Transcriptome Analysis of Subcutaneous Adipose Tissue from Severely Obese Patients Highlights Deregulation Profiles in Coding and Non-Coding Oncogenes
Abstract
1. Introduction
2. Results
2.1. Transcriptional Differences and Substantial Deregulation in Oncogenes in SAT Tissue
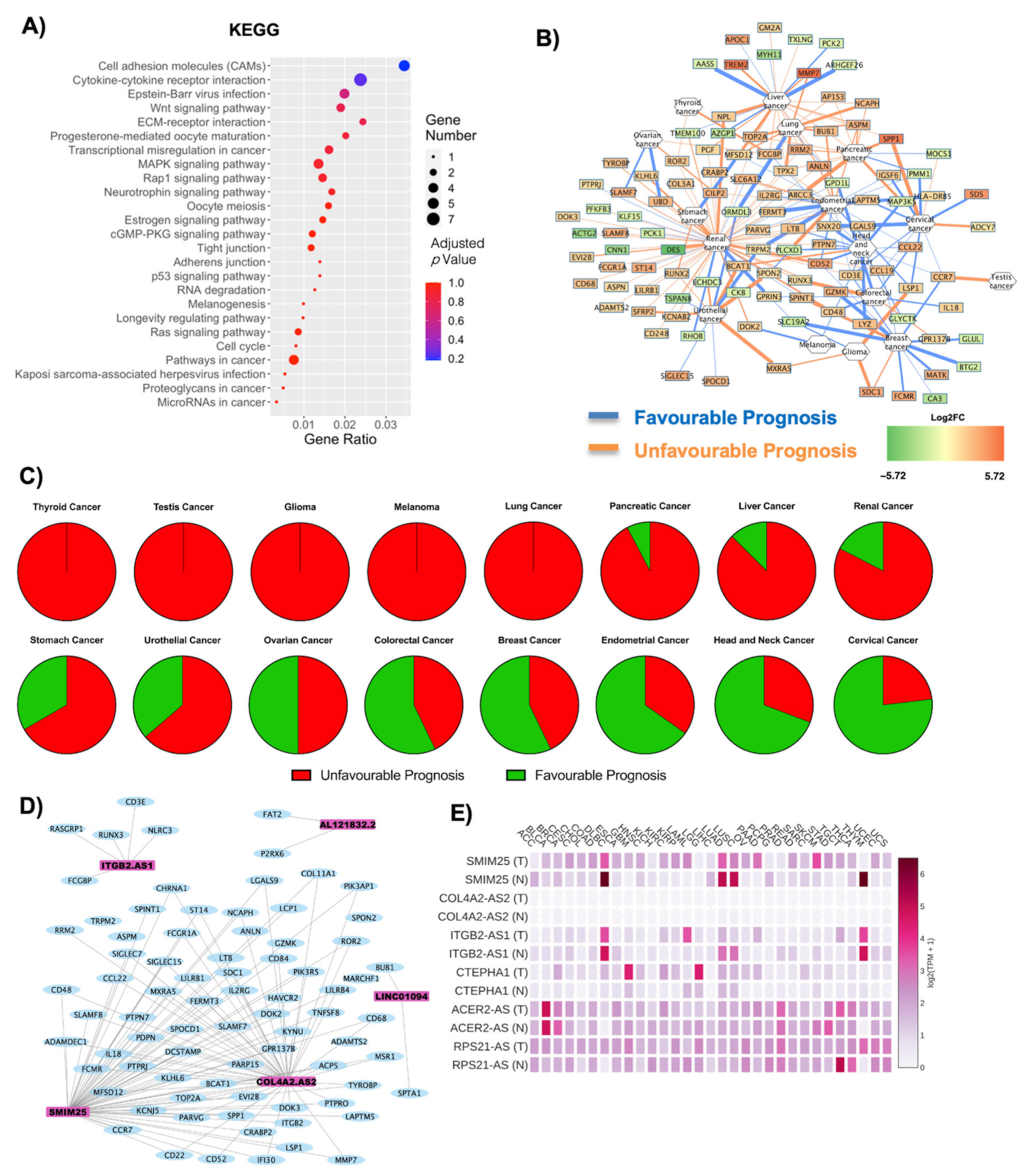
2.2. Both Coding and Non-Coding DE RNAs Are Associated with Cancer in Obese Subjects
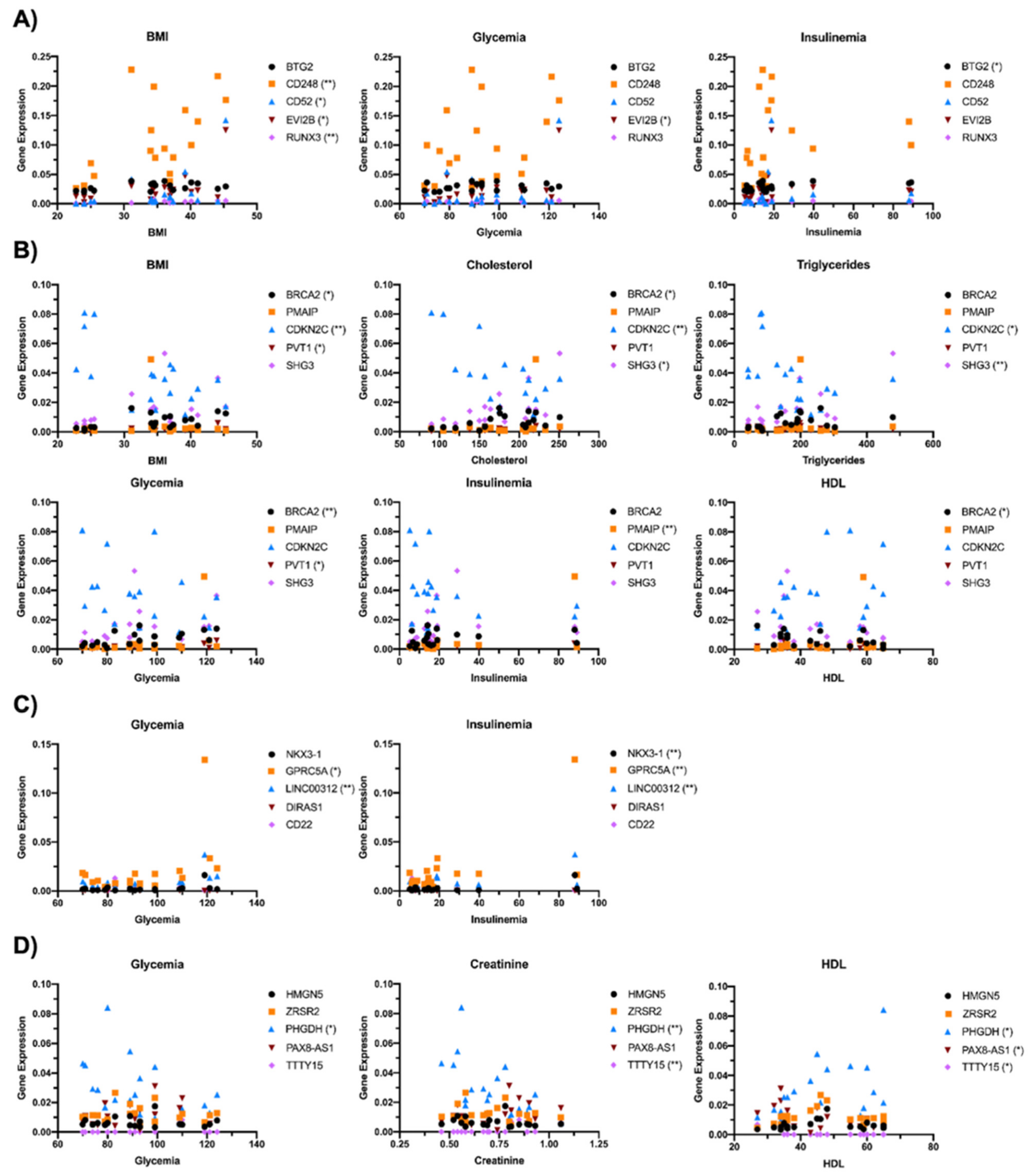
2.3. Correlation of Transcriptional Deregulation with Cancer in Obese Subjects with Type 2 Diabetes
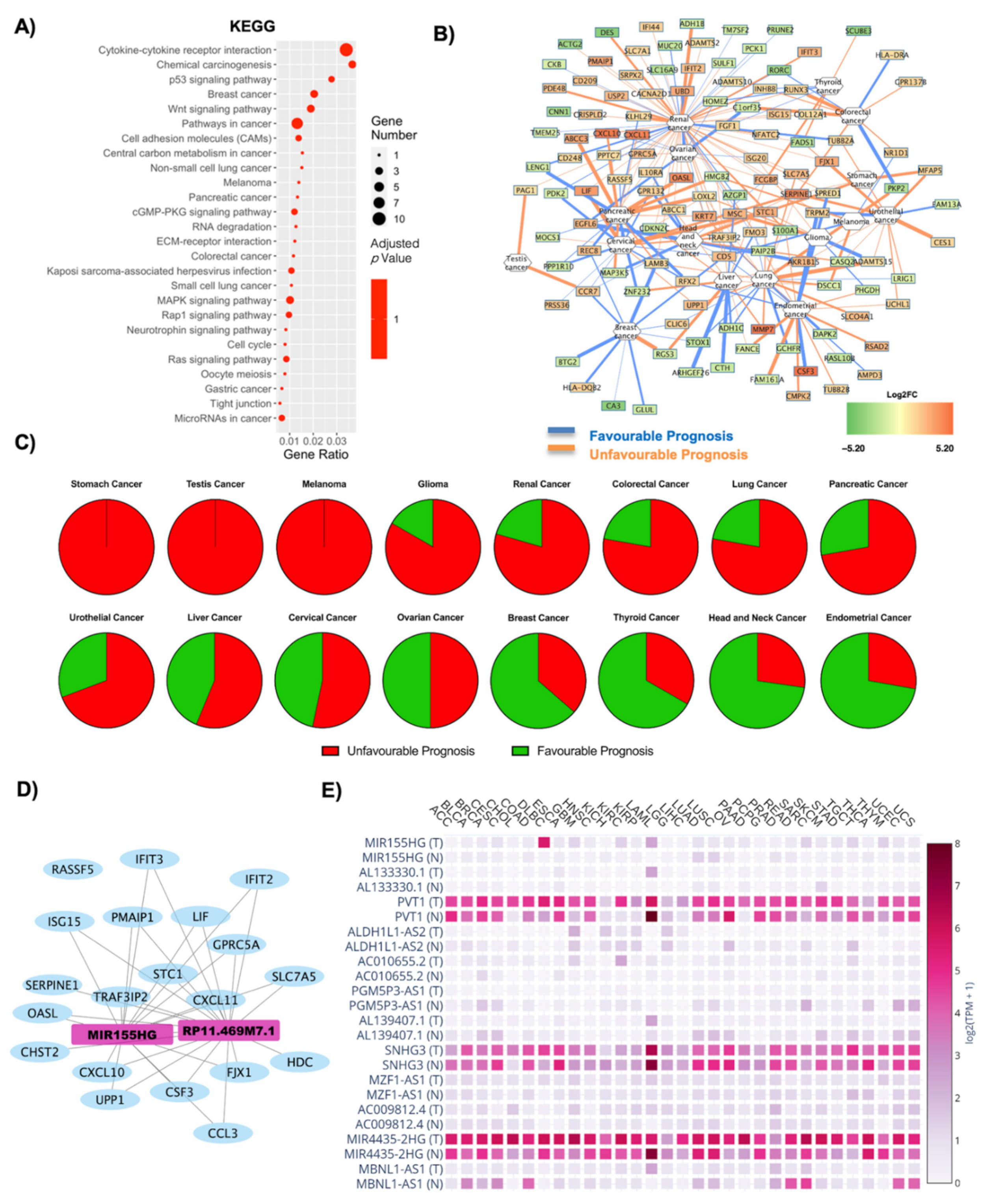
2.4. Influence of T2D on Oncogene Expression: Role of Both Coding and Non-Coding RNAs in OBT2D vs. OBF
2.5. Gender Differences Lead to Alteration in Oncogene Expression in Relation to Obesity
3. Discussion
4. Materials and Methods
4.1. Adult Human Adipose Tissue Collection, Isolation and Differentiation
4.2. SAT RNA Extraction
4.3. Library Preparation for RNA-Seq and Bioinformatic Data Analysis
4.4. RNA Extraction and Real-Time PCR
4.5. Pathway Analysis and Cancer Correlations
4.6. Correlation Analysis
4.7. Coding and ncRNA Co-Expression Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | Adrenocortical carcinoma |
