; Tang, W.-D.; Hou, P.; Zhao, X.-X.; Shi, C.-Y. High level of circulating microRNA-142 is associated with acute myocardial infarction and reduced survival. Ir. J. Med. Sci. 2020, 189, 933–937. [Google Scholar] [CrossRef]
- Nigam, V.; Sievers, H.H.; Jensen, B.C.; Sier, H.A.; Simpson, P.C.; Srivastava, D.; Mohamed, S.A. Altered microRNAs in bicuspid aortic valve: A comparison between stenotic and insufficient valves. J. Heart Valve Dis. 2010, 19, 459–465. [Google Scholar]
- Yanagawa, B.; Lovren, F.; Pan, Y.; Garg, V.; Quan, A.; Tang, G.; Singh, K.K.; Shukla, P.C.; Kalra, N.P.; Peterson, M.D.; et al. miRNA-141 is a novel regulator of BMP-2–mediated calcification in aortic stenosis. J. Thorac. Cardiovasc. Surg. 2012, 144, 256–262.e2. [Google Scholar] [CrossRef]
- Hulanicka, M.; Garncarz, M.; Parzeniecka-Jaworska, M.; Jank, M. Plasma miRNAs as potential biomarkers of chronic degenerative valvular disease in Dachshunds. BMC Vet. Res. 2014, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Freeman, L.M.; Rush, J.E.; Laflamme, D.P. Expression Profiling of Circulating MicroRNAs in Canine Myxomatous Mitral Valve Disease. Int. J. Mol. Sci. 2015, 16, 14098–14108. [Google Scholar] [CrossRef] [PubMed]
- Vatan, M.B.; Yigin, A.K.; Akdemir, R.; Agac, M.T.; Cakar, M.A.; Aksoy, M.; Tatli, E.; Kilic, H.; Gunduz, H.; Guzel, D.; et al. Altered Plasma MicroRNA Expression in Patients with Mitral Chordae Tendineae Rupture. J. Heart Valve Dis. 2016, 25, 580–588. [Google Scholar]
- Sondermeijer, B.M.; Bakker, A.; Halliani, A.; De Ronde, M.W.J.; Marquart, A.A.; Tijsen, A.J.; Mulders, T.A.; Kok, M.G.M.; Battjes, S.; Maiwald, S.; et al. Platelets in Patients with Premature Coronary Artery Disease Exhibit Upregulation of miRNA340* and miRNA624*. PLoS ONE 2011, 6, e25946. [Google Scholar] [CrossRef]
- Yoo, H.; Kim, J.; Lee, A.-R.; Lee, J.-M.; Kim, O.-J.; Kim, J.-K.; Oh, S.-H. Alteration of microRNA 340-5p and Arginase-1 Expression in Peripheral Blood Cells during Acute Ischemic Stroke. Mol. Neurobiol. 2018, 56, 3211–3221. [Google Scholar] [CrossRef]
- Guan, Y.; Song, X.; Sun, W.; Wang, Y.; Liu, B. Effect of Hypoxia-Induced MicroRNA-210 Expression on Cardiovascular Disease and the Underlying Mechanism. Oxidative Med. Cell. Longev. 2019, 2019, 1–12. [Google Scholar] [CrossRef]
- Li, Y.; Xu, J.; Shao, T.; Zhang, Y.; Chen, H.; Li, X. RNA Function Prediction. Methods Mol. Biol. 2017, 1654, 17–28. [Google Scholar] [CrossRef]
- Deroyer, C.; Magne, J.; Moonen, M.; Le Goff, C.; Dupont, L.; Hulin, A.; Radermecker, M.; Colige, A.; Cavalier, E.; Kolh, P.; et al. New biomarkers for primary mitral regurgitation. Clin. Proteom. 2015, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Bohan, A. Genome-wide sequencing and quantification of circulating microRNAs for dogs with congestive heart failure secondary to myxomatous mitral valve degeneration. Am. J. Vet. Res. 2018, 79, 163–169. [Google Scholar] [CrossRef]
- Prot, V.; Skallerud, B. Contributions of prestrains, hyperelasticity, and muscle fiber activation on mitral valve systolic performance. Int. J. Numer. Methods Biomed. Eng. 2016, 33, e2806. [Google Scholar] [CrossRef]
- Krishnamurthy, G.; Ennis, D.B.; Itoh, A.; Bothe, W.; Swanson, J.C.; Karlsson, M.; Kuhl, E.; Miller, D.C.; Ingels, N.B. Material properties of the ovine mitral valve anterior leaflet in vivo from inverse finite element analysis. Am. J. Physiol. Circ. Physiol. 2008, 295, H1141–H1149. [Google Scholar] [CrossRef]
