Long-Range PCR-Based NGS Applications to Diagnose Mendelian Retinal Diseases
Abstract
1. Introduction
2. Results
2.1. First-Tier Assay for Probands without Previous Genetic Testing
2.2. Tackling Missing Heritability
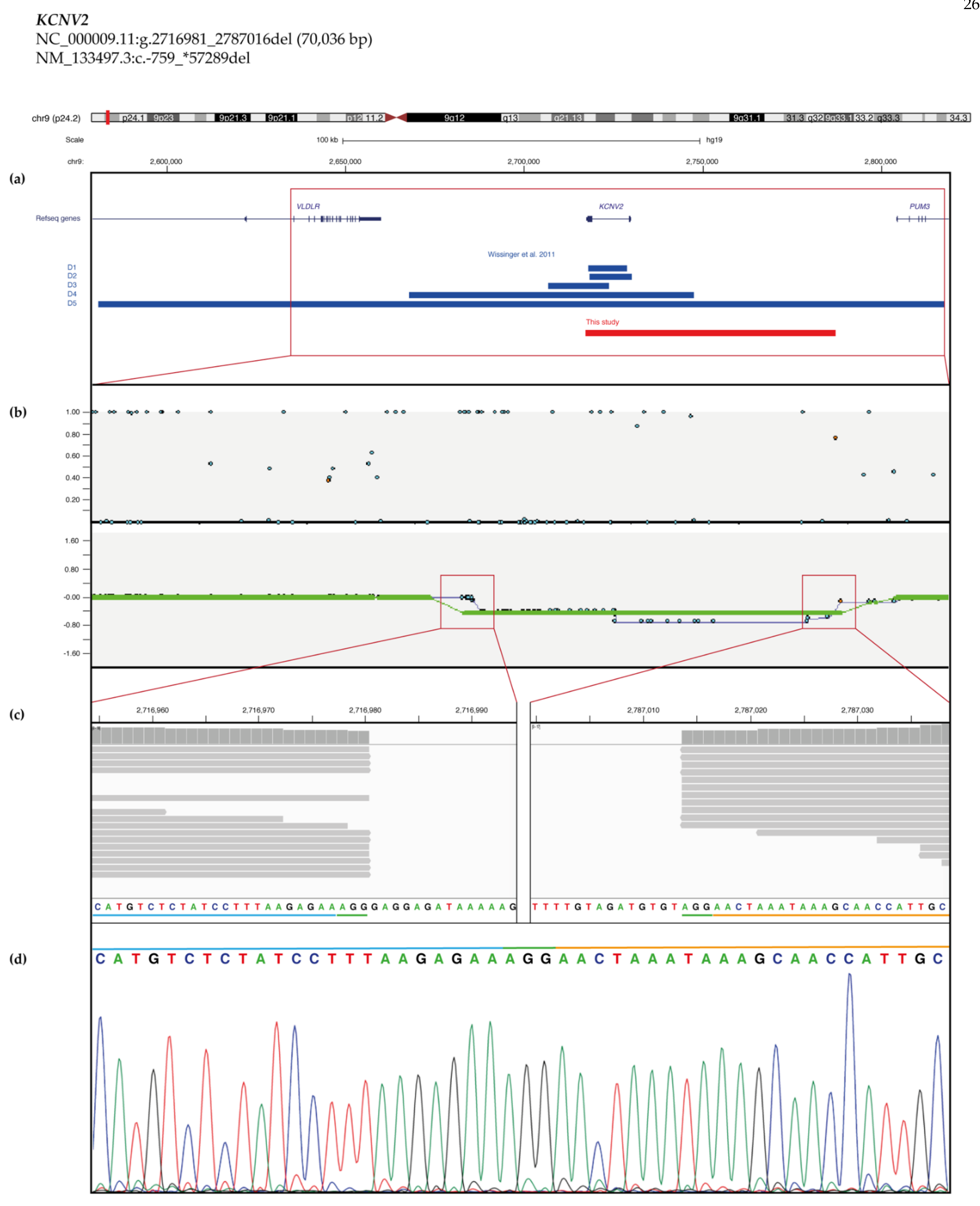
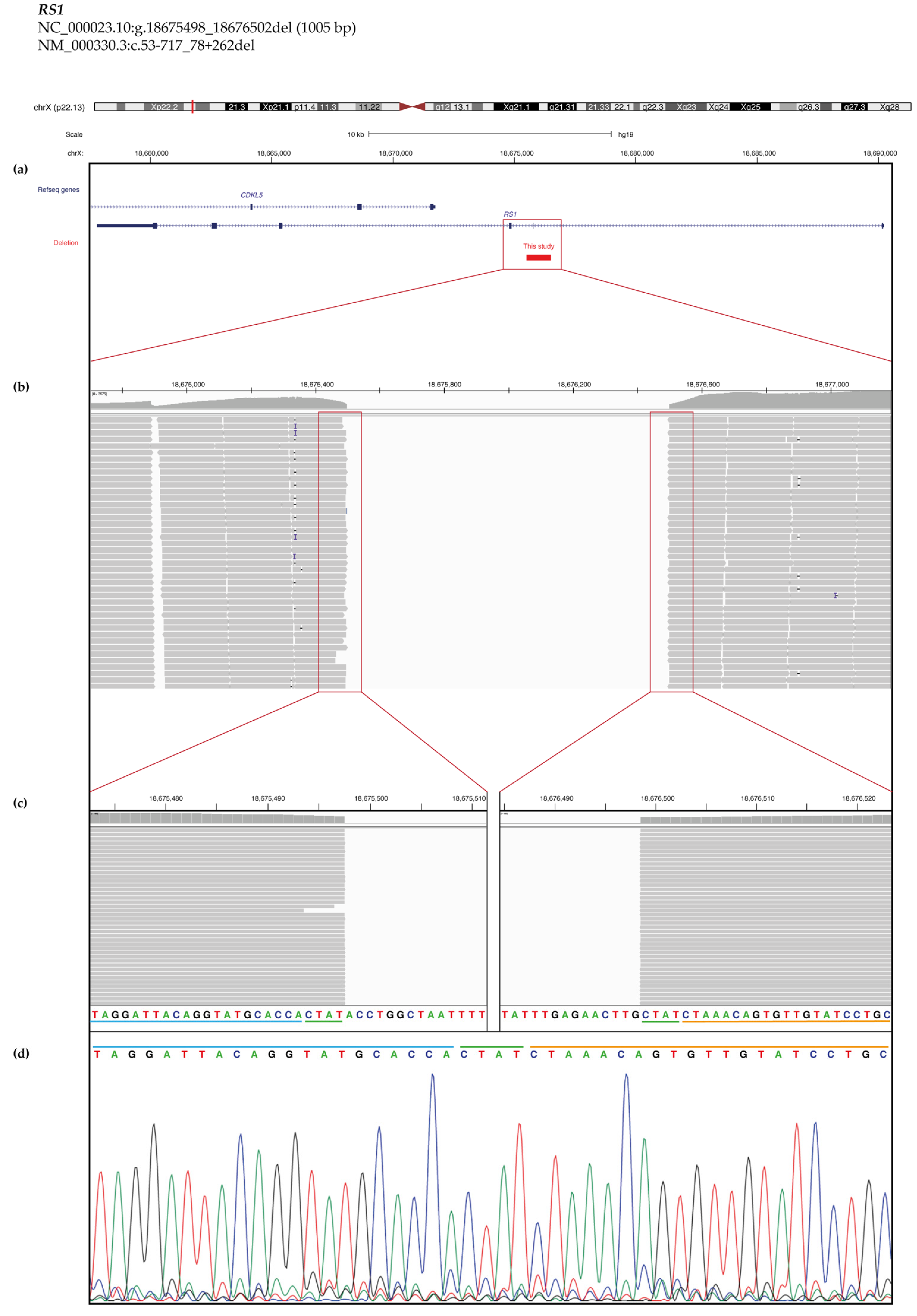
2.3. Copy Number Variant (CNV) Characterization
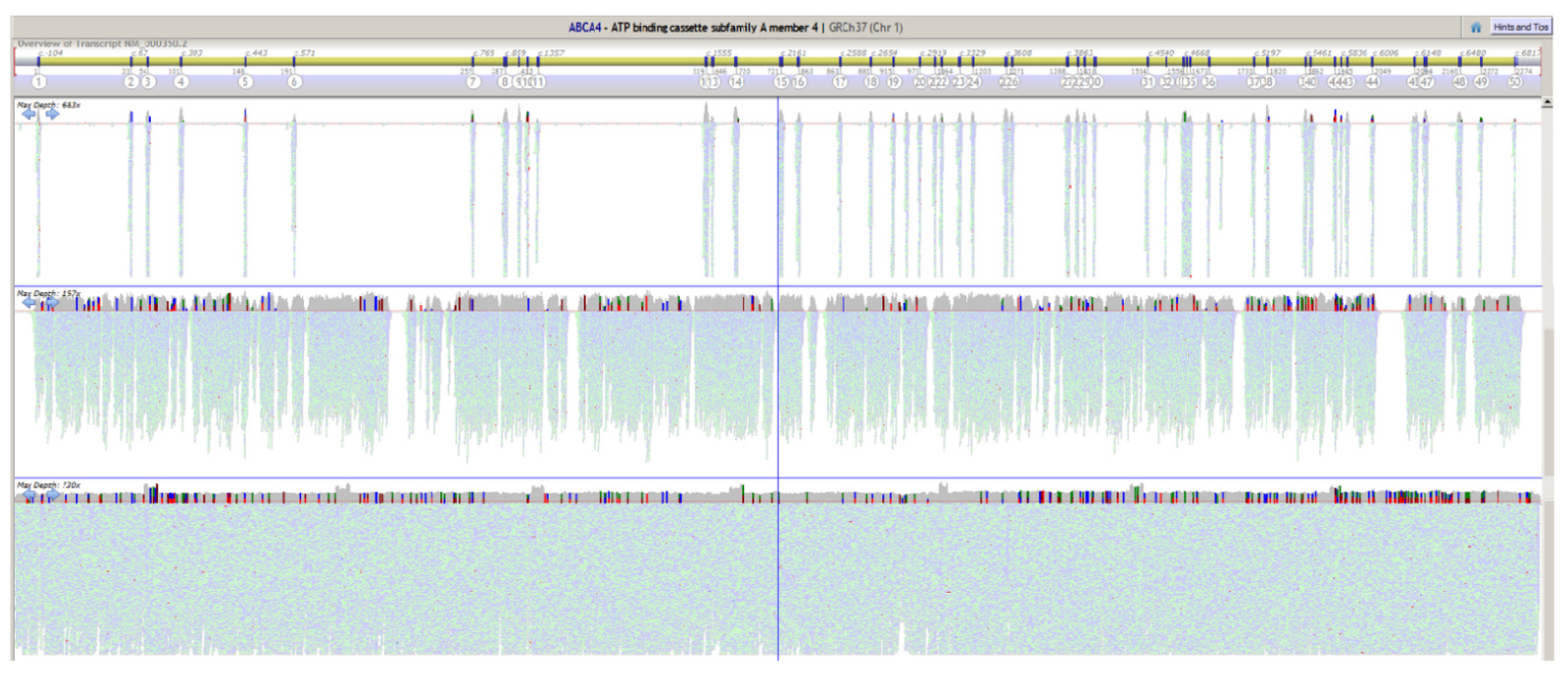
2.4. Exome Spike-In
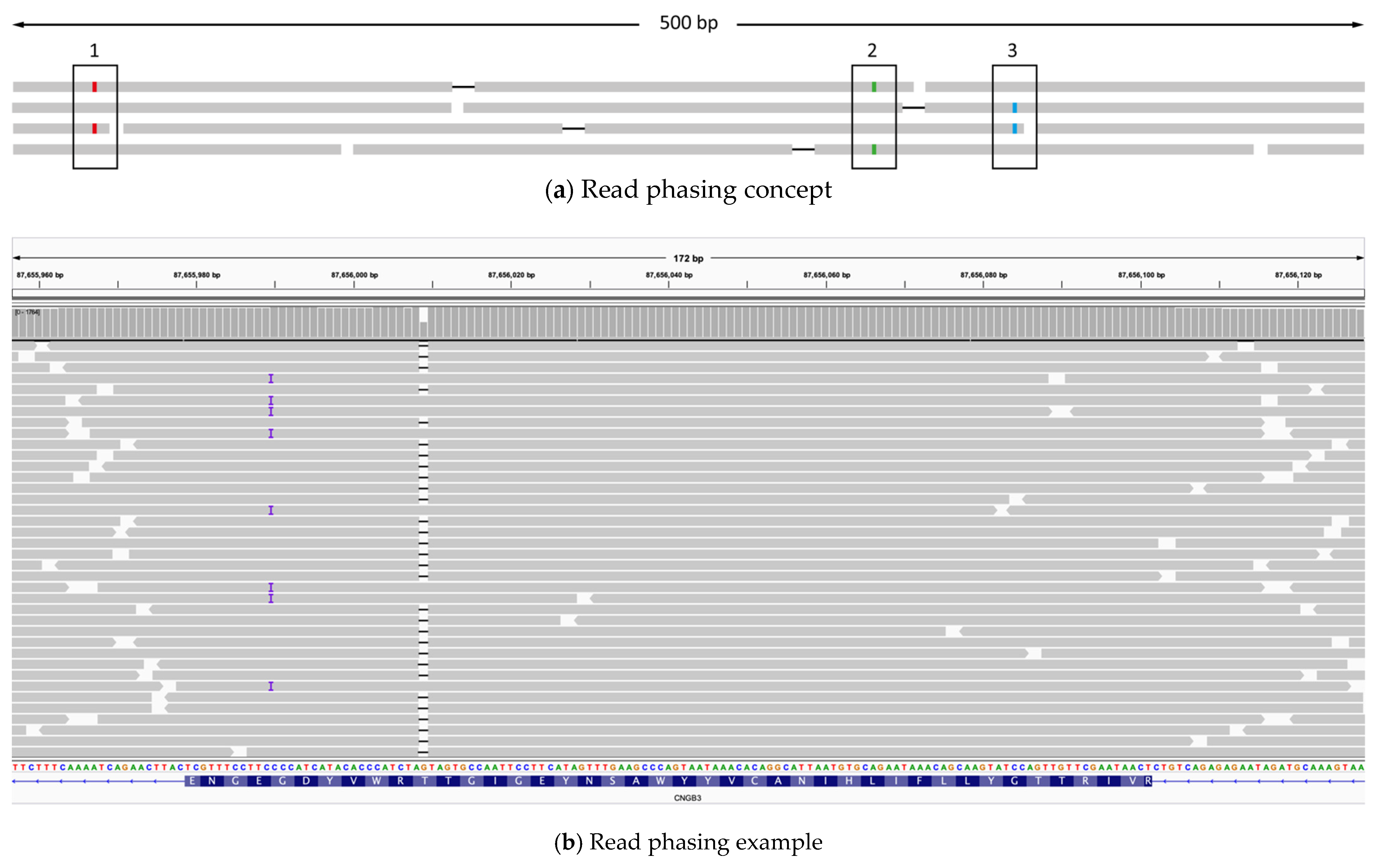
2.5. Read Phasing
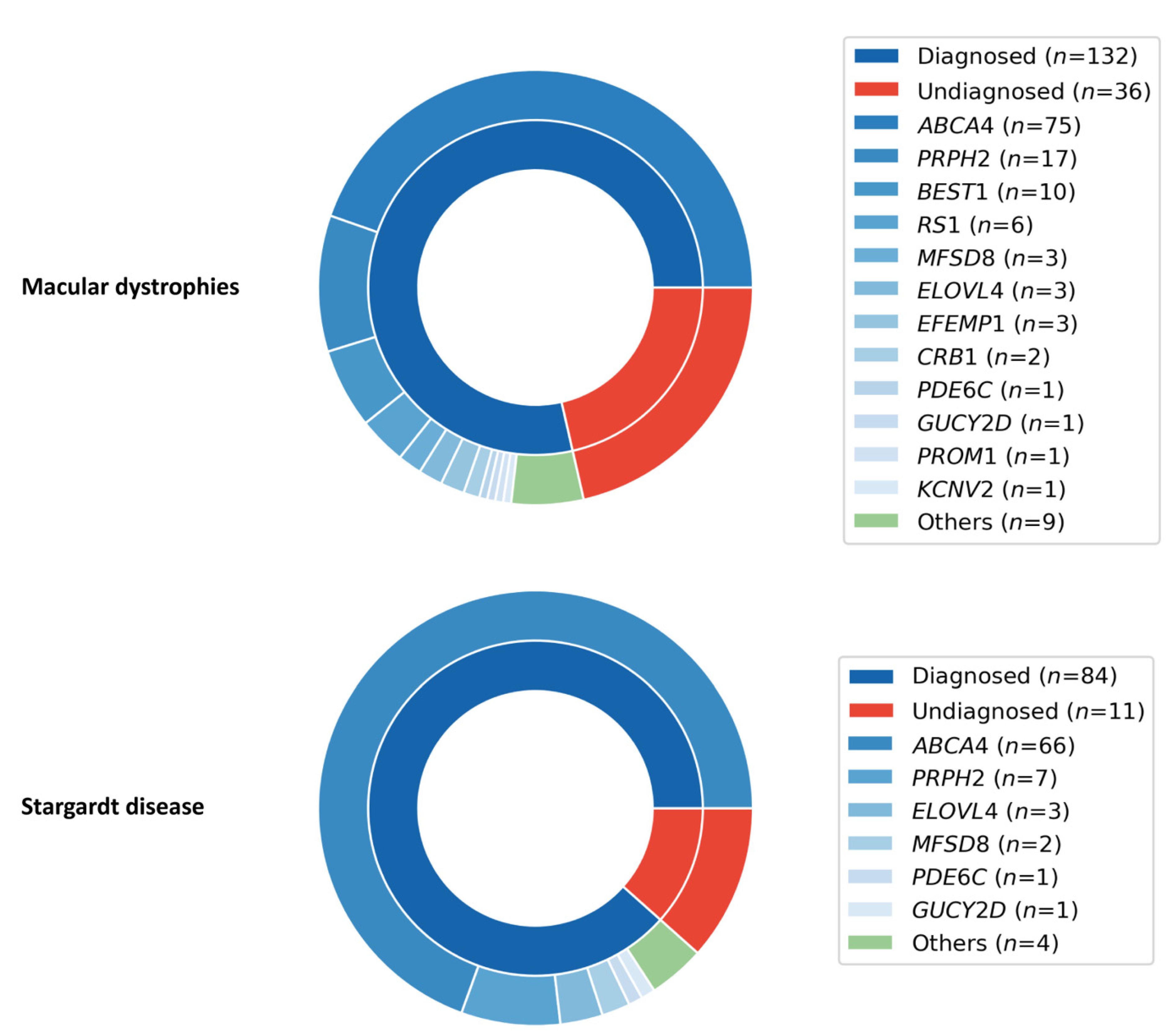
2.6. Macular Dystrophies (MD) Cohort
3. Discussion
4. Materials and Methods
4.1. Patients and Family Members
4.2. Genomic DNA
4.3. Long-Range PCR primers
4.4. Long-Range PCR
4.5. PCR Quality Control and Pooling
4.6. Library Construction and Sequencing
4.7. ABCA4 Capture Sequencing
4.8. Whole-Exome Sequencing
4.9. Exome Spike-In Applications
4.10. Sequencing Data Analysis
4.11. CNV Validation and Breakpoints Characterization
4.12. Chromosomal Phasing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| ID | Diagnosis | Age at Referral | Sex | Locus | Variant 1 cNomen | ACMG | HGMD | Ref | Variant 2 cNomen | ACMG | HGMD | Ref | Phasing |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S100 | LCA | 21–25 | F | GUCY2D | NM_000180.3:c.129_134del | 3 | Pat | [36] | NM_000180.3:c.929C > A | 3 | NA | ||
| S101 | STGD | 26–30 | F | ABCA4 | NM_000350.2:c.[1622T > C;3113C > T] | 4 | Pat | [25] | NM_000350.2:c.4139C > T | 4 | Pat | [37] | Yes (S) |
| S102 | MD | 21–25 | F | CRB1 | NM_201253.2:c.1472A > T | 3 | NA | NM_201253.2:c.2298G > A | 5 | NA | |||
| S103 | STGD | 61–65 | F | ABCA4 | NM_000350.2:c.5381C > A | 3 | Pat | [38] | NM_000350.2:c.6079C > T | 4 | Pat | [25] | |
| S104 | STGD | 46–50 | F | ABCA4 | NM_000350.2:c.5413A > G | 4 | Pat | [39] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S105 | COD | 71–75 | M | PRPH2 | NM_000322.4:c.94A > G | 2 | Pat | [41] | Yes (S) | ||||
| S106 | STGD | 36–40 | M | ABCA4 | NM_000350.2:c.[2588G > C;5603A > T] | 3 | Pat | [25] | NM_000350.2:c.[5714 + 5G > A;5603A > T] | 3 | Pat | [42] | |
| S107 | RP | 16–20 | F | ABCA4 | NM_000350.2:c.6658C > T | 5 | Pat | [43] | NM_000350.2:c.6658C > T | 5 | Pat | [43] | |
| S108 | COD | 11–15 | F | ABCA4 | NM_000350.2:c.571-2A > T | 5 | Pat | [44] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S109 | STGD | 16–20 | F | ABCA4 | NM_000350.2:c.2291G > A | 4 | Pat | [45] | NM_000350.2:c.5714 + 5G > A | 3 | Pat | [42] | |
| S110 | STGD | 21–25 | M | ABCA4 | NM_000350.2:c.[1622T > C;3113C > T] | 4 | Pat | [25] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | Yes (S) |
| S111 | COD | 26–30 | M | BEST1 | NM_004183.3:c.422G > A | 3 | Pat | [46] | NM_004183.3:c.422G > A | 3 | Pat | [46] | Yes (S) |
| S112 | RP | 21–25 | M | RPGR | NM_001034853.1:c.2426_2427del | 3 | Pat | [47] | Yes (S) | ||||
| S113 | MD | 46–50 | F | PRPH2 | NM_000322.4:c.514C > T | 4 | Pat | [48] | |||||
| S114 | MD | 36–40 | M | PRPH2 | NM_000322.4:c.514C > T | 4 | Pat | [48] | |||||
| S115 | STGD | 21–25 | F | ABCA4 | NM_000350.2:c.[5461–10T > C;5603A > T] | 3 | Pat | [49] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | Yes (S) |
| S116 | BEST | 16–20 | M | BEST1 | NM_004183.3:c.658C > T | 5 | Pat | [50] | NM_004183.3:c.658C > T | 5 | Pat | [50] | Yes (S) |
| S117 | STGD | 66–70 | F | ABCA4 | NM_000350.2:c.[2588G > C;5603A > T] | 3 | Pat | [25] | NM_000350.2:c.5714 + 5G > A | 3 | Pat | [42] | Yes (R) |
| S118 | COD | 6–10 | F | KCNV2 | NC_000009.11:g.2716981_2787016del | 5 | NA | NM_133497.3:c.1381G > A | 3 | Pat | [51] | Yes (S) | |
| S119 | MD | 41–45 | F | PRPH2 | NM_000322.4:c.514C > T | 4 | Pat | [48] | Yes (S) | ||||
| S120 | XLRS | 21–25 | M | RS1 | NM_000330.3:c.53–713_78 + 266del | 4 | NA | Yes (S) | |||||
| S121 | STGD | 11–15 | M | ABCA4 | NM_000350.2:c.5942C > G | 5 | Pat | [52] | NM_000350.2:c.6323_6331delinsGGC | 4 | NA | ||
| S122 | STGD | 16–20 | M | ABCA4 | NM_000350.2:c.3323del | 5 | NA | NM_000350.2:c.5377G > A | 3 | P Pat | [12] | ||
| S123 | STGD | 46–50 | F | ABCA4 | NM_000350.2:c.768G > T | 4 | Pat | [13] | NM_000350.2:c.4739T > C | 4 | Pat | [53] | Yes (S) |
| S124 | LCA | 0–5 | M | CEP290 | NM_025114.3:c.2991 + 1655A > G | 3 | Pat | [54] | NM_025114.3:c.6604del | 5 | Pat | [55] | Yes (S) |
| S125 | STGD | 16–20 | F | ABCA4 | NM_000350.2:c.[1610G > A;5603A > T] | 1 | Pat | [56] | NM_000350.2:c.3523-1G > A | 5 | Pat | [21] | Yes (S) |
| S126 | STGD | 26–30 | M | ABCA4 | NM_000350.2:c.5882G > A | 3 | Pat | [40] | NM_000350.2:c.6088C > T | 5 | Pat | [37] | |
| S127 | ML | 46–50 | M | PRPH2 | NM_000322.4:c.514C > T | 4 | Pat | [48] | |||||
| S128 | RP | 6–10 | M | RP2 | NM_006915.2:c.884-9T > A | 3 | Pat | [57] | Yes (S) | ||||
| S129 | STGD | 41–45 | F | ABCA4 | NM_000350.2:c.2291G > A | 4 | Pat | [45] | NM_000350.2:c.5381C > A | 3 | Pat | [38] | |
| S130 | COD | 21–25 | F | CNGB3 | NM_019098.4:c.1148del | 5 | Pat | [58] | NM_019098.4:c.1148del | 5 | Pat | [58] | |
| S131 | BEST | 16–20 | M | BEST1 | NM_004183.3:c.728C > T | 4 | Pat | [59] | |||||
| S132 | STGD | 21–25 | F | ABCA4 | NM_000350.2:c.61C > T | 5 | Pat | [60] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S133 | MD | 21–25 | F | PRPH2 | NM_000322.4:c.514C > T | 4 | Pat | [48] | Yes (S) | ||||
| S134 | MD | 31–35 | F | BEST1 | NM_004183.3:c.584C > T | 3 | Pat | [61] | NM_004183.3:c.584C > T | 3 | Pat | [61] | |
| S135 | MD | 36–40 | M | PRPH2 | NM_000322.4:c.514C > T | 4 | Pat | [48] | |||||
| S136 | MD | 46–50 | M | PRPH2 | NM_000322.4:c.514C > T | 4 | Pat | [48] | |||||
| S137 | BEST | 26–30 | M | BEST1 | NM_004183.3:c.-37 + 1G > T | 5 | Pat | [62] | NM_004183.3:c.-37 + 1G > T | 5 | Pat | [62] | Yes (S) |
| S138 | CRD | 36–40 | M | PROM1 | NM_006017.2:c.380G > A | 3 | Pat | [63] | NM_006017.2:c.380G > A | 3 | Pat | [63] | |
| S139 | RD | 41–45 | F | ABCA4 | NM_000350.2:c.[52C > T;5603A > T] | 4 | Pat | [64] | NM_000350.2:c.[52C > T;5603A > T] | 4 | Pat | [64] | |
| S140 | LCA | 0–5 | F | CRB1 | NM_201253.2:c.2230C > T | 5 | Pat | [65] | NM_201253.2:c.2230C > T | 5 | Pat | [65] | |
| S141 | CRD | 31–35 | M | ABCA4 | NM_000350.2:c.1804C > T | 5 | Pat | [37] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S142 | LCA | 0–5 | F | CRB1 | NM_201253.2:c.547T > C | 3 | Pat | [63] | NM_201253.2:c.2687G > C | 4 | Pat | [63] | |
| S143 | MD | 46–50 | F | ABCA4 | NM_000350.2:c.514G > A | 3 | Pat | [56] | NM_000350.2:c.676C > A | 3 | NA | ||
| S144 | MD | 31–35 | M | ABCA4 | NM_000350.2:c.1957C > T | 5 | Pat | [45] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S145 | RP | 36–40 | F | ABCA4 | NM_000350.2:c.4873C > T | 3 | Pat | [63] | NM_000350.2:c.4873C > T | 3 | Pat | [63] | |
| S146 | MD | 26–30 | F | ABCA4 | NM_000350.2:c.[1610G > A;5603A > T] | 1 | Pat | [56] | NM_000350.2:c.[5461-10T > C;5603A > T] | 3 | Pat | [49] | Yes (S) |
| S147 | CRD | 21–25 | M | ABCA4 | NM_000350.2:c.[5461–10T > C;5603A > T] | 3 | Pat | [49] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S148 | STGD | 46–50 | M | ABCA4 | NM_000350.2:c.4873C > T | 3 | Pat | [63] | NM_000350.2:c.5714 + 5G > A | 3 | Pat | [42] | |
| S149 | RD | 21–25 | M | ABCA4 | NM_000350.2:c.4462T > C | 5 | Pat | [37] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S150 | STGD | 31–35 | F | ABCA4 | NM_000350.2:c.2932G > A | 4 | P Pat | [12] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S151 | RP | 11–15 | F | ABCA4 | NM_000350.2:c.1988G > A | 5 | Pat | [45] | NM_000350.2:c.2160 + 1G > T | 5 | Pat | [63] | Yes (R) |