| BLCA | Bladder urothelial carcinoma |
| BRCA | Breast invasive carcinoma |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma |
| CHOL | Cholangiocarcinoma |
| COAD | Colon adenocarcinoma |
| DLBC | Lymphoid neoplasm diffuse large B-cell lymphoma |
| ESCA | Esophageal Carcinoma |
| GBM | Glioblastoma Multiforme |
| HNSC | Head and neck squamous cell carcinoma |
| KICH | Kidney chromophobe |
| KIRC | Kidney renal clear cell carcinoma |
| KIRP | Kidney renal papillary cell carcinoma |
| LAML | Acute myeloid leukemia |
| LGG | Brain lower grade glioma |
| LIHC | Liver hepatocellular carcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| MESO | Mesothelioma |
| OV | Ovarian serous cystadenocarcinoma |
| PAAD | Pancreatic adenocarcinoma |
| PCPG | Pheochromocytoma and paraganglioma |
| PRAD | Prostate adenocarcinoma |
| READ | Rectum adenocarcinoma |
| SARC | Sarcoma |
| SKCM | Skin cutaneous melanoma |
| STAD | Stomach adenocarcinoma |
| TGCT | Testicular germ cell tumors |
| THYM | Thymoma |
| THCA | Thyroid carcinoma |
| UCS | Uterine carcinosarcoma |
| UCEC | Uterine corpus endometrial carcinoma |
| UVM | Uveal melanoma |
References
- WHO. Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 15 February 2021).
