Modeling and Molecular Dynamics of the 3D Structure of the HPV16 E7 Protein and Its Variants
Abstract
1. Introduction
2. Results and Discussion
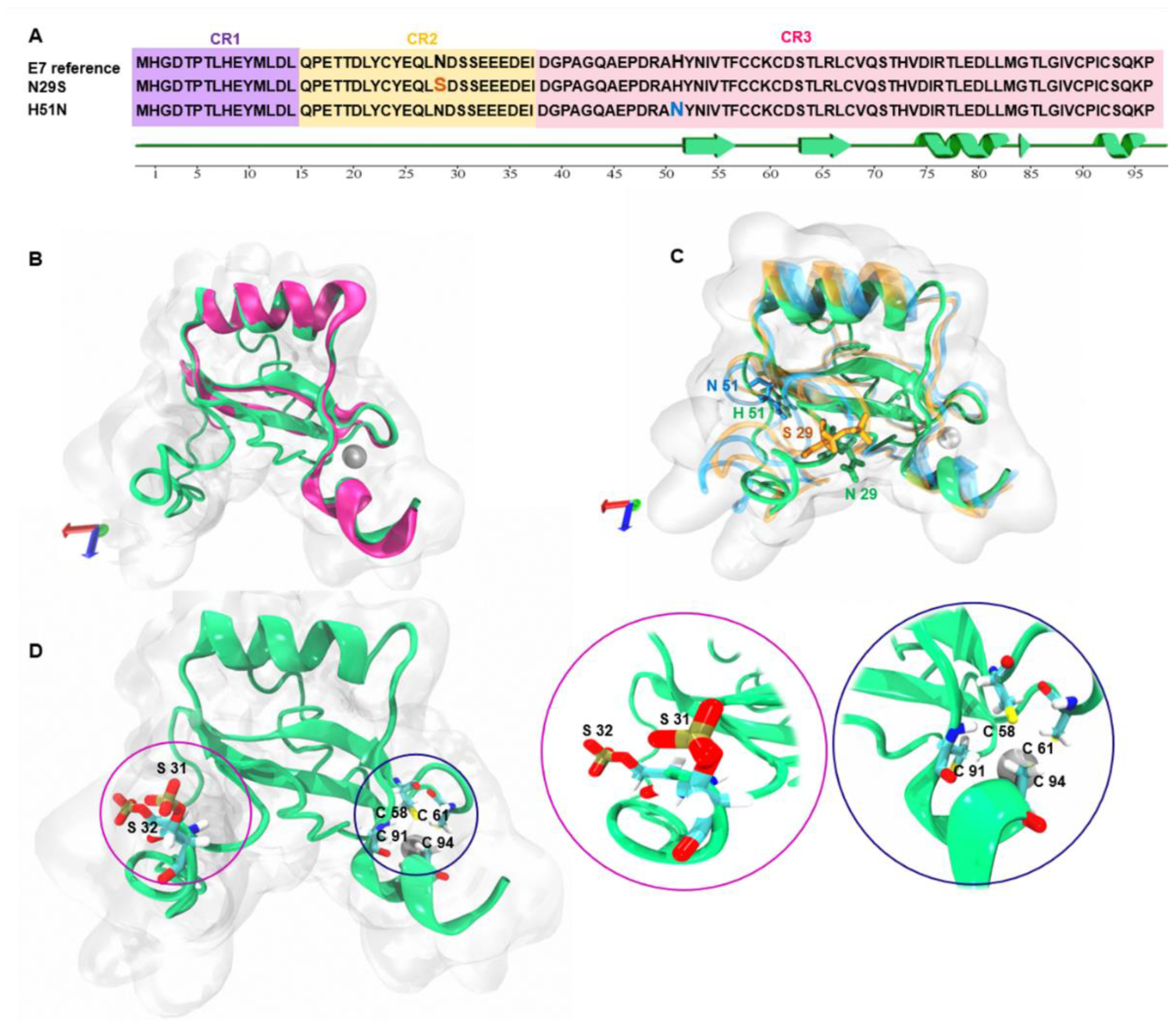
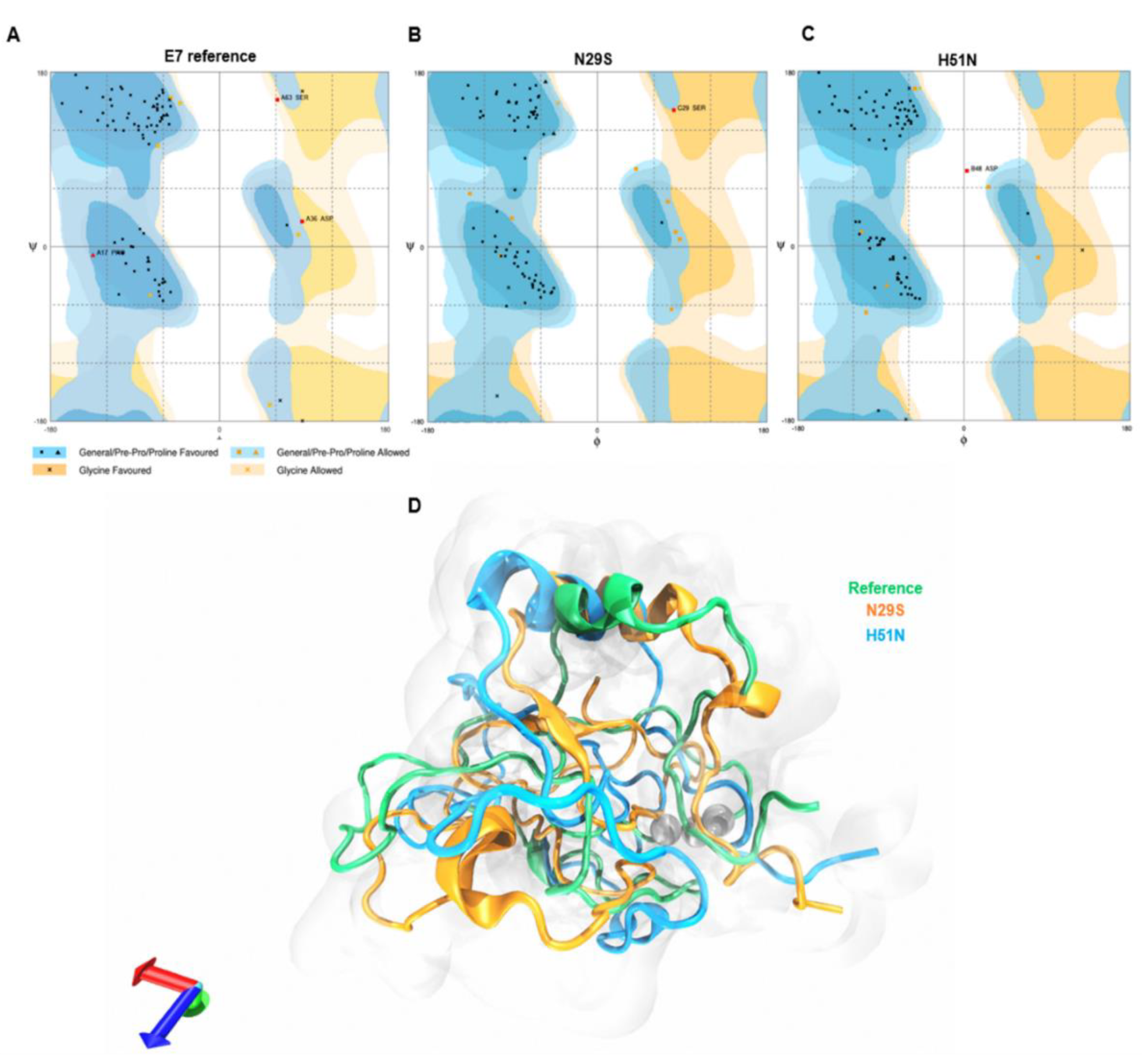
2.1. Modeling and Structural Alignment of the E7 Reference and Its Variants
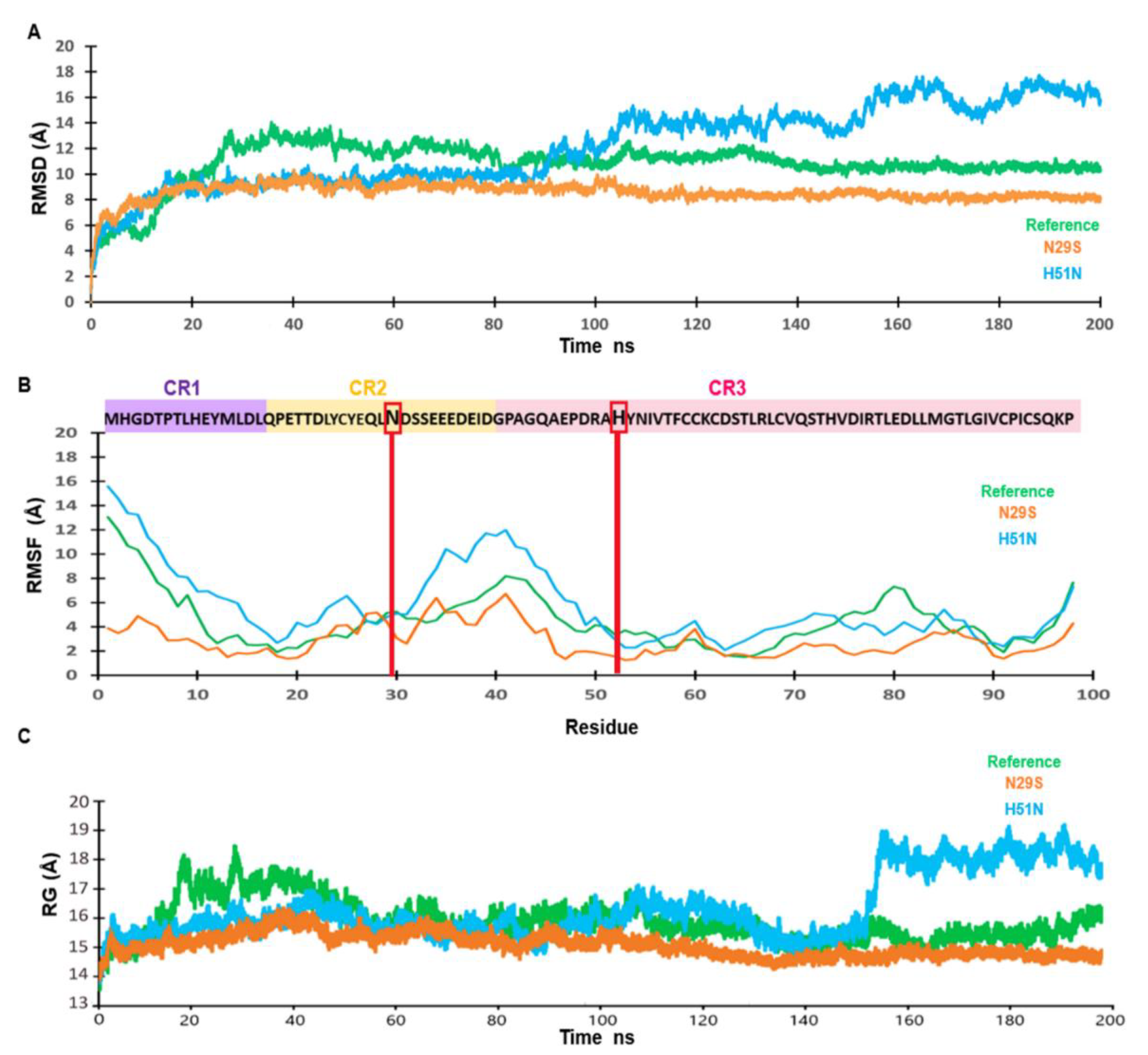
2.2. N29S Presents the Most Stable Trajectory Compared to the E7 Reference and H51N
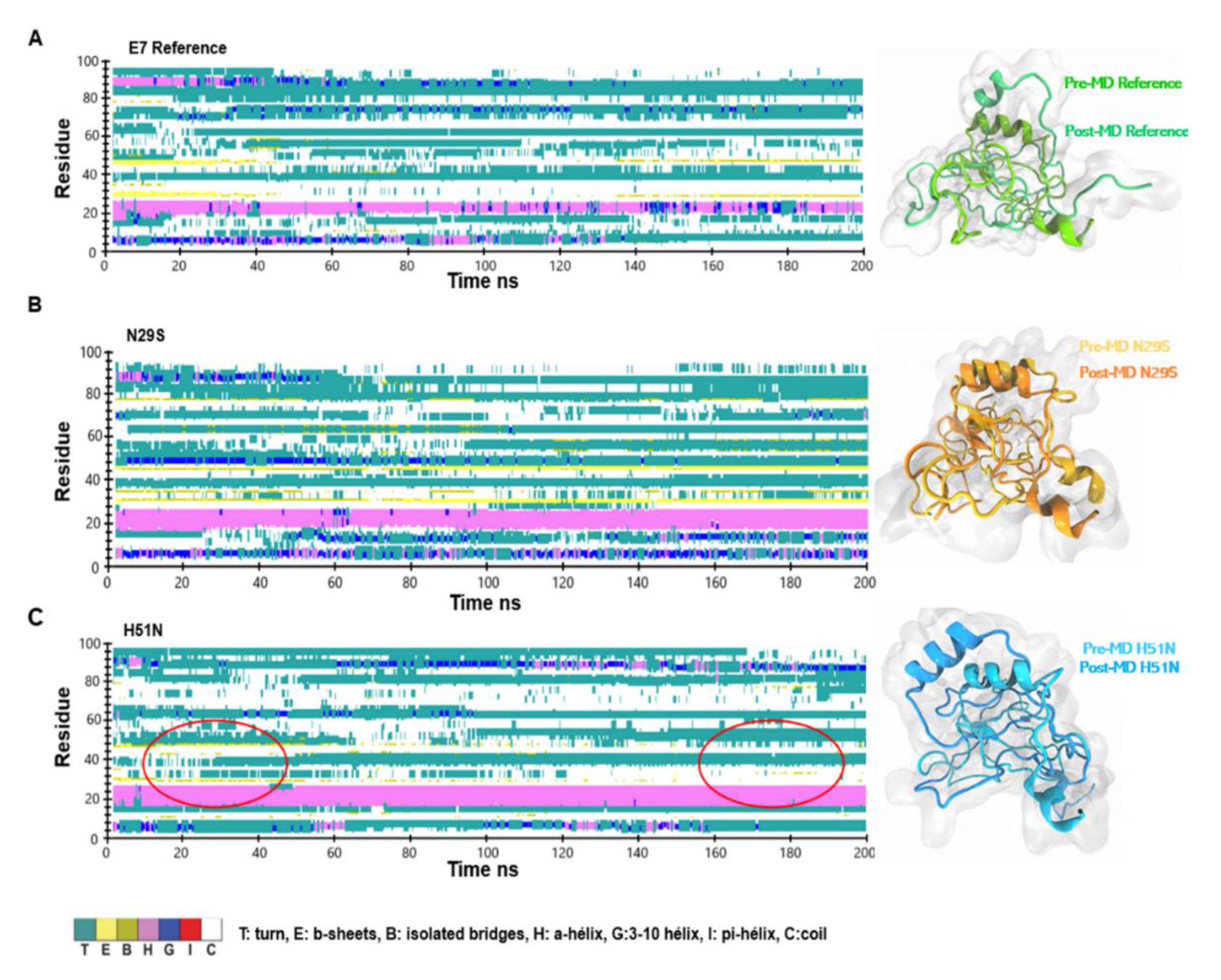
2.3. H51N Loses β-Sheets throughout the Trajectory Simulation
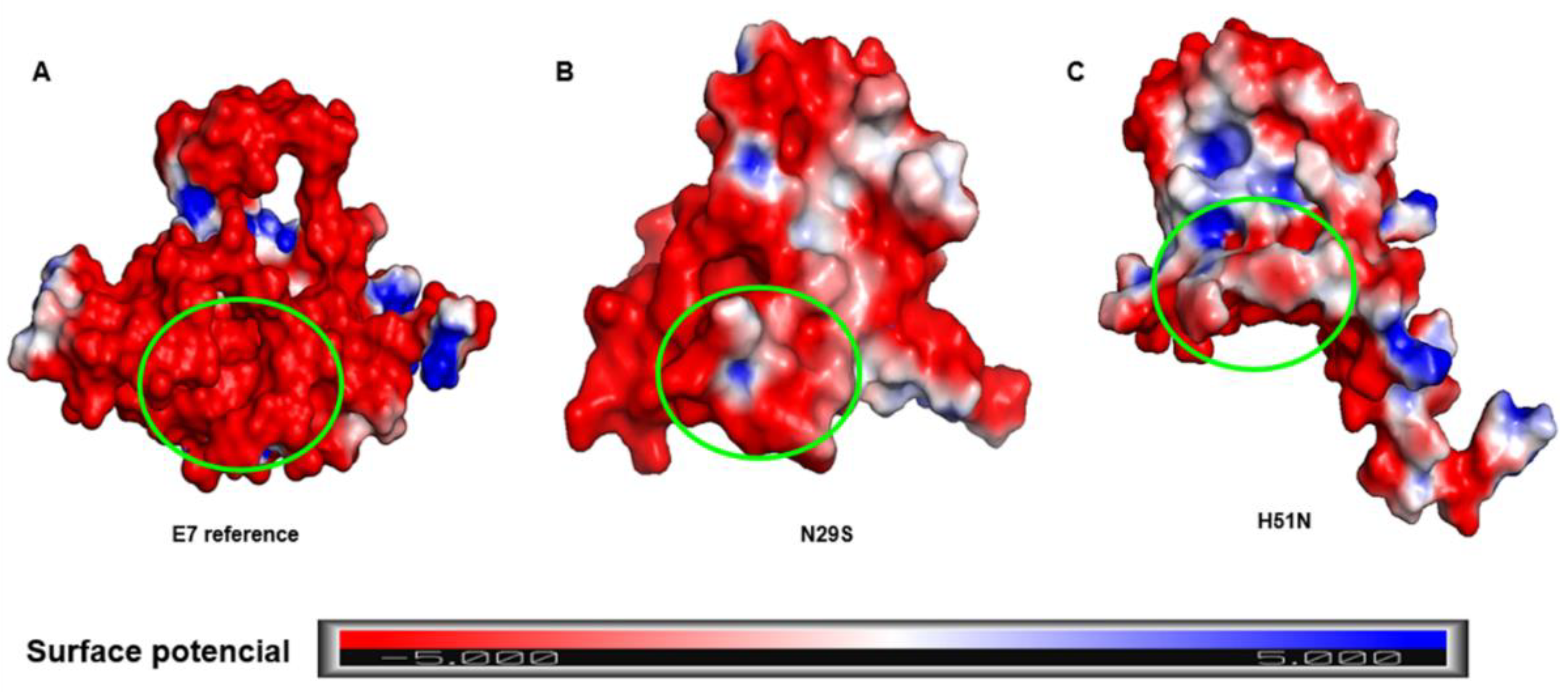
