Genomic Architecture of Phenotypic Plasticity in Response to Water Stress in Tetraploid Wheat
Abstract
1. Introduction
2. Results
2.1. High-Density Genetic Map
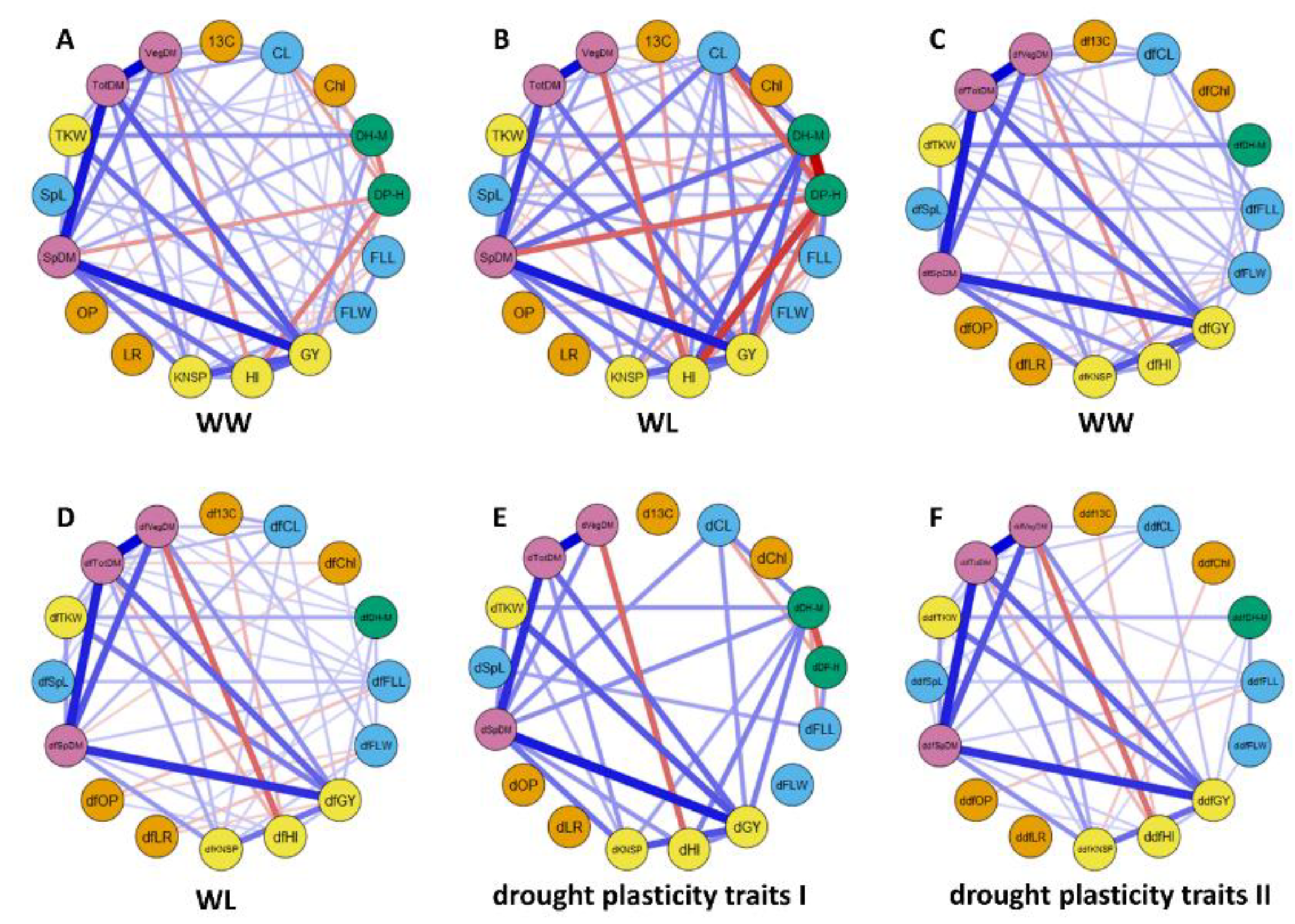
2.2. Relationships between Phenotypic Traits
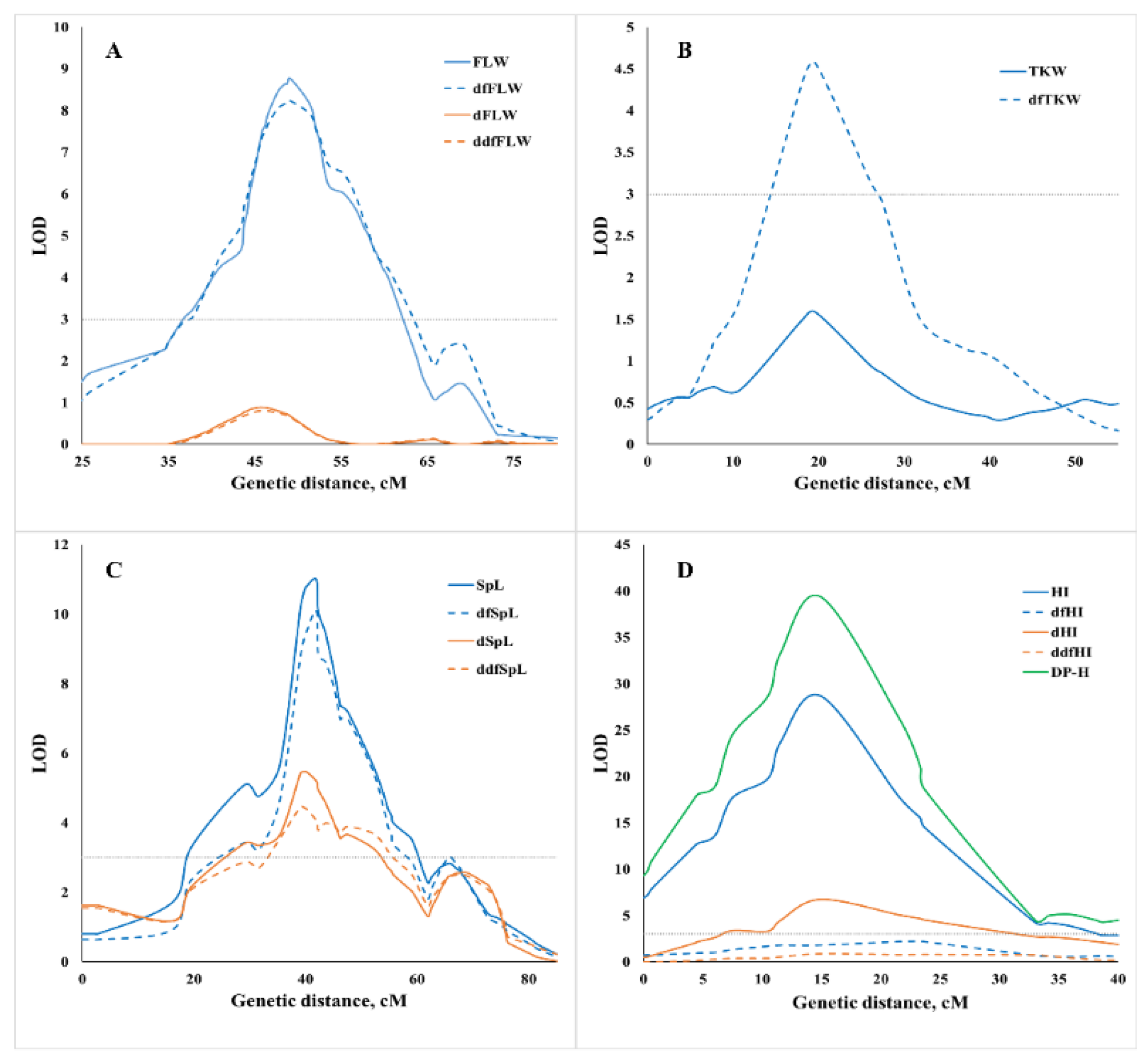
2.3. Genomic Dissection of Initial and Derivative Traits
2.4. Genomic Dissection of Traits Adjusted for Heading Time
2.5. Genomic Dissection of Drought Plasticity Traits
2.6. QTLs Associated with Drought Resistance Strategies
2.7. Candidate Gene Analysis
3. Discussion
3.1. Detection of QTLs Using a High-Density SNP-Based Genetic Map
3.2. Complexity of Quantitative Trait Genetic Architecture and the Interaction with Altered Phenology
3.3. Drought-Plasticity and Drought-Resistance Strategies
3.4. Candidate Genes within QTL Intervals
3.5. Vrn-B3 Is a Candidate Gene for Major Drought Escape QTL
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. DNA Extraction and SNP Genotyping
4.3. Phenotypic Traits
- (1)
- The first derivative set defined here as ‘adjusted phenology traits’ was obtained, for each environment separately, by calculating the residuals of the linear regression between the means of the corresponding observed trait values and DP-H values (Figure 1A) to exclude the effect of differences in flowering phenology on these traits (prefix ’df‘ was added to the observed trait name):
- (2)
- The second derivative set defined here as ‘drought plasticity traits I’ (Figure 1B) was obtained by calculating the residuals of the linear regression between means of the observed trait values in the WW and WL conditions (prefix ’d‘ was added to observed trait name), to get a deviation between trait value in WL stress and WW condition, adjusted for the differences in trait values in the population under normal conditions:
- (3)
- The third derivative set defined here as ‘drought plasticity traits II’ was obtained by calculating the residuals of the linear regression between means of the corresponding observed trait values in the WL treatment and trait values in the WW conditions and DP-H values in the WL (prefix ‘ddf’ was added to the observed trait name), to exclude the effect of drought escape mechanisms in ‘drought plasticity traits I’ by taking into account the effect of heading time:
4.4. Statistical Analysis of Phenotypic Data
4.5. Construction of the High-Density Genetic Map
4.6. QTL Analysis
4.7. Identification of Physical Position of the Mapped SNP Markers and CGs Residing within QTL Intervals
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| WEW | wild emmer wheat |
| QTL | quantitative trait loci |
| DAT | drought adaptive traits |
| MEA | multi-environmental approach |
| DRI | drought-resistant index |
| RIL | recombinant inbred line |
| CG | candidate gene |
| SNP | single nucleotide polymorphism |
| LG | linkage group |
| cM | centimorgan |
| Chr. | chromosome |
| WW | well-watered |
| WL | water-limited |
| GY | grain yield |
| TKW | thousand kernel weight |
| KNSP | kernel number per spike |
| HI | harvest index |
| SpDM | spike dry matter |
| VegDM | vegetative dry matter |
| TotDM | total dry matter |
| CL | culm length |
| SpL | spike length |
| FLL | flag leaf length |