| S152 | STGD | 16–20 | F | ABCA4 | NM_000350.2:c.1957C > T | 5 | Pat | [45] | NM_000350.2:c.5691G > T | 3 | NA | Yes (S) | |
| S153 | STGD | 31–35 | F | ELOVL4 | NM_022726.3:c.810C > G | 5 | Pat | [66] | |||||
| S154 | MD | 31–35 | M | CRB1 | NM_201253.2:c.498_506del | 3 | Pat | [67] | NM_201253.2:c.2290C > T | 4 | Pat | [68] | Yes (S) |
| S155 | RP | 51–55 | M | RP1 | NM_006269.1:c.2613dup | 5 | Pat | [69] | Yes (S) | ||||
| S156 | RP | 21–25 | M | RPGR | NM_001034853.1:c.2586_2587del | 3 | Pat | [70] | |||||
| S157 | RP | 11–15 | F | RPGR | NM_001034853.1:c.2008_2017del | 4 | Pat | [63] | Yes (S) | ||||
| S158 | MD | 36–40 | M | PRPH2 | NM_000322.4:c.514C > T | 4 | Pat | [48] | |||||
| S159 | RD | NA | M | ABCA4 | NM_000350.2:c.4352 + 1G > A | 5 | Pat | [71] | NM_000350.2:c.4919G > A | 4 | Pat | [72] | |
| S160 | STGD | 51–55 | F | PRPH2 | NM_000322.4:c.514C > T | 4 | Pat | [48] | |||||
| S161 | STGD | 46–50 | F | ABCA4 | NM_000350.2:c.2401G > A | 3 | P Pat | [73] | NM_000350.2:c.5381C > A | 3 | Pat | [38] | |
| S162 | STGD | 26–30 | M | ABCA4 | NM_000350.2:c.5882G > A | 3 | Pat | [40] | NM_000350.2:c.6122G > A | 4 | Pat | [74] | |
| S163 | MD | 26–30 | F | ABCA4 | NM_000350.2:c.2041C > T | 5 | Pat | [43] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | Yes (S) |
| S164 | STGD | 41–45 | F | ABCA4 | NM_000350.2:c.5329A > T | 3 | P Pat | [75] | NM_000350.2:c.[5461-10T > C;5603A > T] | 3 | Pat | [49] | Yes (S) |
| S165 | STGD | 56–60 | F | ABCA4 | NM_000350.2:c.4958G > A | 3 | NA | NM_000350.2:c.5882G > A | 3 | Pat | [40] | ||
| S166 | RP | 21–25 | M | RPGR | NM_001034853.1:c.2236_2237del | 4 | Pat | [47] | |||||
| S167 | BEST | 51–55 | M | BEST1 | NM_004183.3:c.388C > A | 4 | Pat | [76] | NM_004183.3:c.638A > G | 4 | Pat | [77] | |
| S168 | STGD | 26–30 | F | ABCA4 | NM_000350.2:c.3179A > C | 4 | NA | NM_000350.2:c.3179A > C | 4 | NA | |||
| S169 | STGD | 6–10 | M | ABCA4 | NM_000350.2:c.160T > G | 4 | Pat | [78] | NM_000350.2:c.160T > G | 4 | Pat | [78] | Yes (S) |
| S170 | BEST | 6–10 | F | BEST1 | NM_004183.3:c.907G > T | 4 | NA | Yes (S) | |||||
| S171 | STGD | 41–45 | M | ELOVL4 | NM_022726.3:c.810C > G | 5 | Pat | [66] | |||||
| S172 | STGD | 26–30 | M | ABCA4 | NM_000350.2:c.5882G > A | 3 | Pat | [40] | NM_000350.2:c.6282 + 1G > C | 5 | Pat | [79] | Yes (S) |
| S173 | STGD | 46–50 | F | ABCA4 | NM_000350.2:c.247_250dup | 5 | Pat | [80] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S174 | STGD | 36–40 | F | ABCA4 | NM_000350.2:c.1742C > A | 3 | NA | NM_000350.2:c.5882G > A | 3 | Pat | [40] | ||
| S175 | RP | 11–15 | M | RPGR | NM_001034853.1:c.2819_2838dup | 3 | NA | ||||||
| S176 | STGD | 21–25 | F | ABCA4 | NM_000350.2:c.4609del | 4 | NA | NM_000350.2:c.5882G > A | 3 | Pat | [40] | ||
| S177 | XLRS | 41–45 | M | RS1 | NM_000330.3:c.304C > T | 4 | Pat | [81] | |||||
| S178 | STGD | 16–20 | M | ABCA4 | NM_000350.2:c.4383G > C | 3 | NA | NM_000350.2:c.5882G > A | 3 | Pat | [40] | Yes (S) | |
| S179 | STGD | 21–25 | F | ABCA4 | NM_000350.2:c.[3322C > T;6320G > A] | 4 | Pat | [82] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | Yes (S) |
| S180 | STGD | 46–50 | M | ELOVL4 | NM_022726.3:c.810C > G | 5 | Pat | [66] | |||||
| S181 | CRD | 31–35 | F | ABCA4 | NM_000350.2:c.727_728dup | 5 | NA | NM_000350.2:c.735T > G | 5 | Pat | [83] | Yes (R) | |
| S182 | STGD | 26–30 | M | ABCA4 | NM_000350.2:c.5413A > G | 4 | Pat | [39] | NM_000350.2:c.6428T > A | 4 | NA | Yes (S) | |
| S183 | STGD | 51–55 | F | CDHR1 | NM_033100.3:c.783G > A | 1 | Pat | [13] | NM_033100.3:c.783G > A | 1 | Pat | [13] | |
| S184 | STGD | 41–45 | F | ABCA4 | NM_000350.2:c.5461-6T > C | 3 | NA | NM_000350.2:c.5882G > A | 3 | Pat | [40] | Yes (S) | |
| S185 | RP | 6–10 | M | SNRNP200 | NM_014014.4:c.1634G > A | 3 | Pat | [84] | NM_014014.4:c.1634G > A | 3 | Pat | [84] | |
| S186 | CRD | 16–20 | F | PROM1 | NM_006017.2:c.1142-1G > A | 5 | P Pat | [13] | NM_006017.2:c.1142–1G > A | 5 | P Pat | [13] | Yes (S) |
| S187 | MD | 16–20 | F | ABCA4 | NM_000350.2:c.5018 + 2T > C | 5 | Pat | [85] | NM_000350.2:c.5018 + 2T > C | 5 | Pat | [85] | |
| S188 | STGD | 26–30 | M | ABCA4 | NM_000350.2:c.454C > T | 5 | Pat | [86] | NM_000350.2:c.[2588G > C;5603A > T] | 3 | Pat | [25] | Yes (S) |
| S189 | MD | 26–30 | M | KCNV2 | NM_133497.3:c.1096del | 4 | NA | NM_133497.3:c.1381G > A | 3 | Pat | [51] | Yes (S) | |
| S190 | STGD | 76–80 | M | ABCA4 | NM_000350.2:c.2041C > T | 5 | Pat | [43] | NM_000350.2:c.5603A > T | 3 | Ass | [18] | |
| S191 | BEST | 56–60 | M | BEST1 | NM_004183.3:c.73C > T | 4 | Pat | [46] | |||||
| S192 | COD | 21–25 | M | CNGB3 | NM_019098.4:c.1578 + 1G > A | 5 | Pat | [87] | NM_019098.4:c.1578 + 1G > A | 5 | Pat | [87] | |
| S193 | BEST | 46–50 | M | BEST1 | NM_004183.3:c.653G > A | 4 | Pat | [61] | |||||
| S194 | STGD | 76–80 | M | ABCA4 | NM_000350.2:c.5714 + 5G > A | 3 | Pat | [42] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S195 | STGD | 46–50 | M | ABCA4 | NM_000350.2:c.1846G > A | 4 | Pat | [53] | NM_000350.2:c.4328G > A | 4 | Pat | [45] | |
| S196 | STGD | 11–15 | F | ABCA4 | NM_000350.2:c.1A > G | 5 | Pat | [88] | NM_000350.2:c.[1622T > C;3113C > T] | 4 | Pat | [25] | |
| S197 | RP | 56–60 | M | RP1L1 | NM_178857.5:c.1024_1026delinsCTCCT | 4 | NA | ||||||
| S198 | RP | 56–60 | F | RP1 | NM_006269.1:c.2613dup | 5 | Pat | [69] | |||||
| S199 | RP | 61–65 | M | PRPH2 | NM_000322.4:c.512T > G | 4 | NA | ||||||
| S200 | ML | 76–80 | M | EFEMP1 | NM_001039348.2:c.1033C > T | 3 | Pat | [89] | |||||
| S201 | RD | 36–40 | M | ABCA4 | NM_000350.2:c.184C > T | 4 | Pat | [21] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | Yes (S) |
| S202 | RP | 46–50 | M | RP1L1 | NM_178857.5:c.196G > C | 3 | NA | ||||||
| S203 | CRD | 51–55 | M | PROM1 | NM_006017.2:c.2476G > C | 3 | NA | NM_006017.2:c.2476G > C | 3 | NA | |||
| S204 | CRD | 41–45 | M | ELOVL4 | NM_022726.3:c.810C > G | 5 | Pat | [66] | |||||
| S205 | RP | 16–20 | F | RP1 | NM_006269.1:c.-12-1431_*286del | 4 | Pat | [27] | NM_006269.1:c.-12-1431_*286del | 4 | Pat | [27] | Yes (S) |
| S206 | RD | 31–35 | M | CRX | NM_000554.5:c.118C > T | 4 | Pat | [90] | |||||
| S207 | STGD | 16–20 | F | ABCA4 | NM_000350.2:c.[5461-10T > C;5603A > T] | 3 | Pat | [49] | NM_000350.2:c.5714 + 5G > A | 3 | Pat | [42] | Yes (S) |
| ID | Diagnosis | Age at Referral | Sex | Locus | Variant 1 cNomen | ACMG | HGMD | Ref | Variant 2 cNomen | ACMG | HGMD | Ref | Phasing |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S209 | STGD | 21–25 | F | KCNJ13 | NM_002242.4:c.484C > T | 3 | Pat | [91] | Yes (S) | ||||
| S210 | ML | 36–40 | F | EFEMP1 | NM_001039348.2:c.1033C > T | 3 | Pat | [89] | Yes (S) | ||||
| S211 | STGD | 56–60 | M | ABCA4 | NM_000350.2:c.3210_3211dup | 5 | Pat | [25] | NM_000350.2:c.5603A > T | 3 | Ass | [18] | |
| S212 | MD | 36–40 | M | OPA1 | NM_130837.2:c.1890_1891del | 5 | Pat | [92] | |||||
| S213 | CRD | 21–25 | M | ABCA4 | NM_000350.2:c.160T > G | 4 | Pat | [78] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S214 | STGD | 46–50 | F | PRPH2 | NM_000322.4:c.424C > T | 4 | Pat | [93] | |||||
| S215 | MD | 36–40 | M | ABCA4 | NM_000350.2:c.4139C > T | 4 | Pat | [37] | NM_000350.2:c.5714 + 5G > A | 3 | Pat | [42] | |
| S216 | MD | 46–50 | F | OPA1 | NM_130837.2:c.2987A > C | 4 | NA | ||||||