- Jaacks, L.M.; Vandevijvere, S.; Pan, A.; McGowan, C.J.; Wallace, C.; Imamura, F.; Mozaffarian, D.; Swinburn, B.; Ezzati, M. The obesity transition: Stages of the global epidemic. Lancet Diabetes Endocrinol. 2019, 7, 231–240. [Google Scholar] [CrossRef]
- Haslam, D.; Sattar, N.; Lean, M. ABC of obesity. Obesity—Time to wake up. BMJ 2006, 333, 640–642. [Google Scholar] [CrossRef]
- Lawrence, V.J.; Kopelman, P.G. Medical consequences of obesity. Clin. Derm. 2004, 22, 296–302. [Google Scholar] [CrossRef] [PubMed]
- De Pergola, G.; Silvestris, F. Obesity as a major risk factor for cancer. J. Obes. 2013, 2013, 291546. [Google Scholar] [CrossRef]
- Onstad, M.A.; Schmandt, R.E.; Lu, K.H. Addressing the Role of Obesity in Endometrial Cancer Risk, Prevention, and Treatment. J. Clin. Oncol. 2016, 34, 4225–4230. [Google Scholar] [CrossRef] [PubMed]
- Picon-Ruiz, M.; Morata-Tarifa, C.; Valle-Goffin, J.J.; Friedman, E.R.; Slingerland, J.M. Obesity and adverse breast cancer risk and outcome: Mechanistic insights and strategies for intervention. CA Cancer J. Clin. 2017, 67, 378–397. [Google Scholar] [CrossRef]
- Leitzmann, M.F.; Koebnick, C.; Danforth, K.N.; Brinton, L.A.; Moore, S.C.; Hollenbeck, A.R.; Schatzkin, A.; Lacey, J.V. Body mass index and risk of ovarian cancer. Cancer 2009, 115, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Freedland, S.J.; Aronson, W.J. Examining the relationship between obesity and prostate cancer. Rev. Urol. 2004, 6, 73–81. [Google Scholar]
- Aleksandrova, K.; Stelmach-Mardas, M.; Schlesinger, S. Obesity and Liver Cancer. Recent Results Cancer Res. 2016, 208, 177–198. [Google Scholar] [CrossRef]
- Larsson, S.C.; Wolk, A. Obesity and the risk of gallbladder cancer: A meta-analysis. Br. J. Cancer 2007, 96, 1457–1461. [Google Scholar] [CrossRef]
- Liu, X.; Sun, Q.; Hou, H.; Zhu, K.; Wang, Q.; Liu, H.; Zhang, Q.; Ji, L.; Li, D. The association between BMI and kidney cancer risk: An updated dose-response meta-analysis in accordance with PRISMA guideline. Medicine 2018, 97, e12860. [Google Scholar] [CrossRef]
- Frezza, E.E.; Wachtel, M.S.; Chiriva-Internati, M. Influence of obesity on the risk of developing colon cancer. Gut 2006, 55, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Avgerinos, K.I.; Spyrou, N.; Mantzoros, C.S.; Dalamaga, M. Obesity and cancer risk: Emerging biological mechanisms and perspectives. Metabolism 2019, 92, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Kompella, P.; Vasquez, K.M. Obesity and cancer: A mechanistic overview of metabolic changes in obesity that impact genetic instability. Mol. Carcinog. 2019, 58, 1531–1550. [Google Scholar] [CrossRef]
- Ungefroren, H.; Gieseler, F.; Fliedner, S.; Lehnert, H. Obesity and cancer. Horm. Mol. Biol. Clin. Investig. 2015, 21, 5–15. [Google Scholar] [CrossRef]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Morley, T.S.; Kim, M.; Clegg, D.J.; Scherer, P.E. Obesity and cancer--mechanisms underlying tumour progression and recurrence. Nat. Rev. Endocrinol. 2014, 10, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.; Lesma, E.; Massihnia, D.; Ciusani, E.; Nava, S.; Vasco, C.; Al Haj, G.; Ghilardi, G.; Opocher, E.; Gorio, A.; et al. Adipose-Derived Stem Cells from Fat Tissue of Breast Cancer Microenvironment Present Altered Adipogenic Differentiation Capabilities. Stem Cells Int. 2019, 2019, 1480314. [Google Scholar] [CrossRef]
- Pallegar, N.K.; Christian, S.L. Adipocytes in the Tumour Microenvironment. Adv. Exp. Med. Biol. 2020, 1234, 1–13. [Google Scholar] [CrossRef]
- Divella, R.; de Luca, R.; Abbate, I.; Naglieri, E.; Daniele, A. Obesity and cancer: The role of adipose tissue and adipo-cytokines-induced chronic inflammation. J. Cancer 2016, 7, 2346–2359. [Google Scholar] [CrossRef]
- Wojciechowska, J.; Krajewski, W.; Bolanowski, M.; Kręcicki, T.; Zatoński, T. Diabetes and Cancer: A Review of Current Knowledge. Exp. Clin. Endocrinol. Diabetes 2016, 124, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Rönn, T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019, 29, 1028–1044. [Google Scholar] [CrossRef]
- Villeneuve, L.M.; Reddy, M.A.; Natarajan, R. Epigenetics: Deciphering its role in diabetes and its chronic complications. Clin. Exp. Pharm. Physiol. 2011, 38, 451–459. [Google Scholar] [CrossRef]
- Gabriele, L.; Buoncervello, M.; Ascione, B.; Bellenghi, M.; Matarrese, P.; Carè, A. The gender perspective in cancer research and therapy: Novel insights and on-going hypotheses. Ann. Ist Super Sanita 2016, 52, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.D.; Oertelt-Prigione, S.; Adjei, A.; Buclin, T.; Cristina, V.; Csajka, C.; Coukos, G.; Dafni, U.; Dotto, G.P.; Ducreux, M.; et al. Gender medicine and oncology: Report and consensus of an ESMO workshop. Ann. Oncol. 2019, 30, 1914–1924. [Google Scholar] [CrossRef] [PubMed]
- Dorak, M.T.; Karpuzoglu, E. Gender differences in cancer susceptibility: An inadequately addressed issue. Front. Genet. 2012, 3, 268. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; He, P.; Yao, H.; Song, R.; Ma, C.; Cao, M.; Cui, B.; Ning, G. Cancer risk among patients with type 2 diabetes: A real-world study in Shanghai, China. J. Diabetes 2019, 11, 878–883. [Google Scholar] [CrossRef] [PubMed]