| FLW | flag leaf width |
| δ13C | carbon isotope ratio |
| OP | osmotic potential |
| Chl | chlorophyll content |
| LR | flag leaf rolling |
| DP-H | days from planting to heading |
| DH-M | days from heading to maturity |
| ANOVA | analysis of variance |
| dtraits | drought plasticity traits I |
| ddftraits | drought plasticity traits II |
| dftraits | traits adjusted for the effect of phenology |
| G18-16 | wild emmer wheat accession G18-16 |
| LDN | cv. Langdon |
| LOD | logarithm of the odds |
| ITV allele | increased trait value allele |
| PEV | proportion of explained variation |
| MIM | multiple interval mapping |
| GWAS | genome-wide association study |
References
- Leng, G.; Tang, Q.; Rayburg, S. Climate change impacts on meteorological, agricultural and hydrological droughts in China. Glob. Planet. Chang. 2015, 126, 23–34. [Google Scholar] [CrossRef]
- Mann, E.M.; Gleick, H.P. Climate change and California drought in the 21st century. Proc. Natl. Acad. Sci. USA 2015, 112, 3858–3859. [Google Scholar] [CrossRef]
- Nam, W.H.; Hayes, M.J.; Svoboda, M.D.; Tadesse, T.; Wilhite, D.A. Drought hazard assessment in the context of climate change for South Korea. Agric. Water Manag. 2015, 160, 106–117. [Google Scholar] [CrossRef]
- FAO, Food and Agriculture Organization of the United Nations. FAOSTAT; FAO: Rome, Italy, 2017. [Google Scholar]
- Kosina, P.; Reynolds, M.; Dixon, J.; Joshi, A. Stakeholder perception of wheat production constraints, capacity building needs, and research partnerships in developing countries. Euphytica 2007, 157, 475–483. [Google Scholar] [CrossRef]
- Henry, R.J.; Nevo, E. Exploring natural selection to guide breeding for agriculture. Plant Biotechnol. J. 2014, 12, 655–662. [Google Scholar] [CrossRef]
- Levy, A.A.; Feldman, M. Increase in grain protein percentage in high-yielding common wheat breeding lines by genes from wild tetraploid wheat. Euphytica 1987, 36, 353–359. [Google Scholar] [CrossRef]
- Nevo, E.; Korol, A.B.; Bailes, A.; Fahima, T. Evolution of Wild Emmer and Wheat Improvement; Springer: Berlin, Germany, 2002. [Google Scholar]
- Peleg, Z.; Fahima, T.; Abbo, S.; Krugman, T.; Nevo, E.; Yakir, D.; Saranga, Y. Genetic diversity for drought resistance in wild wheat and its ecogeographical association. Plant Cell Environ. 2005, 28, 176–191. [Google Scholar] [CrossRef]
- Peleg, Z.; Fahima, T.; Krugman, T.; Abbo, S.; Yakir, D.; Korol, A.B.; Saranga, Y. Genomic dissection of drought resistance in durum wheat × wild emmer wheat recombinant inbreed line population. Plant Cell Environ. 2009, 32, 758–779. [Google Scholar] [CrossRef]
- Huang, L.; Raats, D.; Sela, H.; Klymiuk, V.; Lidzbarsky, G.; Feng, L.; Krugman, T.; Fahima, T. Evolution and adaptation of wild emmer wheat populations to biotic and abiotic stresses. Annu. Rev. Phytopathol. 2016, 54, 279–301. [Google Scholar] [CrossRef]
- Merchuk-Ovnat, L.; Barak, V.; Fahima, T.; Ordon, F.; Lidzbarsky, G.A.; Krugman, T.; Saranga, Y. Ancestral QTL alleles from wild emmer wheat improve drought resistance and productivity in modern wheat cultivars. Front. Plant Sci. 2016, 7, 452. [Google Scholar] [CrossRef]