| S217 | STGD | 26–30 | M | PDE6C | NM_006204.3:c.864 + 1G > A | 5 | Pat | [27] | NM_006204.3:c.864 + 1G > A | 5 | Pat | [27] | |
| S218 | STGD | 6–10 | F | ABCA4 | NM_000350.2:c.6731T > A | 3 | NA | NM_000350.2:c.6731T > A | 3 | NA | Yes (S) | ||
| S219 | STGD | 21–25 | F | ABCA4 | NM_000350.2:c.2401G > A | 3 | P Pat | [73] | NM_000350.2:c.5018 + 2T > C | 5 | Pat | [85] | |
| S220 | ACHR | 0–5 | F | CNGB3 | NM_019098.4:c.1148del | 5 | Pat | [58] | NM_019098.4:c.1167_1168insC | 4 | NA | Yes (R) | |
| S221 | STGD | 41–45 | M | ABCA4 | NM_000350.2:c.428del | 5 | Pat | [94] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S222 | STGD | 36–40 | M | ABCA4 | NM_000350.2:c.(2918 + 765_2918 + 775)_(3328 + 618_3328 + 662)del | 5 | Pat | [18]? | NM_000350.2:c.5603A > T | 3 | Ass | [18] | |
| S223 | ML | 41–45 | M | CNGB1 | NM_001297.4:c.1658C > A | 3 | NA | NM_001297.4:c.2662G > A | 3 | NA | |||
| S224 | STGD | 26–30 | M | PRPH2 | NM_000322.4:c.605G > A | 4 | NA | ||||||
| S225 | BEST | 41–45 | F | BEST1 | NM_004183.3:c.884_886del | 4 | Pat | [95] | |||||
| S226 | STGD | 36–40 | F | ABCA4 | NM_000350.2:c.[1622T > C;3113C > T] | 4 | Pat | [25] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S227 | STGD | 41–45 | M | ABCA4 | NM_000350.2:c.5196 + 2T > C | 5 | Pat | [25] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | Yes (S) |
| S228 | STGD | 11–15 | M | ABCA4 | NM_000350.2:c.5882G > A | 3 | Pat | [40] | NM_000350.2:c.6238_6239del | 5 | Pat | [96] | |
| S229 | STGD | 16–20 | M | MFSD8 | NM_152778.2:c.670A > T | 3 | NA | NM_152778.2:c.670A > T | 3 | NA | |||
| S230 | STGD | 51–55 | F | PRPH2 | NM_000322.4:c.422A > G | 3 | Pat | [97] | |||||
| S231 | MD | 51–55 | F | ABCA4 | NM_000350.2:c.2005_2006del | 5 | Pat | [37] | NM_000350.2:c.6148G > C | 4 | Pat | [25] | |
| S232 | STGD | 21–25 | F | ABCA4 | NM_000350.2:c.[2588G > C;5603A > T] | 3 | Pat | [25] | NM_000350.2:c.5762_5763del | 5 | NA | ||
| S233 | STGD | 6–10 | F | GUCY2D | NM_000180.3:c.2513G > A | 4 | Pat | [98] | |||||
| S234 | STGD | 66–70 | F | PRPH2 | NM_000322.4:c.611_626del | 5 | NA | ||||||
| S235 | STGD | 51–55 | M | PRPH2 | NM_000322.4:c.422A > G | 3 | Pat | [97] | |||||
| S236 | STGD | 46–50 | F | ABCA4 | NM_000350.2:c.(2918 + 765_2918 + 775)_(3328 + 618_3328 + 662)del | 5 | Pat | [18]? | NM_000350.2:c.5603A > T | 3 | Ass | [18] | |
| S237 | STGD | 46–50 | M | ABCA4 | NM_000350.2:c.4958G > A | 3 | NA | NM_000350.2:c.5882G > A | 3 | Pat | [40] | ||
| S238 | STGD | 16–20 | M | ABCA4 | NM_000350.2:c.5714 + 5G > A | 3 | Pat | [42] | NM_000350.2:c.5714 + 5G > A | 3 | Pat | [42] | |
| S239 | STGD | 56–60 | F | PRPH2 | NM_000322.4:c.422A > G | 3 | Pat | [97] | |||||
| S240 | STGD | 16–20 | M | ABCA4 | NM_000350.2:c.2401G > A | 3 | P Pat | [73] | NM_000350.2:c.5018 + 2T > C | 5 | Pat | [85] | |
| S241 | MD | 56–60 | M | PRPH2 | NM_000322.4:c.514C > T | 4 | Pat | [48] | |||||
| S242 | STGD | 41–45 | M | MFSD8 | NM_152778.2:c.754 + 2T > A | 5 | Pat | [99] | NM_152778.2:c.1006G > C | 1 | Pat | [100] | |
| S243 | MD | 36–40 | M | CYP4V2 | NM_207352.3:c.283G > A | 3 | Pat | [101] | NM_207352.3:c.1198C > T | 3 | Pat | [102] | Yes (S) |
| S244 | STGD | 26–30 | F | ABCA4 | NM_000350.2:c.[5461–10T > C;5603A > T] | 3 | Pat | [49] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S245 | MD | 21–25 | F | PROM1 | NM_006017.2:c.1142–1G > A | 5 | P Pat | [13] | NM_006017.2:c.1142-1G > A | 5 | P Pat | [13] | Yes (S) |
| S246 | XLRS | 11–15 | M | RS1 | NM_000330.3:c.598C > T | 4 | Pat | [103] | |||||
| S247 | ML | 26–30 | F | EFEMP1 | NM_001039348.2:c.1033C > T | 3 | Pat | [89] | |||||
| S248 | MD | 36–40 | M | OPA1 | NM_130837.2:c.2873_2876del | 5 | Pat | [104] | |||||
| S249 | STGD | 16–20 | M | ABCA4 | NM_000350.2:c.214G > A | 5 | Pat | [45] | NM_000350.2:c.1819G > A | 5 | Pat | [45] | |
| S250 | MD | 56–60 | F | PRPH2 | NM_000322.4:c.514C > T | 4 | Pat | [48] | |||||
| S251 | MD | 61–65 | M | MFSD8 | NM_152778.2:c.670A > T | 3 | NA | NM_152778.2:c.1006G > C | 1 | Pat | [100] | ||
| S252 | STGD | 26–30 | F | ABCA4 | NM_000350.2:c.5311G > A | 4 | Pat | [6] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S253 | XLRS | 26–30 | M | RS1 | NM_000330.3:c.544C > T | 4 | Pat | [103] | |||||
| S254 | BEST | 41–45 | M | BEST1 | NM_004183.3:c.653G > A | 4 | Pat | [61] | |||||
| S255 | XLRS | 36–40 | M | RS1 | NM_000330.3:c.150G > A | 5 | NA | ||||||
| S256 | STGD | 26–30 | M | ABCA4 | NM_000350.2:c.5603A > T | 3 | Ass | [18] | NM_000350.2:c.5172G > A | 5 | Pat | [26] | |
| S257 | STGD | 36–40 | F | ABCA4 | NM_000350.2:c.5690_5704del | 3 | NA | NM_000350.2:c.5882G > A | 3 | Pat | [40] | Yes (S) | |
| S258 | STGD | 26–30 | M | ABCA4 | NM_000350.2:c.1903C > T | 5 | Pat | [45] | NM_000350.2:c.2401G > A | 3 | P Pat | [73] | |
| S259 | XLRS | 41–45 | M | RS1 | NM_000330.3:c.209G > A | 4 | NA | ||||||
| S260 | COD | 41–45 | F | CDHR1 | NM_033100.3:c.783G > A | 1 | Pat | [13] | NM_033100.3:c.2522_2528del | 4 | Pat | [6] | |
| S261 | STGD | 46–50 | M | CNGA3 | NM_001298.2:c.1126G > A | 4 | Pat | [105] | NM_001298.2:c.1687C > T | 4 | Pat | [106] | |
| S262 | STGD | 26–30 | M | ABCA4 | NM_000350.2:c.[2588G > C;5603A > T] | 3 | Pat | [25] | NM_000350.2:c.4978C > T | 3 | Pat | [107] | |
| S263 | STGD | 51–55 | F | ABCA4 | NM_000350.2:c.[5461-10T > C;5603A > T] | 3 | Pat | [49] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S264 | STGD | 46–50 | F | ABCA4 | NM_000350.2:c.5311G > A | 4 | Pat | [6] | NM_000350.2:c.5603A > T | 3 | Ass | [18] | Yes (S) |
| S265 | MD | 36–40 | M | ABCA4 | NM_000350.2:c.4539 + 2064C > T | 2 | Pat | [20] | NM_000350.2:c.5882G > A | 3 | Pat | [40] | |
| S266 | STGD | 46–50 | M | ABCA4 | NM_000350.2:c.[2588G > C;5603A > T] | 3 | Pat | [25] | NM_000350.2:c.5018 + 2T > C | 5 | Pat | [85] | |
| S267 | STGD | 46–50 | F | ABCA4 | NM_000350.2:c.5316G > A | 5 | Pat | [88] | NM_000350.2:c.5603A > T | 3 | Ass | [18] | Yes (S) |
| S268 | COD | 26–30 | M | RGR | NM_002921.3:c.236G > A | 3 | NA | NM_002921.3:c.236G > A | 3 | NA | |||
| S269 | STGD | 26–30 | M | PCARE | NM_001029883.2:c.3002G > A | 5 | Pat | [108] | NM_001029883.2:c.3002G > A | 5 | Pat | [108] | |
| S270 | STGD | 46–50 | M | ABCA4 | NM_000350.2:c.5882G > A | 3 | Pat | [40] | NM_000350.2:c.5924G > T | 4 | NA | Yes (R) | |
| S271 | COD | 26–30 | M | GUCA1A | NM_000409.4:c.333G > C | 4 | NA | NA | Yes (S) |
| ID | Diagnosis | Age at Referral | Sex | Locus | Variant 1 cNomen | ACMG | HGMD | Ref | Variant 2 cNomen | ACMG | HGMD | Ref | Phasing |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S272 | RD | 46–50 | M | ABCA4 | NM_000350.2:c.[1622T > C;3113C > T] | 4 | Pat | [25] | NM_000350.2:c.5603A > T | 3 | Ass | [18] | |
| S273 | STGD | 6–10 | M | ABCA4 | NM_000350.2:c.634C > T | 4 | Pat | [64] | NM_000350.2:c.5196 + 1056A > G | 3 | P Pat | [64] | Yes (S) |
| S274 | RD | 66–70 | F | ABCA4 | NM_000350.2:c.[1622T > C;3113C > T] | 4 | Pat | [25] | NM_000350.2:c.5603A > T | 3 | Ass | [18] | |
| S275 | MD | 31–35 | M | ABCA4 | NM_000350.2:c.2947A > G | 4 | Pat | [71] | NM_000350.2:c.4253 + 43G > A | 2 | P Pat | [22] | |
| S276 | STGD | 41–45 | M | ABCA4 | NM_000350.2:c.1621_1622del | 4 | NA | NM_000350.2:c.5603A > T | 3 | Ass | [18] | ||
| S277 | STGD | 21–25 | F | ABCA4 | NM_000350.2:c.768G > T | 4 | Pat | [13] | NM_000350.2:c.4539 + 2064C > T | 2 | Pat | [20] |
Appendix B

References