- Saporiti, F.; Piacentini, L.; Alfieri, V.; Bono, E.; Ferrari, F.; Chiesa, M.; Colombo, G.I. Melanocortin-1 Receptor Positively Regulates Human Artery Endothelial Cell Migration. Cell. Physiol. Biochem. 2019, 52, 1339–1360. [Google Scholar] [CrossRef]
- Wylie-Sears, J.; Aikawa, E.; Levine, R.A.; Yang, J.-H.; Bischoff, J. Mitral Valve Endothelial Cells With Osteogenic Differentiation Potential. Arter. Thromb. Vasc. Biol. 2011, 31, 598–607. [Google Scholar] [CrossRef]
- Rabkin, E.; Aikawa, M.; Stone, J.R.; Fukumoto, Y.; Libby, P.; Schoen, F.J. Activated Interstitial Myofibroblasts Express Catabolic Enzymes and Mediate Matrix Remodeling in Myxomatous Heart Valves. Circulation 2001, 104, 2525–2532. [Google Scholar] [CrossRef] [PubMed]
- Roberts, W.C.; Vowels, T.J.; Ko, J.M.; Hebeler, R.F. Gross and Histological Features of Excised Portions of Posterior Mitral Leaflet in Patients Having Operative Repair of Mitral Valve Prolapse and Comments on the Concept of Missing (= Ruptured) Chordae Tendineae. J. Am. Coll. Cardiol. 2014, 63, 1667–1674. [Google Scholar] [CrossRef] [PubMed]
- Sainger, R.; Grau, J.B.; Branchetti, E.; Poggio, P.; Seefried, W.F.; Field, B.C.; Acker, M.A.; Gorman, R.C.; Gorman, J.H.; Hargrove, C.W.; et al. Human myxomatous mitral valve prolapse: Role of bone morphogenetic protein 4 in valvular interstitial cell activation. J. Cell. Physiol. 2011, 227, 2595–2604. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Wang, J.; Wee, A.S.Y.; Yong, Q.-W.; Tay, E.L.-W.; Woo, C.C.; Sorokin, V.; Richards, A.M.; Ling, L.-H. Differential MicroRNA Expression Profile in Myxomatous Mitral Valve Prolapse and Fibroelastic Deficiency Valves. Int. J. Mol. Sci. 2016, 17, 753. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yan, X.; Yang, A.; Xu, A.; Huang, T.; You, H. miRNA-150-5p promotes hepatic stellate cell proliferation and sensitizes hepatocyte apoptosis during liver fibrosis. Epigenomics 2020, 12, 53–67. [Google Scholar] [CrossRef]
- Liu, F.; Di Wang, X. miR-150-5p represses TP53 tumor suppressor gene to promote proliferation of colon adenocarcinoma. Sci. Rep. 2019, 9, 6740. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Han, S. miR-150-5p promotes the proliferation and epithelial-mesenchymal transition of cervical carcinoma cells via targeting SRCIN1. Pathol. Res. Pract. 2019, 215, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Kutmon, M.; Ehrhart, F.; Willighagen, E.L.; Evelo, C.T.; Coort, S.L. CyTargetLinker app update: A flexible solution for network extension in Cytoscape. F1000Research 2018, 7, ELIXIR-743. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2013, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chou, C.-H.; Shrestha, S.; Yang, C.-D.; Chang, N.-W.; Lin, Y.-L.; Liao, K.-W.; Huang, W.-C.; Sun, T.-H.; Tu, S.-J.; Lee, W.-H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J.; et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [CrossRef] [PubMed]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’Ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9, 1366. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Chiesa, M.; Maioli, G.; Colombo, G.I.; Piacentini, L. GARS: Genetic Algorithm for the identification of a Robust Subset of features in high-dimensional datasets. BMC Bioinform. 2020, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