- Merchuk-Ovnat, L.; Fahima, T.; Krugman, T.; Saranga, Y. Ancestral QTL alleles from wild emmer wheat improve grain yield, biomass and photosynthesis across enviroinments in modern wheat. Plant Sci. 2016, 251, 23–34. [Google Scholar] [CrossRef]
- Merchuk-Ovnat, L.; Fahima, T.; Ephrath, E.J.; Krugman, T.; Saranga, Y. Ancestral QTL alleles from wild emmer wheat enhance root development under drought in modern wheat. Front. Plant Sci. 2017, 8, 703–715. [Google Scholar] [CrossRef]
- Krugman, T.; Chagué, V.; Peleg, Z.; Balzergue, S.; Just, J.; Korol, A.B.; Nevo, E.; Saranga, Y.; Chalhoub, B.; Fahima, T. Multilevel regulation and signalling processes associated with adaptation to terminal drought in wild emmer wheat. Funct. Integr. Genom. 2010, 10, 167–186. [Google Scholar] [CrossRef]
- Krugman, T.; Peleg, Z.; Quansah, L.; Chagué, V.; Korol, A.B.; Nevo, E.; Saranga, Y.; Fait, A.; Chalhoub, B.; Fahima, T. Alteration in expression of hormone-related genes in wild emmer wheat roots associated with drought adaptation mechanisms. Funct. Integr. Genom. 2011, 11, 565–583. [Google Scholar] [CrossRef]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Tardieu, F.; Tuberosa, R. Dissection and modeling of abiotic stress tolerance in plants. Curr. Opin. Plant Biol. 2010, 13, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Blum, A. Drought resistance—Is it really a complex trait? Funct. Plant Biol. 2011, 38, 753–757. [Google Scholar] [CrossRef]
- Lopes, M.S.; Rebetzke, G.J.; Reynolds, M. Integration of phenotyping and genetic platforms for a better understanding of wheat performance under drought. J. Exp. Bot. 2014, 65, 6167–6177. [Google Scholar] [CrossRef] [PubMed]
- Rebetzke, G.J.; Condon, A.G.; Farquhar, G.D.; Appels, R.; Richards, R.A. Quantitative trait loci for carbon isotope discrimination are repeatable across environments and wheat mapping populations. Theor. Appl. Genet. 2008, 118, 123–137. [Google Scholar] [CrossRef]
- Adiredjo, A.L.; Navaud, O.; Muños, S.; Langlade, N.B.; Lamaze, T.; Grieu, P. Genetic control of water use efficiency and leaf carbon isotope discrimination in sunflower (Helianthus annuus L.) subjected to two drought scenarios. PLoS ONE 2014, 9, e101218. [Google Scholar] [CrossRef]
- Borrell, A.K.; Mullet, J.E.; George-Jaeggli, B.; van Oosterom, E.J.; Hammer, G.L.; Klein, P.E.; Jordan, D.R. Drought adaptation of stay-green sorghum is associated with canopy development, leaf anatomy, root growth, and water uptake. J. Exp. Bot. 2014, 65, 6251–6263. [Google Scholar] [CrossRef]
- Blum, A. Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant Cell Environ. 2017, 40, 4–10. [Google Scholar] [CrossRef]
- Pinto, R.S.; Reynolds, M.P.; Mathews, K.L.; McIntyre, C.L.; Olivares-Villegas, J.J.; Chapman, S.C. Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theor. Appl. Genet. 2010, 121, 1001–1021. [Google Scholar] [CrossRef]
- Ahmad, M.Q.; Khan, S.H.; Khan, A.S.; Kazi, A.M.; Basra, S. Identification of QTLs for drought tolerance traits on wheat chromosome 2A using association mapping. Int. J. Agric. Biol. 2014, 16, 862–870. [Google Scholar]
- Graziani, M.; Maccaferri, M.; Royo, C.; Salvatorelli, F.; Tuberosa, R. QTL dissection of yield components and morpho-physiological traits in a durum wheat elite population tested in contrasting thermo-pluviometric conditions. Crop Pasture Sci. 2014, 65, 80–95. [Google Scholar] [CrossRef]