- Berger, W.; Kloeckener-Gruissem, B.; Neidhardt, J. The molecular basis of human retinal and vitreoretinal diseases. Prog. Retin. Eye Res. 2010, 29, 335–375. [Google Scholar] [CrossRef]
- Sahel, J.-A.; Marazova, K.; Audo, I. Clinical Characteristics and Current Therapies for Inherited Retinal Degenerations. Cold Spring Harb. Perspect. Med. 2015, 5, a017111. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.; Ratnapriya, R.; Du Plessis, M.; Chaitankar, V.; Ramesar, R.; Swaroop, A. Molecular Diagnosis of Inherited Retinal Diseases in Indigenous African Populations by Whole-Exome Sequencing. Investig. Opthalmol. Vis. Sci. 2016, 57, 6374–6381. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Lemke, J.; Altmueller, J.; Thiele, H.; Glaus, E.; Fleischhauer, J.; Nürnberg, P.; Neidhardt, J.; Berger, W. Identification of Novel and Recurrent Disease-Causing Mutations in Retinal Dystrophies Using Whole Exome Sequencing (WES): Benefits and Limitations. PLoS ONE 2016, 11, e0158692. [Google Scholar] [CrossRef] [PubMed]
- Toulis, V.; Cortés-González, V.; De Castro-Miró, M.; Sallum, J.M.F.; Català-Mora, J.; Villanueva-Mendoza, C.; Ciccioli, M.; Gonzàlez-Duarte, R.; Valero, R.; Marfany, G. Increasing the Genetic Diagnosis Yield in Inherited Retinal Dystrophies: Assigning Pathogenicity to Novel Non-canonical Splice Site Variants. Genes 2020, 11, 378. [Google Scholar] [CrossRef]
- Weisschuh, N.; Mayer, A.K.; Strom, T.M.; Kohl, S.; Glöckle, N.; Schubach, M.; Andreasson, S.; Bernd, A.; Birch, D.G.; Hamel, C.P.; et al. Mutation Detection in Patients with Retinal Dystrophies Using Targeted Next Generation Sequencing. PLoS ONE 2016, 11, e0145951. [Google Scholar] [CrossRef]
- Huang, X.-F.; Huang, F.; Wu, K.-C.; Wu, J.; Chen, J.; Pang, C.-P.; Lu, F.; Qu, J.; Jin, Z.-B. Genotype–phenotype correlation and mutation spectrum in a large cohort of patients with inherited retinal dystrophy revealed by next-generation sequencing. Genet. Med. 2015, 17, 271–278. [Google Scholar] [CrossRef]
- Bravo-Gil, N.; Méndez-Vidal, C.; Romero-Pérez, L.; Gonzáles-del Pozo, M.; Rodríguez-de la Rúa, E.; Dopazo, J.; Borrego, S.; Antiñolo, G. Improving the management of Inherited Retinal Dystrophies by targeted sequencing of a population-specific gene panel. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Ellingford, J.M.; Barton, S.; Bhaskar, S.; O’Sullivan, J.; Williams, S.G.; Lamb, J.A.; Panda, B.; Sergouniotis, P.I.; Gillespie, R.L.; Daiger, S.P.; et al. Molecular findings from 537 individuals with inherited retinal disease. J. Med Genet. 2016, 53, 761–767. [Google Scholar] [CrossRef]
- Kim, M.S.; Joo, K.; Seong, M.-W.; Kim, M.J.; Park, K.H.; Park, S.S.; Woo, S.J. Genetic Mutation Profiles in Korean Patients with Inherited Retinal Diseases. J. Korean Med Sci. 2019, 34, e161. [Google Scholar] [CrossRef]
- Beryozkin, A.; Shevah, E.; Kimchi, A.; Mizrahi-Meissonnier, L.; Khateb, S.; Ratnapriya, R.; Lazar, C.H.; Blumenfeld, A.; Ben-Yosef, T.; Hemo, Y.; et al. Whole Exome Sequencing Reveals Mutations in Known Retinal Disease Genes in 33 out of 68 Israeli Families with Inherited Retinopathies. Sci. Rep. 2015, 5, 13187. [Google Scholar] [CrossRef] [PubMed]
- Stone, E.M.; Andorf, J.L.; Whitmore, S.S.; DeLuca, A.P.; Giacalone, J.C.; Streb, L.M.; Braun, T.A.; Mullins, R.F.; Scheetz, T.E.; Sheffield, V.C.; et al. Clinically Focused Molecular Investigation of 1000 Consecutive Families with Inherited Retinal Disease. Ophthalmol. 2017, 124, 1314–1331. [Google Scholar] [CrossRef] [PubMed]
- Birtel, J.; Eisenberger, T.; Gliem, M.; Müller, P.L.; Herrmann, P.; Betz, C.; Zahnleiter, D.; Neuhaus, C.; Lenzner, S.; Holz, F.G.; et al. Clinical and genetic characteristics of 251 consecutive patients with macular and cone/cone-rod dystrophy. Sci. Rep. 2018, 8, 4824. [Google Scholar] [CrossRef] [PubMed]
- Perea-Romero, I.; Gordo, G.; Iancu, I.F.; Del Pozo-Valero, M.; Almoguera, B.; Blanco-Kelly, F.; Carreño, E.; Jimenez-Rolando, B.; Lopez-Rodriguez, R.; The ESRETNET Study Group; et al. Genetic landscape of 6089 inherited retinal dystrophies affected cases in Spain and their therapeutic and extended epidemiological implications. Sci. Rep. 2021, 11, 1526. [Google Scholar] [CrossRef] [PubMed]
- Maroilley, T.; Tarailo-Graovac, M. Uncovering Missing Heritability in Rare Diseases. Genes 2019, 10, 275. [Google Scholar] [CrossRef]
- Braun, T.A.; Mullins, R.F.; Wagner, A.H.; Andorf, J.L.; Johnston, R.M.; Bakall, B.B.; DeLuca, A.P.; Fishman, G.A.; Lam, B.L.; Weleber, R.G.; et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum. Mol. Genet. 2013, 22, 5136–5145. [Google Scholar] [CrossRef] [PubMed]
- Van Cauwenbergh, C.; Van Schil, K.; Cannoodt, R.; Bauwens, M.; Van Laethem, T.; De Jaegere, S.; Steyaert, W.; Sante, T.; Menten, B.; Leroy, B.P.; et al. arrEYE: A customized platform for high-resolution copy number analysis of coding and noncoding regions of known and candidate retinal dystrophy genes and retinal noncoding RNAs. Genet. Med. 2017, 19, 457–466. [Google Scholar] [CrossRef]
- Bauwens, M.; Garanto, A.; Sangermano, R.; Naessens, S.; Weisschuh, N.; De Zaeytijd, J.; Khan, M.; Sadler, F.; Balikova, I.; Van Cauwenbergh, C.; et al. ABCA4-associated disease as a model for missing heritability in autosomal recessive disorders: Novel noncoding splice, cis-regulatory, structural, and recurrent hypomorphic variants. Genet. Med. 2019, 21, 1761–1771. [Google Scholar] [CrossRef]
- Ellingford, J.M.; Barton, S.; Bhaskar, S.; Williams, S.G.; Sergouniotis, P.I.; O’Sullivan, J.; Lamb, J.A.; Perveen, R.; Hall, G.; Newman, W.G.; et al. Whole Genome Sequencing Increases Molecular Diagnostic Yield Compared with Current Diagnostic Testing for Inherited Retinal Disease. Ophthalmology 2016, 123, 1143–1150. [Google Scholar] [CrossRef]
- Zernant, J.; Xie, Y.A.; Ayuso, C.; Riveiro-Alvarez, R.; Lopez-Martinez, M.-A.; Simonelli, F.; Testa, F.; Gorin, M.B.; Strom, S.P.; Bertelsen, M.; et al. Analysis of the ABCA4 genomic locus in Stargardt disease. Hum. Mol. Genet. 2014, 23, 6797–6806. [Google Scholar] [CrossRef]
- Zernant, J.; Schubert, C.; Im, K.M.; Burke, T.; Brown, C.M.; Fishman, G.A.; Tsang, S.H.; Gouras, P.; Dean, M.; Allikmets, R. Analysis of the ABCA4 Gene by Next-Generation Sequencing. Investig. Opthalmology Vis. Sci. 2011, 52, 8479–8487. [Google Scholar] [CrossRef] [PubMed]
- Sangermano, R.; Garanto, A.; Khan, M.; Runhart, E.H.; Bauwens, M.; Bax, N.M.; Van den Born, L.I.; Khan, M.I.; Cornelis, S.S.; Verheij, J.B.G.M.; et al. Deep-intronic ABCA4 variants explain missing heritability in Stargardt disease and allow correction of splice defects by antisense oligonucleotides. Genet. Med. 2019, 21, 1751–1760. [Google Scholar] [CrossRef] [PubMed]
- Zernant, J.; Lee, W.; Nagasaki, T.; Collison, F.T.; Fishman, G.A.; Bertelsen, M.; Rosenberg, T.; Gouras, P.; Tsang, S.H.; Allikmets, R. Extremely hypomorphic and severe deep intronic variants in the ABCA4 locus result in varying Stargardt disease phenotypes. Cold Spring Harb. Mol. Case Stud. 2018, 4. [Google Scholar] [CrossRef] [PubMed]
- Holtan, J.P.; Selmer, K.K.; Heimdal, K.R.; Bragadóttir, R. Inherited retinal disease in Norway – a characterization of current clinical and genetic knowledge. Acta Ophthalmol. 2020, 98, 286–295. [Google Scholar] [CrossRef]