- Piazza, R.; Ramazzotti, D.; Spinelli, R.; Pirola, A.; de Sano, L.; Ferrari, P.; Magistroni, V.; Cordani, N.; Sharma, N.; Gambacorti-Passerini, C. OncoScore: A novel, Internet-based tool to assess the oncogenic potential of genes. Sci. Rep. 2017, 7, 46290. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Janiszewska, M.; Primi, M.C.; Izard, T. Cell adhesion in cancer: Beyond the migration of single cells. J. Biol. Chem. 2020, 295, 2495–2505. [Google Scholar] [CrossRef]
- Moh, M.C.; Shen, S. The roles of cell adhesion molecules in tumor suppression and cell migration: A new paradox. Cell Adh. Mig.r 2009, 3, 334–336. [Google Scholar] [CrossRef] [PubMed]
- Harjunpää, H.; Llort Asens, M.; Guenther, C.; Fagerholm, S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef]
- Ang, T.L.; Fock, K.M. Clinical epidemiology of gastric cancer. Singap. Med. J. 2014, 55, 621–628. [Google Scholar] [CrossRef]
- Poorolajal, J.; Moradi, L.; Mohammadi, Y.; Cheraghi, Z.; Gohari-Ensaf, F. Risk factors for stomach cancer: A systematic review and meta-analysis. Epidemiol. Health 2020, 42, e2020004. [Google Scholar] [CrossRef]
- Olsen, C.M.; Nagle, C.M.; Whiteman, D.C.; Ness, R.; Pearce, C.L.; Pike, M.C.; Rossing, M.A.; Terry, K.L.; Wu, A.H.; Risch, H.A.; et al. Obesity and risk of ovarian cancer subtypes: Evidence from the Ovarian Cancer Association Consortium. Endocr. Relat. Cancer 2013, 20, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, C.M.; McCullough, M.L.; Franceschi, S.; Rinaldi, S.; Wolk, A.; Neta, G.; Olov Adami, H.; Anderson, K.; Andreotti, G.; Beane Freeman, L.E.; et al. Anthropometric Factors and Thyroid Cancer Risk by Histological Subtype: Pooled Analysis of 22 Prospective Studies. Thyroid 2016, 26, 306–318. [Google Scholar] [CrossRef]
- Schmid, D.; Ricci, C.; Behrens, G.; Leitzmann, M.F. Adiposity and risk of thyroid cancer: A systematic review and meta-analysis. Obes. Rev. 2015, 16, 1042–1054. [Google Scholar] [CrossRef]
- Martinez-Useros, J.; Garcia-Foncillas, J. Obesity and colorectal cancer: Molecular features of adipose tissue. J. Transl. Med. 2016, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.H.; Wu, K.; Ng, K.; Zauber, A.G.; Nguyen, L.H.; Song, M.; He, X.; Fuchs, C.S.; Ogino, S.; Willett, W.C.; et al. Association of Obesity With Risk of Early-Onset Colorectal Cancer Among Women. JAMA Oncol. 2019, 5, 37–44. [Google Scholar] [CrossRef]
- Lee, K.; Kruper, L.; Dieli-Conwright, C.M.; Mortimer, J.E. The Impact of Obesity on Breast Cancer Diagnosis and Treatment. Curr. Oncol. Rep. 2019, 21, 41. [Google Scholar] [CrossRef]
- Poorolajal, J.; Jenabi, E. The association between BMI and cervical cancer risk: A meta-analysis. Eur. J. Cancer Prev. 2016, 25, 232–238. [Google Scholar] [CrossRef]
- Disney-Hogg, L.; Sud, A.; Law, P.J.; Cornish, A.J.; Kinnersley, B.; Ostrom, Q.T.; Labreche, K.; Eckel-Passow, J.E.; Armstrong, G.N.; Claus, E.B.; et al. Influence of obesity-related risk factors in the aetiology of glioma. Br. J. Cancer 2018, 118, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Carreras-Torres, R.; Johansson, M.; Gaborieau, V.; Haycock, P.C.; Wade, K.H.; Relton, C.L.; Martin, R.M.; Davey Smith, G.; Brennan, P. The Role of Obesity, Type 2 Diabetes, and Metabolic Factors in Pancreatic Cancer: A Mendelian Randomization Study. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef]
- Xu, M.; Jung, X.; Hines, O.J.; Eibl, G.; Chen, Y. Obesity and Pancreatic Cancer: Overview of Epidemiology and Potential Prevention by Weight Loss. Pancreas 2018, 47, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Huxley, R.; Ansary-Moghaddam, A.; Berrington de González, A.; Barzi, F.; Woodward, M. Type-II diabetes and pancreatic cancer: A meta-analysis of 36 studies. Br. J. Cancer 2005, 92, 2076–2083. [Google Scholar] [CrossRef]
- Wang, C.; Pan, Y.; Xu, Q.; Li, B.; Kim, K.; Mao, M.; Li, J.; Qin, L.; Li, H.; Han, Z.; et al. Relationship between body mass index and outcomes for patients with oral squamous cell carcinoma. Oral Dis. 2019, 25, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Zheng, W.; Johansson, M.; Lan, Q.; Park, Y.; White, E.; Matthews, C.E.; Sawada, N.; Gao, Y.T.; Robien, K.; et al. Overall and Central Obesity and Risk of Lung Cancer: A Pooled Analysis. J. Natl. Cancer Inst. 2018, 110, 831–842. [Google Scholar] [CrossRef]
- Gild, P.; Ehdaie, B.; Kluth, L.A. Effect of obesity on bladder cancer and renal cell carcinoma incidence and survival. Curr. Opin. Urol. 2017, 27, 409–414. [Google Scholar] [CrossRef]
- Larsson, S.C.; Wolk, A. Overweight, obesity and risk of liver cancer: A meta-analysis of cohort studies. Br. J. Cancer 2007, 97, 1005–1008. [Google Scholar] [CrossRef] [PubMed]
- Bae, W.J.; Choi, J.B.; Moon, H.W.; Park, Y.H.; Cho, H.J.; Hong, S.H.; Lee, J.Y.; Kim, S.W.; Han, K.D.; Ha, U.S. Influence of diabetes on the risk of urothelial cancer according to body mass index: A 10-year nationwide population-based observational study. J. Cancer 2018, 9, 488–493. [Google Scholar] [CrossRef]
- Crosbie, E.J.; Zwahlen, M.; Kitchener, H.C.; Egger, M.; Renehan, A.G. Body mass index, hormone replacement therapy, and endometrial cancer risk: A meta-analysis. Cancer Epidemiol. Biomark. Prev. 2010, 19, 3119–3130. [Google Scholar] [CrossRef]
- Culig, Z. Cytokine disbalance in common human cancers. Biochim. Biophys. Acta 2011, 1813, 308–314. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, M.; Rhee, I. Cytokine Signaling in Tumor Progression. Immune Netw. 2017, 17, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Liu, X.; Qi, Y.; Niu, F.; Niu, Z.; Geng, W.; Zou, Z.; Huang, R.; Wang, J.; Zou, H. BMP3 suppresses colon tumorigenesis via ActRIIB/SMAD2-dependent and TAK1/JNK signaling pathways. J. Exp. Clin. Cancer Res. 2019, 38, 428. [Google Scholar] [CrossRef] [PubMed]