- Mwadzingeni, L.; Shimelis, H.; Tesfay, S.; Tsilo, T.J. Screening of bread wheat genotypes for drought tolerance using phenotypic and proline analyses. Front. Plant Sci. 2016, 7, 1276. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Bloomfield, J.A.; Rose, T.J.; King, G.J. Sustainable harvest: Managing plasticity for resilient crops. Plant Biotechnol. J. 2014, 12, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Messmer, R.; Fracheboud, Y.; Bänziger, M.; Vargas, M.; Stamp, P.; Ribaut, J.M. Drought stress and tropical maize: QTL-by-environment interactions and stability of QTLs across environments for yield components and secondary traits. Theor. Appl. Genet. 2009, 119, 913–930. [Google Scholar] [CrossRef]
- Lacaze, X.; Hayes, P.M.; Korol, A. Genetics of phenotypic plasticity: QTL analysis in barley, Hordeum vulgare. Heredity 2009, 102, 163–173. [Google Scholar] [CrossRef]
- Van Eeuwijk, F.A.; Bink, M.C.A.M.; Chenu, K.; Chapman, S.C. Detection and use of QTL for complex traits in multiple environments. Curr. Opin. Plant Biol. 2010, 13, 193–205. [Google Scholar] [CrossRef]
- Kamran, A.; Iqbal, M.; Spaner, D. Flowering time in wheat (Triticum aestivum L.): A key factor for global adaptability. Euphytica 2014, 197, 1–26. [Google Scholar] [CrossRef]
- Fusco, G.; Minelli, A. Phenotypic plasticity in development and evolution: Facts and concepts. Introd. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2010, 365, 547–556. [Google Scholar] [CrossRef]
- Kazan, K.; Lyons, R. The link between flowering time and stress tolerance. J. Exp. Bot. 2016, 67, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Sabadin, P.K.; Malosetti, M.; Boer, M.P.; Tardin, F.D.; Santos, F.G.; Guimarães, C.T.; Gomide, R.L.; Andrade, C.L.; Albuquerque, P.E.; Caniato, F.F.; et al. Studying the genetic basis of drought tolerance in sorghum by managed stress trials and adjustments for phonological and plant height differences. Theor. Appl. Genet. 2012, 124, 1389–1402. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.B.; Taylor, J.D.; Edwards, J.; Mather, D.; Bacic, A.; Langridge, P.; Roessner, U. Whole-genome mapping of agronomic and metabolic traits to identify novel quantitative trait loci in bread wheat grown in a water-limited environment. Plant Physiol. 2013, 162, 1266–1281. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.; Dreisigacker, S.; Peña, R.; Sukumaran, S.; Reynolds, M. Genetic characterization of the Wheat Association Mapping Initiative (WAMI) panel for dissection of complex traits in spring wheat. Theor. Appl. Genet. 2014, 128, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Onogi, A.; Ideta, O.; Yoshioka, T.; Ebana, K.; Yamasaki, M.; Iwata, H. Uncovering a nuisance influence of a phenological trait of plants using a nonlinear structural equation: Application to days to heading and culm length in Asian cultivated rice (Oryza sativa L.). PLoS ONE 2016, 11, e0148609. [Google Scholar] [CrossRef] [PubMed]
- Bidinger, F.R.; Mahalakshmi, Y.; Talukdar, B.S.; Alagarswamy, G. Improvement of drought resistance in pearl millet. In Drought Resistance in Crops with Emphasis on Rice; IRRI: Los Banos, Philippines, 1982; pp. 357–375. [Google Scholar]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef]