- Allikmets, R.; Singh, N.; Sun, H.; Shroyer, N.F.; Hutchinson, A.; Chidambaram, A.; Gerrard, B.; Baird, L.; Stauffer, D.; Peiffer, A.; et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Starqardt macular dystrophy. Nat. Genet. 1997, 15, 236–246. [Google Scholar] [CrossRef]
- Jiang, F.; Pan, Z.; Xu, K.; Tian, L.; Xie, Y.; Zhang, X.; Chen, J.; Dong, B.; Li, Y. Screening of ABCA4 Gene in a Chinese Cohort with Stargardt Disease or Cone-Rod Dystrophy with a Report on 85 Novel Mutations. Investig. Opthalmol. Vis. Sci. 2016, 57, 145–152. [Google Scholar] [CrossRef]
- Carss, K.J.; Arno, G.; Erwood, M.; Stephens, J.; Sanchis-Juan, A.; Hull, S.; Megy, K.; Grozeva, D.; Dewhurst, E.; Malka, S.; et al. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am. J. Hum. Genet. 2017, 100, 75–90. [Google Scholar] [CrossRef]
- Li, J.; Tang, J.; Feng, Y.; Xu, M.; Chen, R.; Zou, X.; Sui, R.; Chang, E.Y.; Lewis, R.A.; Zhang, V.W.; et al. Improved Diagnosis of Inherited Retinal Dystrophies by High-Fidelity PCR of ORF15 followed by Next-Generation Sequencing. J. Mol. Diagn. 2016, 18, 817–824. [Google Scholar] [CrossRef]
- Maggi, J.; Roberts, L.; Koller, S.; Rebello, G.; Berger, W.; Ramesar, R. De Novo Assembly-Based Analysis of RPGR Exon ORF15 in an Indigenous African Cohort Overcomes Limitations of a Standard Next-Generation Sequencing (NGS) Data Analysis Pipeline. Genes 2020, 11, 800. [Google Scholar] [CrossRef]
- Zhi, D. Sequence correlation between neighboring Alu instances suggests post-retrotransposition sequence exchange due to Alu gene conversion. Gene 2007, 390, 117–121. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Depristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Povysil, G.; Tzika, A.; Vogt, J.; Haunschmid, V.; Messiaen, L.; Zschocke, J.; Klambauer, G.; Hochreiter, S.; Wimmer, K. panelcn.MOPS: Copy-number detection in targeted NGS panel data for clinical diagnostics. Hum. Mutat. 2017, 38, 889–897. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Stone, E.M. Leber Congenital Amaurosis–A Model for Efficient Genetic Testing of Heterogeneous Disorders: LXIV Edward Jackson Memorial Lecture. Am. J. Ophthalmol. 2007, 144, 791–811. [Google Scholar] [CrossRef]
- Lewis, R.A.; Shroyer, N.F.; Singh, N.; Allikmets, R.; Hutchinson, A.; Li, Y.; Lupski, J.R.; Leppert, M.; Dean, M. Genotype/Phenotype Analysis of a Photoreceptor-Specific ATP-Binding Cassette Transporter Gene, ABCR, in Stargardt Disease. Am. J. Hum. Genet. 1999, 64, 422–434. [Google Scholar] [CrossRef]
- Maugeri, A.; Van Driel, M.A.; Van De Pol, D.J.; Klevering, B.J.; Van Haren, F.J.; Tijmes, N.; Bergen, A.A.; Rohrschneider, K.; Blankenagel, A.; Pinckers, A.J.; et al. The 2588G→C Mutation in the ABCR Gene Is a Mild Frequent Founder Mutation in the Western European Population and Allows the Classification of ABCR Mutations in Patients with Stargardt Disease. Am. J. Hum. Genet. 1999, 64, 1024–1035. [Google Scholar] [CrossRef]
- Paloma, E.; Martínez-Mir, A.; Vilageliu, L.; Gonzàlez-Duarte, R.; Balcells, S. Spectrum of ABCA4 (ABCR) gene mutations in Spanish patients with autosomal recessive macular dystrophies. Hum. Mutat. 2001, 17, 504–510. [Google Scholar] [CrossRef]
- Allikmets, R.; Shroyer, N.F.; Singh, N.; Seddon, J.M.; Lewis, R.A.; Bernstein, P.S.; Peiffer, A.; Zabriskie, N.A.; Li, Y.; Hutchinson, A.; et al. Mutation of the Stargardt Disease Gene (ABCR) in Age-Related Macular Degeneration. Science 1997, 277, 1805–1807. [Google Scholar] [CrossRef]
- Barbazetto, I.A.; Yannuzzi, N.A.; Klais, C.M.; Merriam, J.E.; Zernant, J.; Peiretti, E.; Yannuzzi, L.A.; Allikmets, R. Pseudo-Vitelliform Macular Detachment and Cuticular Drusen: Exclusion of 6 Candidate Genes. Ophthalmic Genet. 2007, 28, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Cremers, F.P.M.; Van De Pol, D.J.R.; Van Driel, M.; Den Hollander, A.I.; Van Haren, F.J.J.; Knoers, N.V.A.M.; Tijmes, N.; Bergen, A.A.B.; Rohrschneider, K.; Blankenagel, A.; et al. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt’s disease gene ABCR. Hum. Mol. Genet. 1998, 7, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A.; Klevering, B.J.; Rohrschneider, K.; Blankenagel, A.; Brunner, H.G.; Deutman, A.F.; Hoyng, C.B.; Cremers, F.P.M. Mutations in the ABCA4 (ABCR) Gene Are the Major Cause of Autosomal Recessive Cone-Rod Dystrophy. Am. J. Hum. Genet. 2000, 67, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Testa, F.; Attanasio, M.; Orrico, A.; De Benedictis, A.; Della Corte, M.; Simonelli, F. Subretinal Fibrosis in Stargardt’s Disease with Fundus Flavimaculatus and ABCA4 Gene Mutation. Case Rep. Ophthalmol. 2012, 3, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Rivera, A.; White, K.; Stöhr, H.; Steiner, K.; Hemmrich, N.; Grimm, T.; Jurklies, B.; Lorenz, B.; Scholl, H.P.N.; Apfelstedt-Sylla, E.; et al. A Comprehensive Survey of Sequence Variation in the ABCA4 (ABCR) Gene in Stargardt Disease and Age-Related Macular Degeneration. Am. J. Hum. Genet. 2000, 67, 800–813. [Google Scholar] [CrossRef] [PubMed]
- Krämer, F.; White, K.; Pauleikhoff, D.; Gehrig, A.; Passmore, L.; Rivera, A.; Rudolph, G.; Kellner, U.; Andrassi, M.; Lorenz, B.; et al. Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration. Eur. J. Hum. Genet. 2000, 8, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, R.; Lennon, A.; Bird, A.C.; Tulloch, B.; Axton, R.; Miano, M.G.; Meindl, A.; Meitinger, T.; Ciccodicola, A.; Wright, A.F. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat. Genet. 2000, 25, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.; Wroblewski, J.; Keen, J.; Inglehearn, C.; Jubb, C.; Eckstein, A.; Jay, M.; Arden, G.B.; Bhattacharya, S.; Fitzke, F.; et al. Mutations in the human retinal degeneration slow (RDS) gene can cause either retinitis pigmentosa or macular dystrophy. Nat. Genet. 1993, 3, 213–218. [Google Scholar] [CrossRef]
- Klevering, B.J.; Deutman, A.F.; Maugeri, A.; Cremers, F.P.M.; Hoyng, C.B. The spectrum of retinal phenotypes caused by mutations in the ABCA4 gene. Graefe’s Arch. Clin. Exp. Ophthalmol. 2005, 243, 90–100. [Google Scholar] [CrossRef]
- Introini, U.; Casalino, G.; Khan, K.N.; Eandi, C.; Alovisi, C.; Michaelides, M.; Bandello, F. Clinical Course of Autosomal Recessive Bestrophinopathy Complicated by Choroidal Neovascularization. Ophthal. Surg. Lasers Imaging Retin. 2018, 49, 888–892. [Google Scholar] [CrossRef]
- Thiagalingam, S.; McGee, T.L.; Weleber, R.G.; Sandberg, M.A.; Trzupek, K.M.; Berson, E.L.; Dryja, T.P. Novel Mutations in the KCNV2 Gene in Patients with Cone Dystrophy and a Supernormal Rod Electroretinogram. Ophthalmic Genet. 2007, 28, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Schulz, H.L.; Grassmann, F.; Kellner, U.; Spital, G.; Rüther, K.; Jägle, H.; Hufendiek, K.; Rating, P.; Huchzermeyer, C.; Baier, M.J.; et al. Mutation Spectrum of the ABCA4 Gene in 335 Stargardt Disease Patients from a Multicenter German Cohort—Impact of Selected Deep Intronic Variants and Common SNPs. Investig. Opthalmol. Vis. Sci. 2017, 58, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Passerini, I.; Sodi, A.; Giambene, B.; Mariottini, A.; Menchini, U.; Torricelli, F. Novel mutations in of the ABCR gene in italian patients with Stargardt disease. Eye 2010, 24, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Den Hollander, A.I.; Koenekoop, R.K.; Yzer, S.; Lopez, I.; Arends, M.L.; Voesenek, K.E.J.; Zonneveld, M.N.; Strom, T.M.; Meitinger, T.; Brunner, H.G.; et al. Mutations in the CEP290 (NPHP6) Gene Are a Frequent Cause of Leber Congenital Amaurosis. Am. J. Hum. Genet. 2006, 79, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Perrault, I.; Delphin, N.; Hanein, S.; Gerber, S.; Dufier, J.-L.; Roche, O.; Defoort-Dhellemmes, S.; Dollfus, H.; Fazzi, E.; Munnich, A.; et al. Spectrum of NPHP6/CEP290 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum. Mutat. 2007, 28, 416. [Google Scholar] [CrossRef]