2.4. Surface Electrostatic Potential of E7 and Its Variants
2.5. The CR3 Region Allows Homodimer Formation of the E7 Protein
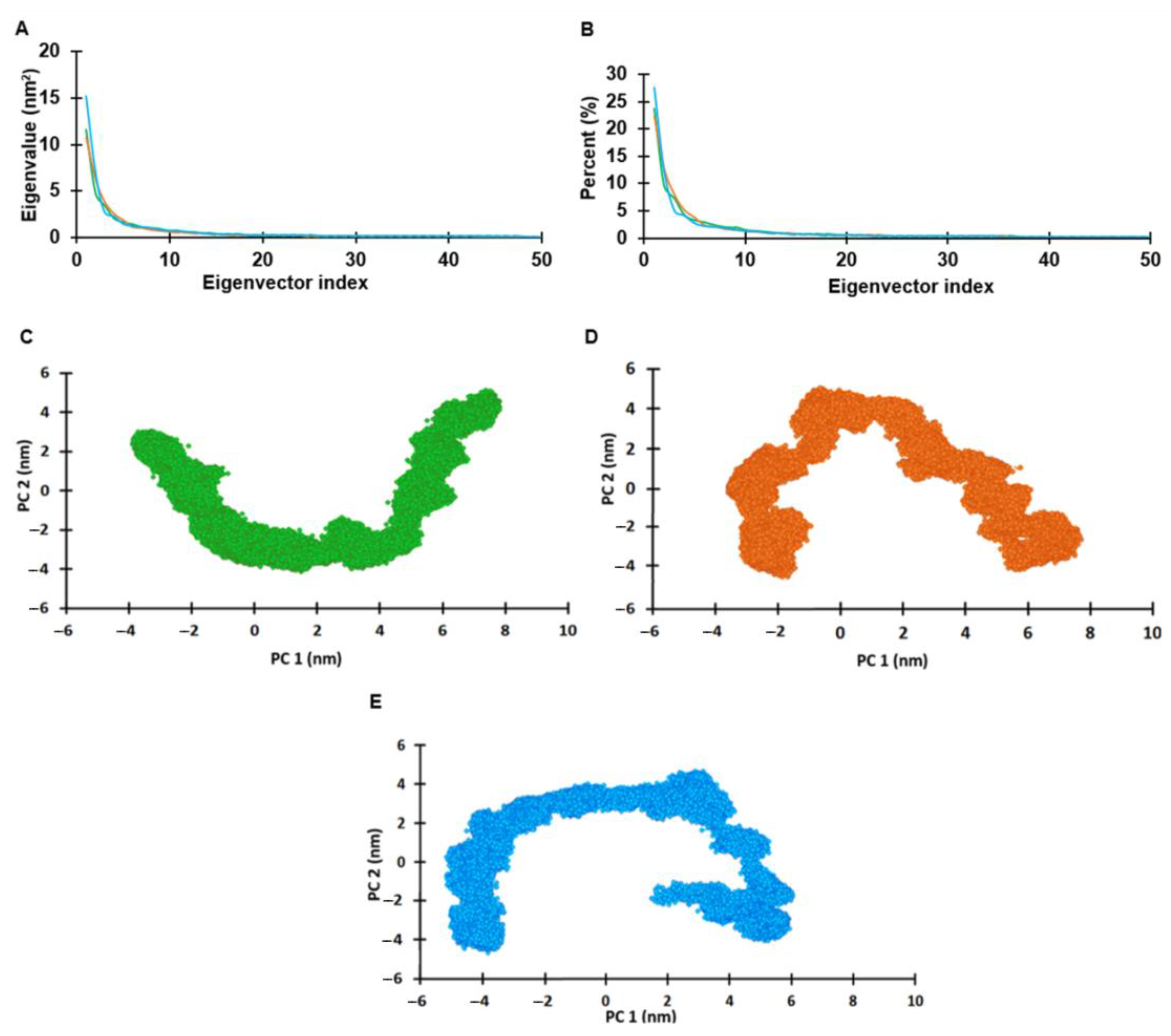
2.6. Principal Component Analysis (PCA)
3. Materials and Methods
3.1. The 3D Structure of E7 and Its Variants
3.2. Molecular Dynamics Simulation
3.3. Trajectory and Secondary Structure Analysis
3.4. Calculation of Surface Electrostatic Potential
3.5. Homodimer Prediction
3.6. Principal Component Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 3D | Three Dimensional |
| CC | Cervical Cancer |
| HPV16 | Human Papillomavirus Type 16 |