- Peleg, Z.; Fahima, T.; Korol, A.B.; Abbo, S.; Saranga, Y. Genetic analysis of wheat domestication and evolution under domestication. J. Exp. Bot. 2011, 62, 5051–5061. [Google Scholar] [CrossRef]
- Matesanz, S.; Milla, R. Differential plasticity to water and nutrients between crops and their wild progenitors. Environ. Exp. Bot. 2018, 145, 54–63. [Google Scholar] [CrossRef]
- Peleg, Z.; Saranga, Y.; Suprunova, T.; Ronin, Y.; Röder, M.; Kilian, A.; Korol, A.; Fahima, T. High-density genetic map of durum wheat × wild emmer wheat based on SSR and DArT markers. Theor. Appl. Genet. 2008, 117, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Peleg, Z.; Cakmak, I.; Ozturk, L.; Yazici, A.; Jun, Y.; Budak, H.; Korol, A.B.; Fahima, T.; Saranga, Y. Quantitative trait loci conferring grain mineral nutrient concentrations in durum wheat × wild emmer wheat RIL population. Theor. Appl. Genet. 2009, 119, 353–369. [Google Scholar] [CrossRef]
- Tzarfati, R.; Saranga, Y.; Barak, V.; Gopher, A.; Korol, A.B.; Abbo, S. Threshing efficiency as an incentive for rapid domestication of emmer wheat. Ann. Bot. 2013, 112, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.; Shrestha, D.; Ludwig, C. Reproductive strategies in Mediterranean legumes: Trade-offs between phenology, seed size and vigor within and between wild and domesticated Lupinus species collected along aridity gradients. Front. Plant Sci. 2017, 8, 548. [Google Scholar] [CrossRef]
- Sadras, V.O.; Richards, R.A. Improvement of crop yield in dry environments: Benchmarks, levels of organisation and the role of nitrogen. J. Exp. Bot. 2014, 65, 1981–1995. [Google Scholar] [CrossRef]
- Tetard-Jones, C.; Kertesz, M.A.; Preziosi, R.F. Quantitative Trait Loci mapping of phenotypic plasticity and genotype–environment interactions in plant and insect performance. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 1368–1379. [Google Scholar] [CrossRef]
- Bac-Molenaar, J.A.; Fradin, E.F.; Becker, F.F.M.; Rienstra, J.A.; van der Schoot, J.; Vreugdenhil, D.; Keurentjes, J.J.B. Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis thaliana. Plant Cell 2015, 27, 1857–1874. [Google Scholar] [CrossRef]
- Scheiner, S.M. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Evol. Syst. 1993, 24, 35–68. [Google Scholar] [CrossRef]
- Gulisija, D.; Plotkin, J.B. Phenotypic plasticity promotes recombination and gene clustering in periodic environments. Nat. Commun. 2017, 8, 2041. [Google Scholar] [CrossRef]
- Levitt, J. Responses of Plants to Environmental Stresses; Academic Press: New York, NY, USA, 1980. [Google Scholar]
- Guo, Y.; Gan, S. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2012, 35, 644–655. [Google Scholar] [CrossRef]
- Tian, F.; Gong, J.; Zhang, J.; Zhang, M.; Wang, G.; Li, A.; Wang, W. Enhanced stability of thylakoid membrane proteins and antioxidant competence contribute to drought stress resistance in the tasg1 wheat stay-green mutant. J. Exp. Bot. 2013, 64, 1509–1520. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Ougham, H. The stay-green trait. J. Exp. Bot. 2014, 65, 3889–3900. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.C. Sustainable production of crops and pastures under drought in a Mediterranean environment. Ann. Appl. Biol. 2004, 144, 139–147. [Google Scholar] [CrossRef]