- Jaakson, K.; Zernant, J.; Külm, M.; Hutchinson, A.; Tonisson, N.; Glavač, D.; Ravnik-Glavač, M.; Hawlina, M.; Meltzer, M.; Caruso, R.C.; et al. Genotyping microarray (gene chip) for the ABCR (ABCA4) gene. Hum. Mutat. 2003, 22, 395–403. [Google Scholar] [CrossRef]
- Pomares, E.; Riera, M.; Castro-Navarro, J.; Andrés-Gutiérrez, Á.; Gonzàlez-Duarte, R.; Marfany, G. Identification of an Intronic Single-Point Mutation in RP2 as the Cause of Semidominant X-linked Retinitis Pigmentosa. Investig. Opthalmol. Vis. Sci. 2009, 50, 5107–5114. [Google Scholar] [CrossRef]
- Sundin, O.H.; Yang, J.-M.; Li, Y.; Zhu, D.; Hurd, J.N.; Mitchell, T.N.; Silva, E.D.; Maumenee, I.H. Genetic basis of total colourblindness among the Pingelapese islanders. Nat. Genet. 2000, 25, 289–293. [Google Scholar] [CrossRef]
- White, K.; Marquardt, A.; Weber, B.H.F. VMD2 mutations in vitelliform macular dystrophy (Best disease) and other macu-lopathies. Hum. Mutat. 2000, 15, 301–308. [Google Scholar] [CrossRef]
- Sodi, A.; Bini, A.; Passerini, I.; Forconi, S.; Menchini, U.; Torricelli, F. Different Patterns of Fundus Autofluorescence Related to ABCA4 Gene Mutations in Stargardt Disease. Ophthal. Surg. Lasers Imaging 2010, 41, 48–53. [Google Scholar] [CrossRef]
- Lotery, A.; Munier, F.L.; A Fishman, G.; Weleber, R.G.; Jacobson, S.G.; Affatigato, L.M.; E Nichols, B.; Schorderet, D.F.; Sheffield, V.C.; Stone, E.M. Allelic variation in the VMD2 gene in best disease and age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2000, 41, 1291–1296. [Google Scholar]
- Boon, C.J.F.; Van Den Born, L.I.; Visser, L.; Keunen, J.E.; Bergen, A.A.B.; Booij, J.C.; Riemslag, F.C.; Florijn, R.J.; Van Schooneveld, M.J. Autosomal Recessive Bestrophinopathy: Differential Diagnosis and Treatment Options. Ophthalmology 2013, 120, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Bahr, A.; Bähr, L.; Fleischhauer, J.; Zinkernagel, M.S.; Winkler, N.; Barthelmes, D.; Berger, L.; Gerth-Kahlert, C.; Neidhardt, J.; et al. Next generation sequencing based identification of disease-associated mutations in Swiss patients with retinal dystrophies. Sci. Rep. 2016, 6, 28755. [Google Scholar] [CrossRef] [PubMed]
- Gerber, S.; Rozet, J.-M.; Van De Pol, T.J.R.; Hoyng, C.B.; Munnich, A.; Blankenagel, A.; Kaplan, J.; Cremers, F.P.M. Complete Exon–Intron Structure of the Retina-Specific ATP Binding Transporter Gene (ABCR) Allows the Identification of Novel Mutations Underlying Stargardt Disease. Genomics 1998, 48, 139–142. [Google Scholar] [CrossRef]
- Kuniyoshi, K.; Ikeo, K.; Sakuramoto, H.; Furuno, M.; Yoshitake, K.; Hatsukawa, Y.; Nakao, A.; Tsunoda, K.; Kusaka, S.; Shimomura, Y.; et al. Novel nonsense and splice site mutations in CRB1 gene in two Japanese patients with early-onset retinal dystrophy. Doc. Ophthalmol. 2015, 130, 49–55. [Google Scholar] [CrossRef]
- Maugeri, A.; Meire, F.; Hoyng, C.B.; Vink, C.; Van Regemorter, N.; Karan, G.; Yang, Z.; Cremers, F.P.M.; Zhang, K. A Novel Mutation in the ELOVL4 Gene Causes Autosomal Dominant Stargardt-like Macular Dystrophy. Investig. Opthalmol. Vis. Sci. 2004, 45, 4263–4267. [Google Scholar] [CrossRef]
- Corton, M.; Tatu, S.D.; Avila-Fernandez, A.; Vallespín, E.; Tapias, I.; Cantalapiedra, D.; Blanco-Kelly, F.; Riveiro-Alvarez, R.; Bernal, S.; García-Sandoval, B.; et al. High frequency of CRB1 mutations as cause of Early-Onset Retinal Dystrophies in the Spanish population. Orphanet J. Rare Dis. 2013, 8, 20. [Google Scholar] [CrossRef]
- Den Hollander, A.I.; Ten Brink, J.B.; De Kok, Y.J.M.; Van Soest, S.; Van Den Born, L.I.; Van Driel, M.A.; Van De Pol, D.J.; Payne, A.M.; Bhattacharya, S.S.; Kellner, U.; et al. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12). Nat. Genet. 1999, 23, 217–221. [Google Scholar] [CrossRef]
- Payne, A.; Vithana, E.; Khaliq, S.; Hameed, A.; Deller, J.; Abu-Safieh, L.; Kermani, S.; Leroy, B.P.; Mehdi, S.Q.; Moore, A.T.; et al. RP1 protein truncating mutations predominate at the RP1 adRP locus. Investig. Ophthalmol. Vis. Sci. 2000, 41, 4069–4073. [Google Scholar]
- Yokoyama, A.; Maruiwa, F.; Hayakawa, M.; Kanai, A.; Vervoort, R.; Wright, A.F.; Yamada, K.; Niikawa, N.; Naōi, N. Three novel mutations of the RPGR gene exon ORF15 in three Japanese families with X-linked retinitis pigmentosa. Am. J. Med Genet. 2001, 104, 232–238. [Google Scholar] [CrossRef]
- Ernest, P.J.G.; Boon, C.J.F.; Klevering, B.J.; Hoefsloot, L.H.; Hoyng, C.B. Outcome of ABCA4 microarray screening in routine clinical practice. Mol. Vis. 2009, 15, 2841–2847. [Google Scholar] [PubMed]
- Simonelli, F.; Testa, F.; De Crecchio, G.; Rinaldi, E.; Hutchinson, A.; Atkinson, A.; Dean, M.; D’Urso, M.; Allikmets, R. New ABCR mutations and clinical phenotype in Italian patients with Stargardt disease. Investig. Ophthalmol. Vis. Sci. 2000, 41, 892–897. [Google Scholar]
- Downs, K.; Zacks, D.N.; Caruso, R.; Karoukis, A.J.; Branham, K.; Yashar, B.M.; Haimann, M.H.; Trzupek, K.; Meltzer, M.; Blain, D.; et al. Molecular Testing for Hereditary Retinal Disease as Part of Clinical Care. Arch. Ophthalmol. 2007, 125, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Stenirri, S.; Alaimo, G.; Manitto, M.P.; Brancato, R.; Ferrari, M.; Cremonesi, L. Are microarrays useful in the screening of ABCA4 mutations in Italian patients affected by macular degenerations? Clin. Chem. Lab. Med. 2008, 46, 1250–1255. [Google Scholar] [CrossRef]
- Abed, E.; Placidi, G.; Campagna, F.; Federici, M.; Minnella, A.; Guerri, G.; Bertelli, M.; Piccardi, M.; Galli-Resta, L.; Falsini, B. Early impairment of the full-field photopic negative response in patients with Stargardt disease and pathogenic variants of the ABCA4 gene. Clin. Exp. Ophthalmol. 2018, 46, 519–530. [Google Scholar] [CrossRef]
- Meunier, I.; Sénéchal, A.; Dhaenens, C.-M.; Arndt, C.; Puech, B.; Defoort-Dhellemmes, S.; Manes, G.; Chazalette, D.; Mazoir, E.; Bocquet, B.; et al. Systematic Screening of BEST1 and PRPH2 in Juvenile and Adult Vitelliform Macular Dystrophies: A Rationale for Molecular Analysis. Ophthalmology 2011, 118, 1130–1136. [Google Scholar] [CrossRef]
- Johnson, A.A.; Lee, Y.-S.; Chadburn, A.J.; Tammaro, P.; Manson, F.D.; Marmorstein, L.Y.; Marmorstein, A.D. Disease-causing mutations associated with four bestrophinopathies exhibit disparate effects on the localization, but not the oligomerization, of Bestrophin-1. Exp. Eye Res. 2014, 121, 74–85. [Google Scholar] [CrossRef]
- Özgül, R.K.; Durukan, H.; Turan, A.; Öner, C.; Öğüş, A.; Farber, D.B. Molecular analysis of the ABCA4 gene in Turkish patients with Stargardt disease and retinitis pigmentosa. Hum. Mutat. 2004, 23, 523. [Google Scholar] [CrossRef]
- Testa, F.; Rossi, S.; Sodi, A.; Passerini, I.; Di Iorio, V.; Della Corte, M.; Banfi, S.; Surace, E.M.; Menchini, U.; Auricchio, A.; et al. Correlation between Photoreceptor Layer Integrity and Visual Function in Patients with Stargardt Disease: Implications for Gene Therapy. Investig. Opthalmol. Vis. Sci. 2012, 53, 4409–4415. [Google Scholar] [CrossRef]