- Loh, K.; Chia, J.A.; Greco, S.; Cozzi, S.J.; Buttenshaw, R.L.; Bond, C.E.; Simms, L.A.; Pike, T.; Young, J.P.; Jass, J.R.; et al. Bone morphogenic protein 3 inactivation is an early and frequent event in colorectal cancer development. Genes Chromosomes Cancer 2008, 47, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.F.; Xu, H.; Xu, Y.Y.; Wang, Z.N.; Zhao, T.T.; Song, Y.X.; Xu, H.M. Diabetes mellitus and the risk of gastric cancer: A meta-analysis of cohort studies. Oncotarget 2017, 8, 44881–44892. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Jeon, I.; Kim, J.W.; Song, Y.S.; Yoon, J.M.; Park, S.M. Diabetes mellitus and ovarian cancer risk: A systematic review and meta-analysis of observational studies. Int. J. Gynecol Cancer 2013, 23, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Phillips, L.; Liu, S.; Wactawski-Wende, J.; Margolis, K.L. Diabetes, Diabetes Treatment, and Risk of Thyroid Cancer. J. Clin. Endocrinol. Metab. 2016, 101, 1243–1248. [Google Scholar] [CrossRef]
- Seo, Y.G.; Choi, H.C.; An, A.R.; Park, D.J.; Park, Y.J.; Lee, K.E.; Park, S.K.; Hwang, Y.; Cho, B. The Association between Type 2 Diabetes Mellitus and Thyroid Cancer. J. Diabetes Res. 2017, 2017, 5850879. [Google Scholar] [CrossRef]
- Peeters, P.J.; Bazelier, M.T.; Leufkens, H.G.; de Vries, F.; de Bruin, M.L. The risk of colorectal cancer in patients with type 2 diabetes: Associations with treatment stage and obesity. Diabetes Care 2015, 38, 495–502. [Google Scholar] [CrossRef]
- Eketunde, A.O. Diabetes as a Risk Factor for Breast Cancer. Cureus 2020, 12, e8010. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.N.; Torres, M.A.; Troeschel, A.N.; He, J.; Gogineni, K.; McCullough, L.E. Type 2 diabetes, breast cancer specific and overall mortality: Associations by metformin use and modification by race, body mass, and estrogen receptor status. PLoS One 2020, 15, e0232581. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Kar, S.; Carter, P.; Vithayathil, M.; Mason, A.M.; Burgess, S.; Larsson, S.C. Is Type 2 Diabetes Causally Associated With Cancer Risk? Evidence From a Two-Sample Mendelian Randomization Study. Diabetes 2020, 69, 1588–1596. [Google Scholar] [CrossRef]
- Barami, K.; Lyon, L.; Conell, C. Type 2 Diabetes Mellitus and Glioblastoma Multiforme-Assessing Risk and Survival: Results of a Large Retrospective Study and Systematic Review of the Literature. World Neurosurg. 2017, 106, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Qi, X.; Xiong, H.; Liu, Q.; Li, J.; Zhang, Y.; Ma, X.; Wu, N.; Feng, L. Type 2 diabetes mellitus and risk of malignant melanoma: A systematic review and meta-analysis of cohort studies. Iran J. Public Health 2014, 43, 857–866. [Google Scholar] [PubMed]
- Tsang, N.M.; Pai, P.C.; Chuang, C.C.; Chuang, W.C.; Tseng, C.K.; Chang, K.P.; Yen, T.C.; Lin, J.D.; Chang, J.T. Overweight and obesity predict better overall survival rates in cancer patients with distant metastases. Cancer Med. 2016, 5, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Tseng, K.S.; Lin, C.; Lin, Y.S.; Weng, S.F. Risk of head and neck cancer in patients with diabetes mellitus: A retrospective cohort study in Taiwan. JAMA Otolaryngol. Head Neck Surg. 2014, 140, 746–753. [Google Scholar] [CrossRef]
- Hall, G.C.; Roberts, C.M.; Boulis, M.; Mo, J.; MacRae, K.D. Diabetes and the risk of lung cancer. Diabetes Care 2005, 28, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Graff, R.E.; Sanchez, A.; Tobias, D.K.; Rodríguez, D.; Barrisford, G.W.; Blute, M.L.; Li, Y.; Sun, Q.; Preston, M.A.; Wilson, K.M.; et al. Type 2 Diabetes in Relation to the Risk of Renal Cell Carcinoma Among Men and Women in Two Large Prospective Cohort Studies. Diabetes Care 2018, 41, 1432–1437. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; Shu, X.O.; Gao, J.; Li, H.L.; Cai, H.; Yang, G.; Ji, B.T.; Rothman, N.; Gao, Y.T.; Zheng, W.; et al. Prospective evaluation of type 2 diabetes mellitus on the risk of primary liver cancer in Chinese men and women. Ann. Oncol. 2013, 24, 1679–1685. [Google Scholar] [CrossRef]
- Xu, Y.; Huo, R.; Chen, X.; Yu, X. Diabetes mellitus and the risk of bladder cancer: A PRISMA-compliant meta-analysis of cohort studies. Medicine 2017, 96, e8588. [Google Scholar] [CrossRef]
- Fang, H.; Yao, B.; Yan, Y.; Xu, H.; Liu, Y.; Tang, H.; Zhou, J.; Cao, L.; Wang, W.; Zhang, J.; et al. Diabetes mellitus increases the risk of bladder cancer: An updated meta-analysis of observational studies. Diabetes Technol. Ther. 2013, 15, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Jonklaas, J.; Nogueras-Gonzalez, G.; Munsell, M.; Litofsky, D.; Ain, K.B.; Bigos, S.T.; Brierley, J.D.; Cooper, D.S.; Haugen, B.R.; Ladenson, P.W.; et al. The impact of age and gender on papillary thyroid cancer survival. J. Clin. Endocrinol. Metab. 2012, 97, E878–E887. [Google Scholar] [CrossRef]
- Greif, J.M.; Pezzi, C.M.; Klimberg, V.S.; Bailey, L.; Zuraek, M. Gender differences in breast cancer: Analysis of 13,000 breast cancers in men from the National Cancer Data Base. Ann. Surg. Oncol. 2012, 19, 3199–3204. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Kinnersley, B.; Wrensch, M.R.; Eckel-Passow, J.E.; Armstrong, G.; Rice, T.; Chen, Y.; Wiencke, J.K.; McCoy, L.S.; Hansen, H.M.; et al. Sex-specific glioma genome-wide association study identifies new risk locus at 3p21.31 in females, and finds sex-differences in risk at 8q24.21. Sci. Rep. 2018, 8, 7352. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Saba, N.F.; Goodman, M.; Ward, K.; Flowers, C.; Ramalingam, S.; Owonikoko, T.; Chen, A.; Grist, W.; Wadsworth, T.; Beitler, J.J.; et al. Gender and ethnic disparities in incidence and survival of squamous cell carcinoma of the oral tongue, base of tongue, and tonsils: A surveillance, epidemiology and end results program-based analysis. Oncology 2011, 81, 12–20. [Google Scholar] [CrossRef]