- Thudi, M.; Gaur, P.M.; Krishnamurthy, L.; Mir, R.R.; Kudapa, H.; Fikre, A.; Kimurto, P.; Tripathi, S.; Soren, K.R.; Mulwa, R.; et al. Genomics-assisted breeding for drought tolerance in chickpea. Funct. Plant Biol. 2014, 41, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Mwadzingeni, L.; Shimelis, H.; Dube, E.; Laing, M.D.; Tsilo, T.J. Breeding wheat for drought tolerance: Progress and technologies. J. Integr. Agric. 2016, 15, 935–943. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, H.; Chu, S.; Li, H.; Chi, Y.; Triebwasser-Freese, D.; Lv, H.; Yu, D. Integrating QTL mapping and transcriptomics identifies candidate genes underlying QTLs associated with soybean tolerance to low-phosphorus stress. Plant Mol. Biol. 2017, 93, 137–150. [Google Scholar] [CrossRef]
- Milner, S.G.; Maccaferri, M.; Huang, B.E.; Mantovani, P.; Massi, A.; Frascaroli, E.; Tuberosa, R.; Salvi, S. A multiparental cross population for mapping QTL for agronomic traits in durum wheat (Triticum turgidum ssp. durum). Plant Biotechnol. J. 2016, 14, 735–748. [Google Scholar] [CrossRef]
- Beales, J.; Turner, A.; Griffiths, S.; Snape, J.W.; Laurie, D.A. A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 115, 721–733. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Liu, M.S.; Li, J.R.; Guan, C.M.; Zhang, X.S. The wheat TaGI1, involved in photoperiodic flowering, encodes an Arabidopsis GI ortholog. Plant Mol. Biol. 2005, 58, 53–64. [Google Scholar] [CrossRef]
- Halliwell, J.; Borrill, P.; Gordon, A.; Kowalczyk, R.; Pagano, M.L.; Saccomanno, B.; Bentley, A.R.; Uauy, C.; Cockram, J.; Bentley, A.R. Systematic investigation of FLOWERING LOCUS T-like poaceae gene families identifies the short-day expressed flowering pathway gene, TaFT3 in wheat (Triticum aestivum L.). Front. Plant Sci. 2016, 7, 857. [Google Scholar] [CrossRef]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef]
- Pearce, S.; Huttly, A.K.; Prosser, I.M.; Li, Y.D.; Vaughan, S.P.; Gallova, B.; Patil, A.; Coghill, J.A.; Dubcovsky, J.; Hedden, P.; et al. Heterologous expression and transcript analysis of gibberellin biosynthetic genes of grasses reveals novel functionality in the GA3ox family. BMC Plant Biol. 2015, 15, 130. [Google Scholar] [CrossRef]
- Shu, K.; Zhou, W.; Chen, F.; Luo, X.; Yang, W. Abscisic acid and gibberellins antagonistically mediate plant development and abiotic stress responses. Front. Plant Sci. 2018, 9, 416. [Google Scholar] [CrossRef]
- Dubois, M.; Van den Broeck, L.; Inze, D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef]
- Dey, S.; Corina Vlot, A. Ethylene responsive factors in the orchestration of stress responses in monocotyledonous plants. Front. Plant Sci. 2015, 6, 640. [Google Scholar] [CrossRef]
- Yan, L.; Fu, D.; Li, C.; Blechl, A.; Tranquilli, G.; Bonafede, M.; Sanchez, A.; Valarik, M.; Yasuda, S.; Dubcovsky, J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 2006, 103, 19581–19586. [Google Scholar] [CrossRef]
- Lv, B.; Nitcher, R.; Han, X.; Wang, S.; Ni, F.; Li, K.; Pearce, S.; Wu, J.; Dubcovsky, J.; Fu, D. Characterization of FLOWERING LOCUS T1 (FT1) gene in Brachypodium and wheat. PLoS ONE 2014, 9, e94171. [Google Scholar] [CrossRef] [PubMed]