| FED vs. BW | ||
| miRNAs | logFC | p-value |
| miR-140-3p | 0.12 | 0.74 |
| miR-150-5p | −1.2 | 0.03 |
| miR-210-3p | 1 | 0.1 |
| miR-223-3p | 0.29 | 0.39 |
| miR-27a-3p | 0.11 | 0.87 |
| miR-30c-5p | 0.22 | 0.56 |
| miR-323a-3p | −0.59 | 0.48 |
| miR-340-5p | −0.16 | 0.71 |
| miR-361-5p | −0.42 | 0.43 |
| miR-451a | −1.07 | 0.1 |
| miR-487a-3p | 0.41 | 0.6 |
| Variables | CTRL (n = 34) | MVP (n = 43) | p-Value |
|---|---|---|---|
| Age (years) | 53.5 ± 9.4 | 54.3 ± 9.9 | 0.701 |
| Male subjects, n (%) | 16 (47) | 34 (79) | 0.004 |
| BMI | 25.2 ± 4.9 | 24.4 ± 3.3 | 0.390 |
| Diabetes, n (%) | 2 (6) | - | - |
| Hypertension, n (%) | 5 (15) | 16 (37) | 0.039 |
| Dysplidemia n (%) | 15 (44) | 19 (44) | 1.000 |
| Smokers | 5 (15) | 13 (30) | 0.175 |
| Total Cholesterol (mg/dL) | 215.4 ± 45.9 | 202.5 ± 34.2 | 0.203 |
| Triglycerides (mg/dL) | 114.9 ± 50.6 | 102.4 ± 52.5 | 0.317 |
| HDL (mg/dL) | 65.3 ± 36.0 | 58.0 ± 13.5 | 0.315 |
| LDL (mg/dL) | 129.4 ± 38.5 | 124.3 ± 32.5 | 0.568 |
| Drug Therapies | |||
| Antiplatelets, n (%) | 2 (6) | 6 (14) | 0.291 |
| Angiotensin II receptor blockers, n (%) | 1 (3) | 5 (12) | 0.220 |
| Angiotensin-converting enzyme inhibitors, n (%) | 1 (3) | 13 (30) | 0.002 |
| Calcium channel blockers, n (%) | 1 (3) | 1 (2) | 1.000 |
| Beta-blockers, n (%) | 3 (9) | 13 (30) | 0.026 |
| Statins, n (%) | 3 (9) | 6 (14) | 0.723 |
| Echocardiographic data | |||
| LVEF (%) | 62.5 ± 8.3 | 63.8 ± 6.2 | 0.465 |
| Left Ventricular Diastolic Volume (mL) | 96.8 ± 28.5 | 146.0 ± 52.5 | <0.001 |
| Left Ventricular Systolic Volume (mL) | 37.0 ± 17.3 | 41.9 ± 16.2 | <0.001 |
| Left Atrial Area (cm2) | 18.2 ± 3.9 | 28.0 ± 7.0 | <0.001 |
| PAPs | 26.4 ± 3.7 | 34.5 ± 7.9 | <0.001 |
| EROA (cm2) | - | 0.5 ± 0.2 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Songia, P.; Chiesa, M.; Alfieri, V.; Massaiu, I.; Moschetta, D.; Myasoedova, V.; Valerio, V.; Fusini, L.; Gripari, P.; Zanobini, M.; et al. Putative Circulating MicroRNAs Are Able to Identify Patients with Mitral Valve Prolapse and Severe Regurgitation. Int. J. Mol. Sci. 2021, 22, 2102. https://doi.org/10.3390/ijms22042102
Songia P, Chiesa M, Alfieri V, Massaiu I, Moschetta D, Myasoedova V, Valerio V, Fusini L, Gripari P, Zanobini M, et al. Putative Circulating MicroRNAs Are Able to Identify Patients with Mitral Valve Prolapse and Severe Regurgitation. International Journal of Molecular Sciences. 2021; 22(4):2102. https://doi.org/10.3390/ijms22042102
Chicago/Turabian StyleSongia, Paola, Mattia Chiesa, Valentina Alfieri, Ilaria Massaiu, Donato Moschetta, Veronika Myasoedova, Vincenza Valerio, Laura Fusini, Paola Gripari, Marco Zanobini, and et al. 2021. "Putative Circulating MicroRNAs Are Able to Identify Patients with Mitral Valve Prolapse and Severe Regurgitation" International Journal of Molecular Sciences 22, no. 4: 2102. https://doi.org/10.3390/ijms22042102
APA StyleSongia, P., Chiesa, M., Alfieri, V., Massaiu, I., Moschetta, D., Myasoedova, V., Valerio, V., Fusini, L., Gripari, P., Zanobini, M., & Poggio, P. (2021). Putative Circulating MicroRNAs Are Able to Identify Patients with Mitral Valve Prolapse and Severe Regurgitation. International Journal of Molecular Sciences, 22(4), 2102. https://doi.org/10.3390/ijms22042102