- Hefaiedh, R.; Ennaifer, R.; Romdhane, H.; Ben Nejma, H.; Arfa, N.; Belhadj, N.; Gharbi, L.; Khalfallah, T. Gender difference in patients with hepatocellular carcinoma. Tunis Med. 2013, 91, 505–508. [Google Scholar] [PubMed]
- Tait, S.; Baldassarre, A.; Masotti, A.; Calura, E.; Martini, P.; Varì, R.; Scazzocchio, B.; Gessani, S.; del Cornò, M. Integrated Transcriptome Analysis of Human Visceral Adipocytes Unravels Dysregulated microRNA-Long Non-coding RNA-mRNA Networks in Obesity and Colorectal Cancer. Front. Oncol. 2020, 10, 1089. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Xu, H.; Li, D.; Li, H.; Lu, D. A novel long noncoding RNA, AC092834.1, regulates the adipogenic differentiation of human adipose-derived mesenchymal stem cells via the DKK1/Wnt/β-catenin signaling pathway. Biochem. Biophys. Res. Commun. 2020, 525, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Squillaro, T.; Peluso, G.; Galderisi, U.; di Bernardo, G. Long non-coding RNAs in regulation of adipogenesis and adipose tissue function. Elife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Kerr, A.; Jiao, H.; Hon, C.C.; Rydén, M.; Dahlman, I.; Arner, P. Long Non-Coding RNAs Associated with Metabolic Traits in Human White Adipose Tissue. EBioMedicine 2018, 30, 248–260. [Google Scholar] [CrossRef]
- Latorre, J.; Fernández-Real, J.M. LncRNAs in Adipose Tissue from Obese and Insulin-Resistant Subjects: New Targets for Therapy? EBioMedicine 2018, 30, 10–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ji, Y.; Li, M.; Wang, M.; Yi, X.; Yin, C.; Wang, S.; Zhang, M.; Zhao, Z.; Xiao, Y. Integrated analysis of long noncoding RNA and mRNA expression profile in children with obesity by microarray analysis. Sci. Rep. 2018, 8, 8750. [Google Scholar] [CrossRef]
- Lo, K.A.; Huang, S.; Walet, A.C.E.; Zhang, Z.C.; Leow, M.K.; Liu, M.; Sun, L. Adipocyte Long-Noncoding RNA Transcriptome Analysis of Obese Mice Identified Lnc-Leptin, Which Regulates Leptin. Diabetes 2018, 67, 1045–1056. [Google Scholar] [CrossRef]
- Wei, S.; Du, M.; Jiang, Z.; Hausman, G.J.; Zhang, L.; Dodson, M.V. Long noncoding RNAs in regulating adipogenesis: New RNAs shed lights on obesity. Cell Mol. Life Sci. 2016, 73, 2079–2087. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Goff, L.A.; Trapnell, C.; Alexander, R.; Lo, K.A.; Hacisuleyman, E.; Sauvageau, M.; Tazon-Vega, B.; Kelley, D.R.; Hendrickson, D.G.; et al. Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3387–3392. [Google Scholar] [CrossRef]
- Rey, F.; Urrata, V.; Gilardini, L.; Bertoli, S.; Calcaterra, V.; Zuccotti, G.V.; Cancello, R.; Carelli, S. Role of long non-coding RNAs in adipogenesis: State of the art and implications in obesity and obesity-associated diseases. Obes. Rev. 2021. [Google Scholar] [CrossRef] [PubMed]
- Ringel, A.E.; Drijvers, J.M.; Baker, G.J.; Catozzi, A.; García-Cañaveras, J.C.; Gassaway, B.M.; Miller, B.C.; Juneja, V.R.; Nguyen, T.H.; Joshi, S.; et al. Obesity Shapes Metabolism in the Tumor Microenvironment to Suppress Anti-Tumor Immunity. Cell 2020. [Google Scholar] [CrossRef]
- Petrelli, F.; Cortellini, A.; Indini, A.; Tomasello, G.; Ghidini, M.; Nigro, O.; Salati, M.; Dottorini, L.; Iaculli, A.; Varricchio, A.; et al. Obesity paradox in patients with cancer: A systematic review and meta-analysis of 6,320,365 patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Montemurro, N.; Perrini, P.; Rapone, B. Clinical Risk and Overall Survival in Patients with Diabetes Mellitus, Hyperglycemia and Glioblastoma Multiforme. A Review of the Current Literature. Int. J. Environ. Res. Public Health 2020, 17, 8501. [Google Scholar] [CrossRef]
- Shlomai, G.; Neel, B.; LeRoith, D.; Gallagher, E.J. Type 2 Diabetes Mellitus and Cancer: The Role of Pharmacotherapy. J. Clin. Oncol. 2016, 34, 4261–4269. [Google Scholar] [CrossRef] [PubMed]
- Saraei, P.; Asadi, I.; Kakar, M.A.; Moradi-Kor, N. The beneficial effects of metformin on cancer prevention and therapy: A comprehensive review of recent advances. Cancer Manag Res. 2019, 11, 3295–3313. [Google Scholar] [CrossRef] [PubMed]
- Kasznicki, J.; Sliwinska, A.; Drzewoski, J. Metformin in cancer prevention and therapy. Ann. Transl. Med. 2014, 2, 57. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Pratt, D.; Chen, J.; Welker, D.; Rivas, R.; Pillich, R.; Rynkov, V.; Ono, K.; Miello, C.; Hicks, L.; Szalma, S.; et al. NDEx, the Network Data Exchange. Cell Syst. 2015, 1, 302–305. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Hounkpe, B.W.; Chenou, F.; de Lima, F.; de Paula, E.V. HRT Atlas v1.0 database: Redefining human and mouse housekeeping genes and candidate reference transcripts by mining massive RNA-seq datasets. Nucleic Acids Res. 2021, 49, D947–D955. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, D.; Qiu, W.; Shi, Y.; Yang, J.-J.; Chen, S.; Wang, Q.; Pan, H. Application of Weighted Gene Co-expression Network Analysis for Data from Paired Design. Sci. Rep. 2018, 8, 622. [Google Scholar] [CrossRef] [PubMed]

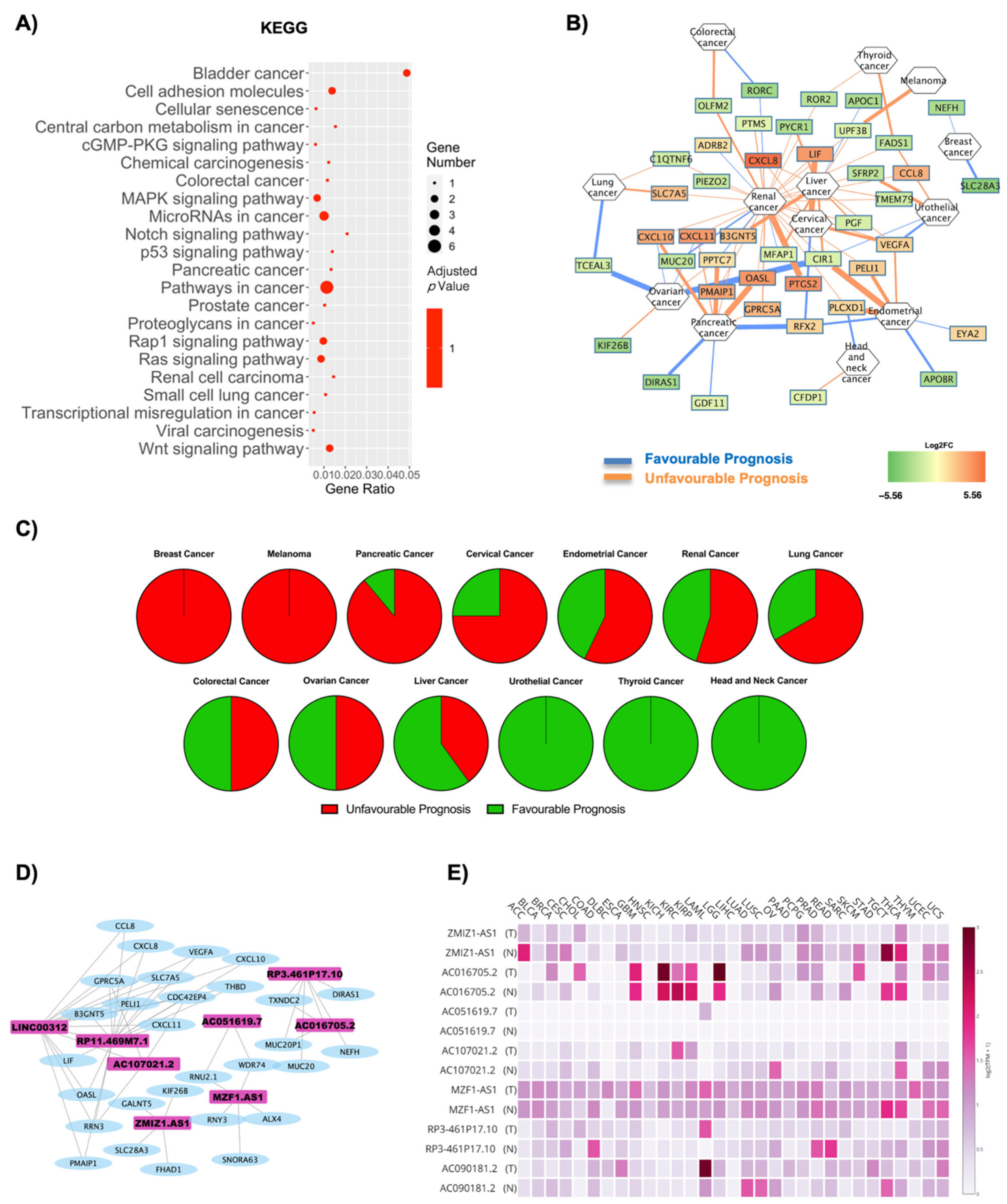