- Nitcher, R.; Distelfeld, A.; Tan, C.; Yan, L.; Dubcovsky, J. Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol. Genet. Genom. 2013, 288, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Pin, P.A.; Nilsson, O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 2012, 35, 1742–1755. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, F.; Chiozzotto, R.; Locatelli, F.; Spada, A.; Genga, A.; Fornara, F. Hd3a, RFT1 and Ehd1 integrate photoperiodic and drought stress signals to delay the floral transition in rice. Plant Cell Environ. 2016, 39, 1982–1993. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J. DNA protocols for plants. In Molecular Techniques in Taxonomy. NATO ASI Series (Series H: Cell Biology) 57; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; Springer: Berlin, Germany, 1991; pp. 283–293. [Google Scholar]
- Muqaddasi, Q.H.; Brassac, J.; Börner, A.; Pillen, K.; Röder, M.S. Genetic architecture of anther extrusion in spring and winter wheat. Front. Plant Sci. 2017, 8, 754. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Ronin, Y.I.; Mester, D.I.; Minkov, D.G.; Akhunov, E.; Korol, A.B. Building ultra-high-density linkage maps based on efficient filtering of trustable markers. Genetics 2017, 206, 1285–1295. [Google Scholar] [CrossRef]
- Mester, D.; Ronin, Y.; Minkov, D.; Nevo, E.; Korol, A. Constructing large-scale genetic maps using an evolutionary strategy algorithm. Genetics 2003, 165, 2269–2282. [Google Scholar]
- Korol, A.B.; Mester, D.; Frenkel, Z.; Ronin, Y.I. Methods for genetic analysis in the Triticeae. In Genetics and Genomics of the Triticeae. Plant Genetics and Genomics: Crops and Models 7; Feuillet, C., Muehlbauer, G.J., Eds.; Springer Business Media LLC: New York, NY, USA, 2009; pp. 163–199. [Google Scholar]
- Maccaferri, M.; Ricci, A.; Salvi, S.; Milner, S.G.; Noli, E.; Martelli, P.L.; Casadio, R.; Akhunov, E.; Scalabrin, S.; Vendramin, V.; et al. A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 2014, 13, 648–663. [Google Scholar] [CrossRef]
- Kao, C.H.; Zeng, Z.B.; Teasdale, R.D. Multiple interval mapping for quantitative trait loci. Genetics 1999, 152, 1203–1216. [Google Scholar]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]


| Trait | # QTLs | LOD | ITV Allele | Environmental Specificity | Relation to Phenology | Relation to Drought | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Multi-trait | Single-trait | G18-16 | LDN | WL | WW | Associated | Non-Plastic | Plastic | Non-Plastic | Plastic | ||
| Yield related traits | |||||||||||||
| GY | 8 | 8 | 0 | 2.1–11.2 | 2 | 6 | 1 | 0 | 2 | 5 | 1 | 7 | 1 |
| TKW | 16 | 9 | 7 | 2.3–9.2 | 9 | 7 | 4 | 2 | 4 | 8 | 4 | 12 | 4 |
| KNSP | 11 | 10 | 1 | 2.3–8.9 | 2 | 9 | 1 | 0 | 4 | 4 | 3 | 9 | 2 |
| HI | 13 | 10 | 3 | 2.2–23.9 | 7 | 6 | 2 | 1 | 4 | 7 | 2 | 7 | 6 |
| Biomass related traits | |||||||||||||
| SpDM | 10 | 10 | 0 | 2.0–13.9 | 4 | 6 | 0 | 0 | 3 | 5 | 2 | 6 | 4 |
| VegDM | 6 | 5 | 1 | 2.2–4.4 | 1 | 4 | 0 | 0 | 0 | 5 | 0 | 1 | 4 |
| TotDM | 6 | 6 | 0 | 2.0–5.4 | 1 | 5 | 1 | 0 | 1 | 5 | 0 | 5 | 1 |
| Morphological traits | |||||||||||||