- Webster, A.R.; Héon, E.; Lotery, A.; Vandenburgh, K.; Casavant, T.L.; Oh, K.T.; Beck, G.; Fishman, G.A.; Lam, B.L.; Levin, A.; et al. An analysis of allelic variation in the ABCA4 gene. Investig. Ophthalmol. Vis. Sci. 2001, 42, 1179–1189. [Google Scholar]
- Sauer, C.G.; Gehrig, A.; Warneke-Wittstock, R.; Marquardt, A.; Ewing, C.C.; Gibson, A.; Lorenz, B.; Jurklies, B.; Weber, B.H. Positional cloning of the gene associated with X-linked juvenile retinoschisis. Nat. Genet. 1997, 17, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Rozet, J.-M.; Gerber, S.; Souied, E.; Perrault, I.; Châtelin, S.; Ghazi, I.; Leowski, C.; Dufier, J.-L.; Munnich, A.; Kaplan, J. Spectrum of ABCR gene mutations in autosomal recessive macular dystrophies. Eur. J. Hum. Genet. 1998, 6, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Stenirri, S.; Fermo, I.; Battistella, S.; Galbiati, S.; Soriani, N.; Paroni, R.; Manitto, M.P.; Martina, E.; Brancato, R.; Allikmets, R.; et al. Denaturing HPLC Profiling of the ABCA4 Gene for Reliable Detection of Allelic Variations. Clin. Chem. 2004, 50, 1336–1343. [Google Scholar] [CrossRef] [PubMed]
- Gerth-Kahlert, C.; Koller, S.; Hanson, J.V.M.; Baehr, L.; Tiwari, A.; Kivrak-Pfiffner, F.; Bahr, A.; Berger, W. Genotype-Phenotype Analysis of a Novel Recessive and a Recurrent Dominant SNRNP200 Variant Causing Retinitis Pigmentosa. Investig. Opthalmol. Vis. Sci. 2019, 60, 2822–2835. [Google Scholar] [CrossRef]
- Cideciyan, A.V.; Swider, M.; Aleman, T.S.; Tsybovsky, Y.; Schwartz, S.B.; Windsor, E.A.; Roman, A.J.; Sumaroka, A.; Steinberg, J.D.; Jacobson, S.G.; et al. ABCA4 disease progression and a proposed strategy for gene therapy. Hum. Mol. Genet. 2009, 18, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Souied, E.H.; Ducroq, D.; Rozet, J.M.; Gerber, S.; Perrault, I.; Sterkers, M.; Benhamou, N.; Munnich, A.; Coscas, G.; Soubrane, G.; et al. A novel ABCR nonsense mutation responsible for late-onset fundus flavimaculatus. Investig. Ophthalmol. Vis. Sci. 1999, 40, 2740–2744. [Google Scholar]
- Kohl, S.; Baumann, B.; Broghammer, M.; Jägle, H.; Sieving, P.; Kellner, U.; Spegal, R.; Anastasi, M.; Zrenner, E.; Sharpe, L.T.; et al. Mutations in the CNGB3 gene encoding the β-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum. Mol. Genet. 2000, 9, 2107–2116. [Google Scholar] [CrossRef]
- Briggs, C.E.; Rucinski, D.; Rosenfeld, P.J.; Hirose, T.; Berson, E.L.; Dryja, T.P. Mutations in ABCR (ABCA4) in patients with Stargardt macular degeneration or cone-rod degeneration. Investig. Ophthalmol. Vis. Sci. 2001, 42, 2229–2236. [Google Scholar]
- Stone, E.M.; Lotery, A.J.; Munier, F.L.; Héon, E.; Piguet, B.; Guymer, R.H.; Vandenburgh, K.; Cousin, P.; Nishimura, D.; Swiderski, R.E.; et al. A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat. Genet. 1999, 22, 199–202. [Google Scholar] [CrossRef]
- Arai, Y.; Maeda, A.; Hirami, Y.; Ishigami, C.; Kosugi, S.; Mandai, M.; Kurimoto, Y.; Takahashi, M. Retinitis Pigmentosa with EYS Mutations Is the Most Prevalent Inherited Retinal Dystrophy in Japanese Populations. J. Ophthalmol. 2015, 2015, 1–10. [Google Scholar] [CrossRef]
- Hejtmancik, J.F.; Jiao, X.; Li, A.; Sergeev, Y.V.; Ding, X.; Sharma, A.K.; Chan, C.-C.; Medina, I.; Edwards, A.O. Mutations in KCNJ13 Cause Autosomal-Dominant Snowflake Vitreoretinal Degeneration. Am. J. Hum. Genet. 2008, 82, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Barboni, P.; Savini, G.; Cascavilla, M.L.; Caporali, L.; Milesi, J.; Borrelli, E.; La Morgia, C.; Valentino, M.L.; Triolo, G.; Lembo, A.; et al. Early Macular Retinal Ganglion Cell Loss in Dominant Optic Atrophy: Genotype-Phenotype Correlation. Am. J. Ophthalmol. 2014, 158, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Smailhodzic, D.; Fleckenstein, M.; Theelen, T.; Boon, C.J.F.; Van Huet, R.A.C.; Van De Ven, J.P.H.; Den Hollander, A.I.; Schmitz-Valckenberg, S.; Hoyng, C.B.; Weber, B.H.F.; et al. Central Areolar Choroidal Dystrophy (CACD) and Age-Related Macular Degeneration (AMD): Differentiating Characteristics in Multimodal Imaging. Investig. Opthalmol. Vis. Sci. 2011, 52, 8908–8918. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.-Y.; Li, J.-K.; Gao, F.-J.; Qi, Y.-H.; Xu, P.; Zhang, Y.-J.; Wang, D.-D.; Wang, L.-S.; Li, W.; Wang, M.; et al. ABCA4 Gene Screening in a Chinese Cohort with Stargardt Disease: Identification of 37 Novel Variants. Front. Genet. 2019, 10, 773. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, A.; Stöhr, H.; Passmore, L.A.; Krämer, F.; Rivera, A.; Weber, B.H.F. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best’s disease). Hum. Mol. Genet. 1998, 7, 1517–1525. [Google Scholar] [CrossRef]
- Yatsenko, A.N.; Shroyer, N.F.; Lewis, R.A.; Lupski, J.R. Late-onset Stargardt disease is associated with missense mutations that map outside known functional regions of ABCR (ABCA4). Hum. Genet. 2001, 108, 346–355. [Google Scholar] [CrossRef]
- Sohocki, M.M.; Daiger, S.P.; Bowne, S.J.; Rodriquez, J.A.; Northrup, H.; Heckenlively, J.R.; Birch, D.G.; Mintz-Hittner, H.; Ruiz, R.S.; Lewis, R.A.; et al. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum. Mutat. 2001, 17, 42–51. [Google Scholar] [CrossRef]
- Payne, A.M.; Morris, A.G.; Downes, S.M.; Johnson, S.; Bird, A.C.; Moore, A.T.; Bhattacharya, S.S.; Hunt, D.M. Clustering and frequency of mutations in the retinal guanylate cyclase (GUCY2D) gene in patients with dominant cone-rod dystrophies. J. Med Genet. 2001, 38, 611–614. [Google Scholar] [CrossRef]
- Siintola, E.; Topcu, M.; Aula, N.; Lohi, H.; Minassian, B.A.; Paterson, A.D.; Liu, X.-Q.; Wilson, C.; Lahtinen, U.; Anttonen, A.-K.; et al. The Novel Neuronal Ceroid Lipofuscinosis Gene MFSD8 Encodes a Putative Lysosomal Transporter. Am. J. Hum. Genet. 2007, 81, 136–146. [Google Scholar] [CrossRef]
- Roosing, S.; Van Den Born, L.I.; Sangermano, R.; Banfi, S.; Koenekoop, R.K.; Zonneveld-Vrieling, M.N.; Klaver, C.C.W.; Van Lith-Verhoeven, J.J.; Cremers, F.P.M.; Den Hollander, A.I.; et al. Mutations in MFSD8, Encoding a Lysosomal Membrane Protein, Are Associated with Nonsyndromic Autosomal Recessive Macular Dystrophy. Ophthalmology 2015, 122, 170–179. [Google Scholar] [CrossRef]
- Shan, M.; Dong, B.; Zhao, X.; Wang, J.; Li, G.; Yang, Y.; Li, Y. Novel mutations in the CYP4V2 gene associated with Bietti crystalline corneoretinal dystrophy. Mol. Vis. 2005, 11, 738–743. [Google Scholar] [PubMed]
- Lai, T.Y.Y.; Ng, T.K.; Tam, P.O.S.; Yam, G.H.F.; Ngai, J.W.S.; Chan, W.-M.; Liu, D.T.L.; Lam, D.S.C.; Pang, C.P. Genotype–Phenotype Analysis of Bietti’s Crystalline Dystrophy in Patients with CYP4V2 Mutations. Investig. Opthalmol. Vis. Sci. 2007, 48, 5212–5220. [Google Scholar] [CrossRef] [PubMed]
- Den Dunnen, J.T.; Kraayenbrink, T.; Van Schooneveld, M.; Van de Vosse, E.; Ten Brink, J.B.; Schuurman, E.; Tijmes, N.; Van Ommen, G.; Bergen, A.A.; Andolfi, G.; et al. Functional Implications of the Spectrum of Mutations Found in 234 Cases With X-linked Juvenile Retinoschisis (XLRS). Hum. Mol. Genet. 1998, 7, 1185–1192. [Google Scholar] [CrossRef]
- Delettre, C.; Lenaers, G.; Griffoin, J.-M.; Gigarel, N.; Lorenzo, C.; Belenguer, P.; Pelloquin, L.; Grosgeorge, J.; Turc-Carel, C.; Perret, E.; et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000, 26, 207–210. [Google Scholar] [CrossRef]