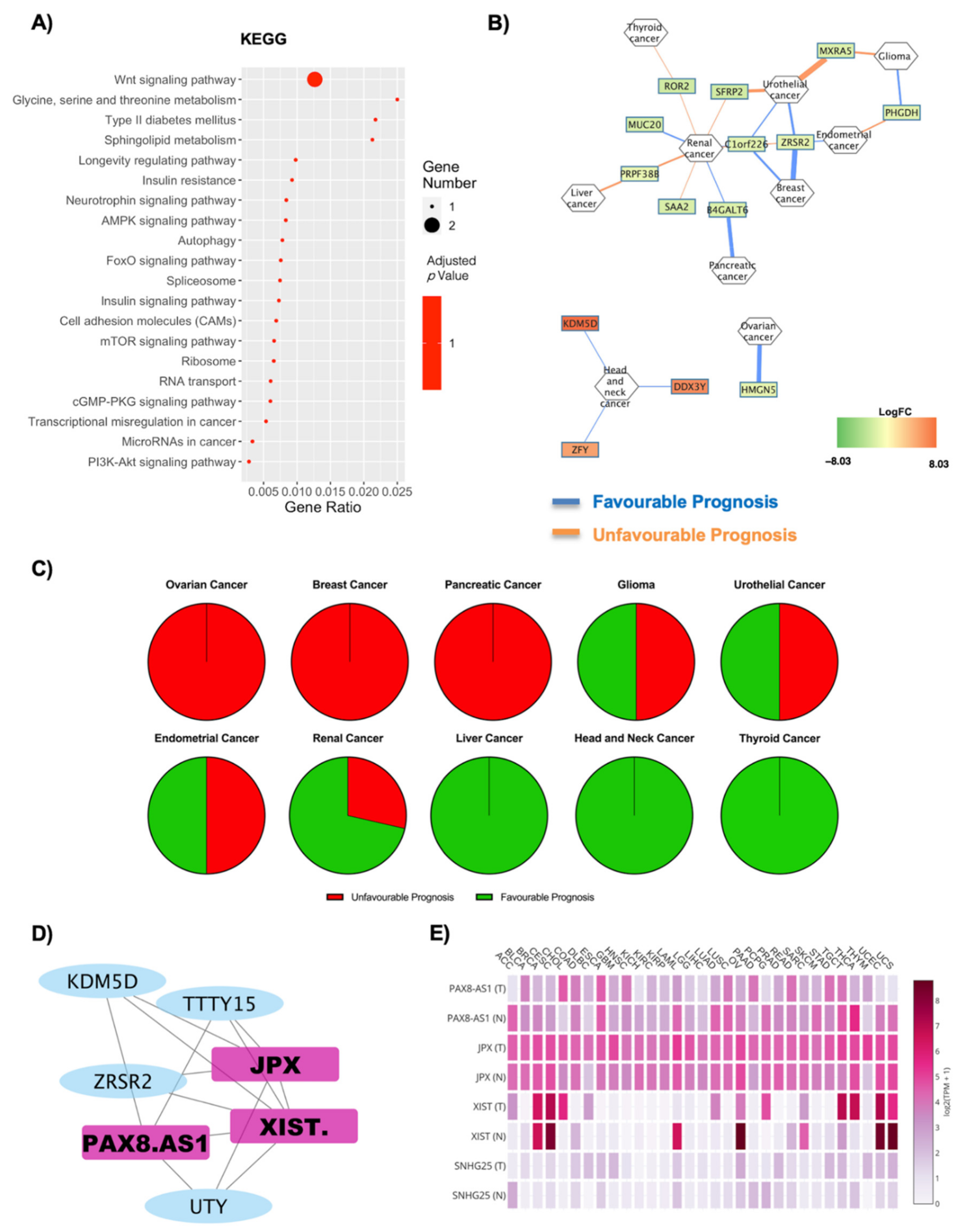
| Cancer | Unfavourable Prognosis | Favourable Prognosis | Literature Evidence for Obesity Correlation | ||
|---|---|---|---|---|---|
| Upregulated Unfavourable Prognosis | Downregulated Favourable Prognosis | Downregulated Unfavourable Prognosis | Upregulated Unfavourable Prognosis | ||
| Stomach Cancer | 2 | 0 | 0 | 1 | Negatively correlated [34,35] |
| Ovarian Cancer | 2 | 0 | 0 | 2 | Positively correlated [36] |
| Thyroid Cancer | 2 | 0 | 0 | 0 | Positively correlated [37,38] |
| Colorectal Cancer | 2 | 1 | 0 | 4 | Positively correlated [39,40] |
| Breast Cancer | 2 | 4 | 0 | 8 | Positively correlated [7,41] |
| Cervical Cancer | 2 | 1 | 1 | 9 | Positively correlated [8,42] |
| Glioma | 3 | 1 | 0 | 0 | Negatively correlated [43] |
| Pancreatic Cancer | 10 | 2 | 1 | 0 | Positively correlated [44,45] |
| Melanoma | 0 | 0 | 0 | 2 | Positive correlation amongst men [46] |
| Head and Neck Cancer | 1 | 3 | 0 | 9 | Positively correlated [47] |
| Lung Cancer | 9 | 2 | 0 | 0 | Negatively correlated [48] |
| Renal Cancer | 44 | 8 | 4 | 7 | Positively correlated [49] |
| Liver Cancer | 15 | 6 | 2 | 1 | Positively correlated [50] |
| Urothelial Cancer | 5 | 2 | 1 | 3 | Positively correlated [51] |
| Endometrial Cancer | 7 | 1 | 1 | 14 | Positively correlated [52] |
| Gene Symbol | ACER2-AS | COL4A2-AS | CTEPHA1 | ITGB2-AS1 | RPS21-AS | SMIM25 |
|---|---|---|---|---|---|---|
| Tumoral Tissue | ||||||
| ACC | ns | ns | ns | ns | ns | + |
| BRCA | ns | ns | + | ns | ns | ns |
| CESC | ns | ns | ns | ns | ns | + |
| COAD | ns | ns | ns | ns | ns | + |
| DLBC | ns | ns | ns | + | + | + |
| GBM | ns | ns | + | ns | ns | + |
| KIRC | ns | ns | + | + | ns | + |
| KIRP | ns | ns | + | ns | ns | + |
| LAML | ns | ns | ns | ns | + | ns |
| LGG | ns | ns | + | ns | ns | ns |
| LUAD | ns | ns | ns | + | ns | + |
| LUSC | ns | ns | ns | + | ns | + |
| OV | ns | ns | + | ns | ns | + |
| PAAD | ns | ns | + | + | ns | + |
| READ | ns | ns | ns | ns | ns | + |
| SKCM | + | ns | ns | ns | ns | + |
| STAD | ns | ns | ns | ns | ns | + |
| TGCT | + | ns | ns | + | + | + |
| THYM | + | ns | ns | + | + | + |
| UCEC | ns | ns | ns | ns | ns | + |
| Cancer | Unfavourable Prognosis | Favourable Prognosis | Column Name Literature Evidence | ||
|---|---|---|---|---|---|
| Upregulated Unfavourable Prognosis | Downregulated Favourable Prognosis | Downregulated Unfavourable Prognosis | Upregulated Unfavourable Prognosis | ||
| Stomach Cancer | 4 | 0 | 0 | 0 | Negatively correlated [57] |
| Ovarian Cancer | 3 | 2 | 0 | 5 | Positively correlated [58] |
| Thyroid Cancer | 1 | 1 | 2 | 2 | Negatively correlated [59,60] |
| Colorectal Cancer | 5 | 2 | 1 | 1 | Positively correlated [61] |
| Breast Cancer | 0 | 4 | 1 | 6 | Positively correlated [62,63] |
| Cervical Cancer | 6 | 2 | 1 | 6 | Positively correlated [64] |
| Glioma | 4 | 1 | 0 | 1 | Negatively correlated [65] |
| Pancreatic Cancer | 9 | 4 | 2 | 3 | Positively correlated [46] |
| Melanoma | 0 | 0 | 2 | 2 | Positively correlated [66] |
| Head and Neck Cancer | 2 | 1 | 1 | 7 | Positively correlated [67,68] |
| Lung Cancer | 9 | 5 | 2 | 2 | Negatively correlated [69] |
| Renal Cancer | 45 | 13 | 10 | 5 | Positively correlated [70] |
| Liver Cancer | 5 | 4 | 6 | 1 | Positively correlated [71] |
| Urothelial Cancer | 8 | 1 | 3 | 1 | Positevly correlated in men [72] |
| Endometrial Cancer | 5 | 0 | 6 | 7 | Positively correlated [64] |
| Gene Symbol → | AC009812.4 | ACO10655.2 | AL133330.1 | AL139407.1 | ALDH1L1-AS2 | MBNL1-AS1 | MIR155HG | MIR4435-2HG | MZF1-AS1 | PGM5P3-AS1 | PVT1 | SNHG3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumoral Tissue ↓ | ||||||||||||
| ACC | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | + |
| BLCA | ns | ns | ns | ns | ns | + | ns | + | ns | ns | ns | ns |
| BRCA | ns | ns | ns | + | ns | + | ns | ns | ns | + | ns | ns |
| CESC | ns | ns | ns | + | ns | + | ns | + | ns | + | ns | ns |
| COAD | ns | ns | ns | ns | ns | + | + | + | ns | ns | + | ns |
| DLBC | ns | ns | ns | ns | ns | ns | ns | + | + | ns | + | + |
| ESCA | ns | ns | ns | ns | ns | ns | ns | + | ns | ns | ns | ns |
| GBM | ns | + | ns | ns | ns | ns | + | + | ns | ns | + | ns |
| HNSC | ns | ns | ns | ns | ns | ns | ns | + | ns | ns | ns | ns |
| KICH | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | + |
| KIRC | ns | + | ns | ns | ns | ns | + | + | ns | ns | + | ns |
| KIRP | ns | ns | ns | ns | ns | ns | ns | + | ns | ns | + | ns |
| LAML | + | ns | + | + | ns | + | + | + | ns | ns | + | ns |
| LIHC | ns | ns | ns | ns | ns | ns | ns | + | ns | ns | + | ns |
| LUAD | ns | ns | ns | + | ns | + | ns | ns | ns | ns | + | + |
| LUSC | ns | ns | ns | + | ns | ns | ns | ns | ns | + | + | + |
| OV | ns | ns | ns | + | + | ns | ns | + | ns | ns | + | ns |
| PAAD | ns | ns | ns | ns | ns | ns | ns | + | ns | ns | + | ns |
| READ | ns | ns | ns | ns | ns | + | ns | + | ns | ns | + | ns |
| SKCM | ns | ns | + | + | ns | ns | ns | + | ns | ns | ns | ns |
| STAD | ns | ns | ns | ns | ns | ns | ns | + | ns | ns | + | ns |
| TGCT | ns | ns | ns | + | + | ns | ns | + | + | ns | ns | + |
| THCA | ns | ns | ns | ns | ns | ns | ns | + | ns | + | + | + |
| THYM | + | ns | ns | ns | ns | ns | + | ns | + | ns | + | + |
| UCEC | ns | ns | ns | + | ns | + | ns | + | ns | + | ns | ns |
| UCS | ns | ns | ns | + | ns | + | ns | + | ns | + | ns | ns |
| Cancer | Unfavourable Prognosis | Favourable Prognosis | Literature Evidence | ||