| CL | 15 | 12 | 3 | 2.5–9.5 | 4 | 11 | 0 | 1 | 6 | 7 | 2 | 10 | 5 |
| SpL | 10 | 10 | 0 | 2.0–10.8 | 6 | 4 | 1 | 1 | 2 | 7 | 1 | 7 | 3 |
| FLL | 9 | 7 | 2 | 2.0–7.7 | 3 | 6 | 0 | 0 | 4 | 5 | 0 | 6 | 3 |
| FLW | 17 | 13 | 4 | 2.1–32.4 | 8 | 9 | 0 | 2 | 5 | 9 | 3 | 14 | 3 |
| Drought adaptive physiological traits | |||||||||||||
| δ13C | 8 | 6 | 2 | 2.2–9.3 | 7 | 1 | 1 | 0 | 1 | 7 | 0 | 4 | 4 |
| OP | 9 | 7 | 2 | 2.3–4.9 | 1 | 8 | 0 | 3 | 0 | 7 | 1 | 7 | 2 |
| Chl | 13 | 10 | 3 | 2.0–7.0 | 7 | 6 | 0 | 2 | 1 | 7 | 5 | 11 | 2 |
| LR | 5 | 3 | 2 | 2.1–5.9 | 3 | 2 | 0 | 0 | 1 | 4 | 0 | 3 | 2 |
| Phenology related traits | |||||||||||||
| DP-H | 10 | 8 | 2 | 2.4–40.8 | 3 | 7 | 0 | 0 | 10 | 0 | 0 | 5 | 5 |
| DP-M | 8 | 8 | 0 | 3.0–13.6 | 4 | 4 | 1 | 0 | 5 | 3 | 0 | 6 | 2 |
| All | 78 | 44 | 34 | 2.0–40.8 | -- | -- | -- | -- | 15 | 41 | 21 | 46 | 33 |
| Trait | ITV Allele of LDN | ITV Allele of G18-16 | ||
|---|---|---|---|---|
| Number of QTLs | Total PEV * | Number of QTLs | Total PEV * | |
| GY | 0 | 0 | 1 | 0.21 |
| TKW | 1 | 0.13 | 3 | 0.37 |
| KNSP | 0 | 0 | 2 | 0.26 |
| HI | 3 | 0.33 | 3 | 0.39 |
| SpDM | 2 | 0.22 | 2 | 0.29 |
| VegDM | 3 | 0.31 | 1 | 0.13 |
| TotDM | 1 | 0.14 | 0 | 0 |
| CL | 4 | 0.46 | 1 | 0.11 |
| SpL | 2 | 0.17 | 1 | 0.17 |
| FLL | 2 | 0.21 | 1 | 0.14 |
| FLW | 1 | 0.12 | 2 | 0.21 |
| δ13C | 0 | 0 | 4 | 0.53 |
| OP ** | 2 | 0.30 | 0 | 0 |
| Chl | 1 | 0.09 | 1 | 0.09 |
| LR | 2 | 0.24 | 0 | 0 |
| DH-M | 1 | 0.12 | 2 | 0.32 |
| Drought Strategy | # QTLs | # QTLs Plastic to Drought | Allele Responsible for Drought Resistance | Effect on Productivity *** | |||
|---|---|---|---|---|---|---|---|
| G18-16 | LDN | − | 0 | + | |||
| Escape | 4 | 4 | 2 (2) | 2 (2) | 0 | 0 | 4 |
| Avoidance | 13 | 5 | 10 (4) | 3 (1) | 2 | 9 | 2 |
| Tolerance | 22 | 8 | 15 (5) * | 7 (3) | 1 | 13 | 8 |
| Total | 33 ** | 17 | 26 (11) | 11 (6) | 3 | 22 | 13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatiukha, A.; Deblieck, M.; Klymiuk, V.; Merchuk-Ovnat, L.; Peleg, Z.; Ordon, F.; Fahima, T.; Korol, A.; Saranga, Y.; Krugman, T. Genomic Architecture of Phenotypic Plasticity in Response to Water Stress in Tetraploid Wheat. Int. J. Mol. Sci. 2021, 22, 1723. https://doi.org/10.3390/ijms22041723
Fatiukha A, Deblieck M, Klymiuk V, Merchuk-Ovnat L, Peleg Z, Ordon F, Fahima T, Korol A, Saranga Y, Krugman T. Genomic Architecture of Phenotypic Plasticity in Response to Water Stress in Tetraploid Wheat. International Journal of Molecular Sciences. 2021; 22(4):1723. https://doi.org/10.3390/ijms22041723
Chicago/Turabian StyleFatiukha, Andrii, Mathieu Deblieck, Valentyna Klymiuk, Lianne Merchuk-Ovnat, Zvi Peleg, Frank Ordon, Tzion Fahima, Abraham Korol, Yehoshua Saranga, and Tamar Krugman. 2021. "Genomic Architecture of Phenotypic Plasticity in Response to Water Stress in Tetraploid Wheat" International Journal of Molecular Sciences 22, no. 4: 1723. https://doi.org/10.3390/ijms22041723
APA StyleFatiukha, A., Deblieck, M., Klymiuk, V., Merchuk-Ovnat, L., Peleg, Z., Ordon, F., Fahima, T., Korol, A., Saranga, Y., & Krugman, T. (2021). Genomic Architecture of Phenotypic Plasticity in Response to Water Stress in Tetraploid Wheat. International Journal of Molecular Sciences, 22(4), 1723. https://doi.org/10.3390/ijms22041723