- Koeppen, K.; Reuter, P.; Ladewig, T.; Kohl, S.; Baumann, B.; Jacobson, S.G.; Plomp, A.S.; Hamel, C.P.; Janecke, A.R.; Wissinger, B. Dissecting the pathogenic mechanisms of mutations in the pore region of the human cone photoreceptor cyclic nucleotide-gated channel. Hum. Mutat. 2010, 31, 830–839. [Google Scholar] [CrossRef]
- Koeppen, K.; Reuter, P.; Kohl, S.; Baumann, B.; Ladewig, T.; Wissinger, B. Functional analysis of human CNGA3 mutations associated with colour blindness suggests impaired surface expression of channel mutants A3R427C and A3R563C. Eur. J. Neurosci. 2008, 27, 2391–2401. [Google Scholar] [CrossRef]
- Cideciyan, A.V.; Swider, M.; Aleman, T.S.; Feuer, W.J.; Schwartz, S.B.; Russell, R.C.; Steinberg, J.D.; Stone, E.M.; Jacobson, S.G. Macular Function in Macular Degenerations: Repeatability of Microperimetry as a Potential Outcome Measure for ABCA4-Associated Retinopathy Trials. Investig. Opthalmol. Vis. Sci. 2012, 53, 841–852. [Google Scholar] [CrossRef]
- Audo, I.; Lancelot, M.-E.; Mohand-Saïd, S.; Antonio, A.; Germain, A.; Sahel, J.-A.; Bhattacharya, S.S.; Zeitz, C. Novel C2orf71 mutations account for ∼1% of cases in a large French arRP cohort. Hum. Mutat. 2011, 32, E2091–E2103. [Google Scholar] [CrossRef]
- Wissinger, B.; Schaich, S.; Baumann, B.; Bonin, M.; Jägle, H.; Friedburg, C.; Varsányi, B.; Hoyng, C.B.; Dollfus, H.; Heckenlively, J.R.; et al. Large deletions of the KCNV2 gene are common in patients with cone dystrophy with supernormal rod response. Hum. Mutat. 2011, 32, 1398–1406. [Google Scholar] [CrossRef]
| Locus | Number of PCRs Needed | Genomic Target Size (bp) |
|---|---|---|
| PPT1 | 2 | 26,746 |
| ABCA4 | 8 | 137,748 |
| CRB1 | 14 | 212,898 |
| PCARE | 1 | 18,628 |
| EFEMP1 | 5 | 60,569 |
| IMPG2 | 6 | 94,768 |
| PROM1 | 7 | 117,712 |
| MFSD8 | 3 | 49,747 |
| CTNNA1 | 13 | 182,694 |
| GUCA1A | 2 | 27,191 |
| GUCA1B | 1 | 13,548 |
| PRPH2 | 2 | 32,242 |
| IMPG1 | 11 | 144,700 |
| ELOVL4 | 2 | 34,792 |
| DHS6S1 | 1 | 13,473 |
| RP1L1 | 3 | 51,877 |
| RP1 | 1 | 16,930 |
| CNGB3 | 12 | 172,052 |
| KCNV2 | 1 | 15,214 |
| ATOH7 | 1 | 9275 |
| PDE6C | 3 | 53,794 |
| BEST1 | 1 | 17,272 |
| C1QTNF5 | 1 | 10,536 |
| PDE6H | 1 | 11,084 |
| RDH5 | 1 | 6122 |
| OTX2 | 1 | 12,758 |
| NR2E3 | 1 | 11,097 |
| RLBP1 | 1 | 13,667 |
| GUCY2D | 1 | 18,939 |
| FSCN2 | 1 | 11,574 |
| RAX2 | 1 | 5845 |
| TIMP3 | 5 | 67,146 |
| RS1 | 3 | 34,731 |
| RPGR | 4 | 60,808 |
| RP2 | 3 | 46,521 |
| Category | Total | Total Diagnosed | Total Undiagnosed | Mean Age at Referral (Years) | Diagnostic Yield (%) |
|---|---|---|---|---|---|
| Overall | 168 | 132 | 36 | 37.86 | 78.6 |
| Stratification group | |||||
| Validation | 70 | 70 | 0 | 35.60 | 100.0 |
| No previous testing | 85 | 58 | 27 | 40.14 | 68.2 |
| Missing heritability | 13 | 4 | 9 | 35.08 | 30.8 |
| Clinical diagnosis | |||||
| STGD | 95 | 84 | 11 | 35.88 | 88.4 |
| Unspecified MD | 45 | 28 | 17 | 40.11 | 62.2 |
| BEST | 11 | 9 | 2 | 35.73 | 81.8 |
| ML | 9 | 5 | 4 | 47.89 | 55.6 |
| XLRS | 6 | 6 | 0 | 32.00 | 100.0 |
| SFD | 1 | 0 | 1 | 63.00 | 0.0 |
| Locus | cNomen | pNomen | GnomAD Overall (%) | GnomAD Max. (%) | ACMG Class | CADD Score |
|---|---|---|---|---|---|---|
| ABCA4 | NM_000350.2:c.6731T > A | p.Val2244Glu | 0 | 0 | 3 | 27.3 |
| ABCA4 | NM_000350.2:c.6428T > A | p.Met2143Lys | 0 | 0 | 4 | 32.0 |
| ABCA4 | NM_000350.2:c.6323_6331delinsGGC | p.Met2108_Asn2111delinsArgHis | 0 | 0 | 4 | 35 |
| ABCA4 | NM_000350.2:c.5924G > T | p.Gly1975Val | 0 | 0 | 4 | 28.8 |
| ABCA4 | NM_000350.2:c.5690_5704del | p.Gln1897_Phe1901del | 0 | 0 | 3 | 22.2 |
| ABCA4 | NM_000350.2:c.5691G > T | p.Gln1897His | 0 | 0 | 3 | 23.8 |
| ABCA4 | NM_000350.2:c.5461–6T > C | p.? | 0 | 0 | 3 | 14.93 |
| ABCA41 | NM_000350.2:c.4958G > A | p.Gly1653Glu | 0 | 0 | 3 | 28.1 |
| ABCA4 | NM_000350.2:c.4609del | p.Thr1537ArgfsTer6 | 0 | 0 | 4 | 33 |
| ABCA4 | NM_000350.2:c.4383G > C | p.Trp1461Cys | 0.0004 | 0.003 | 3 | 32 |
| ABCA4 | NM_000350.2:c.3323del | p.Arg1108ProfsTer40 | 0 | 0 | 5 | 34 |
| ABCA4 | NM_000350.2:c.3179A > C | p.Gln1060Pro | 0.0008 | 0.0065 | 4 | 23.7 |
| ABCA42 | NM_000350.2:c.(2918 + 765_2918 + 775)_(3328 + 618_3328 + 662)del | p.Leu973_Asp2273delinsPheMetAlaArgValGluArgSerLeuGlyAsn | 0 | 0 | 5 | |
| ABCA4 | NM_000350.2:c.1742C > A | p.Thr581Asn | 0 | 0 | 3 | 26.7 |
| ABCA4 | NM_000350.2:c.1621_1622del | p.Leu541ThrfsTer14 | 0 | 0 | 4 | 32 |
| ABCA4 | NM_000350.2:c.727_728dup | p.Tyr245CysfsTer18 | 0 | 0 | 5 | 26.2 |
| ABCA4 | NM_000350.2:c.676C > A | p.Arg226Ser | 0.0068 | 0.0163 | 3 | 14.13 |
| CRB1 | NM_201253.2:c.1472A > T | p.Asp491Val | 0 | 0 | 3 | 15.86 |
| CRB1 | NM_201253.2:c.2298G > A | p.Trp766Ter | 0 | 0 | 5 | 36 |
| OPA1 | NM_130837.2:c.2987A > C | p.Lys996Thr | 0 | 0 | 4 | 23.8 |
| PROM1 | NM_006017.2:c.2476G > C | p.Asp826His | 0 | 0 | 3 | 32 |
| MFSD81 | NM_152778.2:c.670A > T | p.Asn224Tyr | 0 | 0 | 3 | 23.9 |
| GUCA1A | NM_000409.4:c.333G > C | p.Glu111Asp | 0 | 0 | 4 | 22.9 |
| PRPH2 | NM_000322.4:c.611_626del | p.Tyr204SerfsTer47 | 0 | 0 | 5 | 33 |
| PRPH2 | NM_000322.4:c.605G > A | p.Gly202Glu | 0 | 0 | 4 | 29.8 |
| PRPH2 | NM_000322.4:c.512T > G | p.Phe171Cys | 0 | 0 | 4 | 27.3 |
| RP1L1 | NM_178857.5:c.1024_1026delinsCTCCT | p.Arg342LeufsTer22 | 0 | 0 | 4 | 22 |
| RP1L1 | NM_178857.5:c.196G > C | p.Asp66His | 0 | 0 | 3 | 26.6 |
| KCNV23 | NM_133497.3:c.-759_*57289del | p.? | 0 | 0 | 5 | |
| KCNV21 | NM_133497.3:c.1096del | p.Val366TrpfsTer88 | 0 | 0 | 4 | 13.41 |
| RGR | NM_002921.3:c.236G > A | p.Arg79His | 0.0032 | 0.0141 | 3 | 37 |
| BEST1 | NM_004183.3:c.907G > T | p.Asp303Tyr | 0 | 0 | 5 | 28.5 |
| CNGB1 | NM_001297.4:c.2662G > A | p.Ala888Thr | 0.0225 | 0.1145 | 3 | 18.28 |
| CNGB1 | NM_001297.4:c.1658C > A | p.Ala553Glu | 0.0004 | 0.0009 | 3 | 21.1 |
| GUCY2D | NM_000180.3:c.929C > A | p.Thr310Asn | 0 | 0 | 3 | 24.5 |
| RS1 | NM_000330.3:c.209G > A | p.Gly70Asp | 0 | 0 | 4 | 26.7 |
| RS1 | NM_000330.3:c.150G > A | p.Trp50Ter | 0 | 0 | 5 | 35 |
| RS14 | NM_000330.3:c.53–717_78 + 262del | p.Ala18_Glu26delinsGluProGlyGlnHisSerLysThrLeu | 0 | 0 | 5 | |
| RPGR | NM_001034853.1:c.2819_2838dup | p.Glu947LysfsTer149 | 0 | 0 | 3 | 22.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maggi, J.; Koller, S.; Bähr, L.; Feil, S.; Kivrak Pfiffner, F.; Hanson, J.V.M.; Maspoli, A.; Gerth-Kahlert, C.; Berger, W. Long-Range PCR-Based NGS Applications to Diagnose Mendelian Retinal Diseases. Int. J. Mol. Sci. 2021, 22, 1508. https://doi.org/10.3390/ijms22041508
Maggi J, Koller S, Bähr L, Feil S, Kivrak Pfiffner F, Hanson JVM, Maspoli A, Gerth-Kahlert C, Berger W. Long-Range PCR-Based NGS Applications to Diagnose Mendelian Retinal Diseases. International Journal of Molecular Sciences. 2021; 22(4):1508. https://doi.org/10.3390/ijms22041508
Chicago/Turabian StyleMaggi, Jordi, Samuel Koller, Luzy Bähr, Silke Feil, Fatma Kivrak Pfiffner, James V. M. Hanson, Alessandro Maspoli, Christina Gerth-Kahlert, and Wolfgang Berger. 2021. "Long-Range PCR-Based NGS Applications to Diagnose Mendelian Retinal Diseases" International Journal of Molecular Sciences 22, no. 4: 1508. https://doi.org/10.3390/ijms22041508
APA StyleMaggi, J., Koller, S., Bähr, L., Feil, S., Kivrak Pfiffner, F., Hanson, J. V. M., Maspoli, A., Gerth-Kahlert, C., & Berger, W. (2021). Long-Range PCR-Based NGS Applications to Diagnose Mendelian Retinal Diseases. International Journal of Molecular Sciences, 22(4), 1508. https://doi.org/10.3390/ijms22041508