|---|---|---|---|---|---|
| Upregulated Unfavourable Prognosis | Downregulated Favourable Prognosis | Downregulated Unfavourable Prognosis | Upregulated Unfavourable Prognosis | ||
| Ovarian Cancer | 1 | 2 | 1 | 2 | Positively correlated with obesity and diabetes [36,58] |
| Thyroid Cancer | 0 | 0 | 2 | 0 | Positively correlated in obesity [37,38]; negatively correlated in diabetes [59,60] |
| Colorectal Cancer | 0 | 1 | 1 | 0 | Positively correlated with obesity and diabetes [39,61] |
| Breast Cancer | 0 | 2 | 0 | 0 | Positively correlated with obesity and diabetes [7,41,62,63] |
| Cervical Cancer | 3 | 0 | 0 | 1 | Positively correlated with obesity and diabetes [8,42,64] |
| Pancreatic Cancer | 6 | 2 | 0 | 1 | Positively correlated with obesity and diabetes [44,45,46] |
| Melanoma | 0 | 0 | 1 | 0 | Positively correlated with obesity and diabetes [46,66] |
| Head and Neck Cancer | 0 | 0 | 1 | 1 | Positively correlated with obesity and diabetes [47,67,68] |
| Lung Cancer | 1 | 1 | 1 | 0 | Negatively correlated with obesity and diabetes [48,69] |
| Renal Cancer | 14 | 3 | 12 | 2 | Positively correlated with obesity and diabetes [49,70] |
| Liver Cancer | 3 | 1 | 6 | 0 | Positively correlated with obesity and diabetes [50,71] |
| Urothelial Cancer | 0 | 0 | 2 | 1 | Positively correlated in men both in obesity and diabetes [51,72] |
| Endometrial Cancer | 3 | 1 | 1 | 2 | Positively correlated with obesity and diabetes [52,64] |
| Gene Symbol → | AC016705.2 | AC051619.7 | AC090181.2 | AC107021.2 | MZF1-AS1 | RP3-461P17.10 | ZMIZ1-AS1 |
|---|---|---|---|---|---|---|---|
| Tumoral Tissue ↓ | |||||||
| ACC | ns | ns | ns | ns | ns | ns | + |
| COAD | ns | ns | ns | ns | ns | + | ns |
| DLBC | ns | ns | ns | ns | + | ns | ns |
| KIRC | + | ns | ns | + | ns | ns | ns |
| LAML | ns | ns | + | ns | ns | + | ns |
| LGG | + | ns | ns | ns | ns | ns | ns |
| OV | ns | ns | ns | + | ns | ns | ns |
| PRAD | ns | ns | ns | ns | ns | + | ns |
| READ | ns | ns | ns | ns | ns | + | ns |
| SKCM | + | ns | ns | ns | ns | ns | ns |
| TGCT | + | ns | ns | ns | + | ns | + |
| THYM | ns | ns | ns | ns | + | ns | ns |
| Cancer | Unfavourable Prognosis | Favourable Prognosis | Literature Evidence | ||
|---|---|---|---|---|---|
| Upregulated Unfavourable Prognosis | Downregulated Favourable Prognosis | Downregulated Unfavourable Prognosis | Upregulated Unfavourable Prognosis | ||
| Ovarian Cancer | 0 | 1 | 0 | 0 | Positively correlated in women [36] |
| Thyroid Cancer | 0 | 0 | 1 | 0 | Positively correlated in women [74] |
| Breast Cancer | 0 | 2 | 0 | 0 | Positively correlated both in men and in women [75] |
| Glioma | 0 | 1 | 1 | 0 | Positively correlated in men [43,76] |
| Pancreatic Cancer | 0 | 1 | 0 | 0 | Positively correlated in women [77] |
| Head and Neck Cancer | 0 | 0 | 0 | 3 | Negatively correlated [78] |
| Renal Cancer | 0 | 2 | 0 | 5 | Positively correlated in women [70] |
| Liver Cancer | 0 | 0 | 1 | 0 | Positively correlated in men [79] |
| Urothelial Cancer | 0 | 2 | 2 | 0 | Positively correlated in men [72] |
| Endometrial Cancer | 0 | 1 | 1 | 0 | Positively correlated in women [52] |
| Gene Symbol → | JPX | PAX8-AS1 | SNHG25 | XIST |
|---|---|---|---|---|
| Tumoral Tissue ↓ | ||||
| ACC | ns | + | + | + |
| BRCA | ns | + | ns | ns |
| CESC | ns | ns | ns | + |
| COAD | ns | ns | ns | + |
| DLBC | + | ns | + | + |
| ESCA | ns | ns | ns | ns |
| GBM | + | ns | + | ns |
| KICH | ns | + | ns | ns |
| LUAD | ns | + | ns | + |
| OV | ns | + | ns | + |
| PAAD | + | + | + | ns |
| READ | ns | ns | ns | + |
| SKCM | ns | + | ns | ns |
| STAD | ns | ns | + | + |
| TGCT | + | ns | ns | + |
| THCA | ns | + | + | + |
| THYM | + | + | + | ns |
| UCEC | ns | + | ns | + |
| UCS | ns | + | ns | + |
| UVM | ns | ns | ns | ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rey, F.; Messa, L.; Pandini, C.; Launi, R.; Barzaghini, B.; Micheletto, G.; Raimondi, M.T.; Bertoli, S.; Cereda, C.; Zuccotti, G.V.; et al. Transcriptome Analysis of Subcutaneous Adipose Tissue from Severely Obese Patients Highlights Deregulation Profiles in Coding and Non-Coding Oncogenes. Int. J. Mol. Sci. 2021, 22, 1989. https://doi.org/10.3390/ijms22041989
Rey F, Messa L, Pandini C, Launi R, Barzaghini B, Micheletto G, Raimondi MT, Bertoli S, Cereda C, Zuccotti GV, et al. Transcriptome Analysis of Subcutaneous Adipose Tissue from Severely Obese Patients Highlights Deregulation Profiles in Coding and Non-Coding Oncogenes. International Journal of Molecular Sciences. 2021; 22(4):1989. https://doi.org/10.3390/ijms22041989
Chicago/Turabian StyleRey, Federica, Letizia Messa, Cecilia Pandini, Rossella Launi, Bianca Barzaghini, Giancarlo Micheletto, Manuela Teresa Raimondi, Simona Bertoli, Cristina Cereda, Gian Vincenzo Zuccotti, and et al. 2021. "Transcriptome Analysis of Subcutaneous Adipose Tissue from Severely Obese Patients Highlights Deregulation Profiles in Coding and Non-Coding Oncogenes" International Journal of Molecular Sciences 22, no. 4: 1989. https://doi.org/10.3390/ijms22041989
APA StyleRey, F., Messa, L., Pandini, C., Launi, R., Barzaghini, B., Micheletto, G., Raimondi, M. T., Bertoli, S., Cereda, C., Zuccotti, G. V., Cancello, R., & Carelli, S. (2021). Transcriptome Analysis of Subcutaneous Adipose Tissue from Severely Obese Patients Highlights Deregulation Profiles in Coding and Non-Coding Oncogenes. International Journal of Molecular Sciences, 22(4), 1989. https://doi.org/10.3390/ijms22041989