| PDB | Protein Data Bank |
| MD | molecular dynamics |
| HPV | Human Papillomavirus |
| dPCA | Dihedralangles PCA |
References
- Jemal, A.; Siegel, R.; Xu, J.; Ward, E. Cancer Statistics, 2010. CA A Cancer J. Clin. 2011, 60, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Kudela, E.; Holubekova, V.; Farkasova, A.; Danko, J. Determination of malignant potential of cervical intraepithelial neoplasia. Tumor Biol. 2015, 37, 1521–1525. [Google Scholar] [CrossRef] [PubMed]
- Burd, E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Woodman, C.B.J.; Collins, S.I.; Young, L.S. The natural history of cervical HPV infection: Unresolved issues. Nat. Rev. Cancer 2007, 7, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Yugawa, T.; Kiyono, T. Molecular mechanisms of cervical carcinogenesis by high-risk human papillomaviruses: Novel functions of E6 and E7 oncoproteins. Rev. Med. Virol. 2009, 19, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Baseman, J.G.; Koutsky, L.A. The epidemiology of human papillomavirus infections. J. Clin. Virol. 2005, 32, 16–24. [Google Scholar] [CrossRef]
- Schiffman, M.; Doorbar, J.; Wentzensen, N.; de Sanjosé, S.; Fakhry, C.; Monk, B.J.; Stanley, M.A.; Franceschi, S. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Primers 2016, 2, 16086. [Google Scholar] [CrossRef]
- Shanmugasundaram, S.; You, J. Targeting Persistent Human Papillomavirus Infection. Viruses 2017, 9, 229. [Google Scholar] [CrossRef]
- Hoppe-Seyler, K.; Bossler, F.; Braun, J.A.; Herrmann, A.L.; Hoppe-Seyler, F. The HPV E6/E7 Oncogenes: Key Factors for Viral Carcinogenesis and Therapeutic Targets. Trends Microbiol. 2018, 26, 158–168. [Google Scholar] [CrossRef]
- Thomas, J.T.; Hubert, W.G.; Ruesch, M.N.; Laimins, L.A. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. USA 1999, 96, 8449–8454. [Google Scholar] [CrossRef]
- Flores, E.R.; Allen-Hoffmann, B.L.; Lee, D.; Lambert, P.F. The Human Papillomavirus Type 16 E7 Oncogene Is Required for the Productive Stage of the Viral Life Cycle. J. Virol. 2000, 74, 6622. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.T.; Longworth, M.S.; Laimins, L.A. Roles of the E6 and E7 proteins in the life cycle of low-risk human papillomavirus type 11. J. Virol. 2004, 78, 2620–2626. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin-Drubin, M.E.; Münger, K. The human papillomavirus E7 oncoprotein. Virology 2009, 384, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Disbrow, G.L.; Yuan, H.; Tomaić, V.; Schlegel, R. Myc and Human Papillomavirus Type 16 E7 Genes Cooperate To Immortalize Human Keratinocytes. J. Virol. 2007, 81, 12689. [Google Scholar] [CrossRef]
- Hellner, K.; Mar, J.; Fang, F.; Quackenbush, J.; Münger, K. HPV16 E7 oncogene expression in normal human epithelial cells causes molecular changes indicative of an epithelial to mesenchymal transition. Virology 2009, 391, 57–63. [Google Scholar] [CrossRef]
- Zhou, X.; Münger, K. Expression of the human papillomavirus type 16 E7 oncoprotein induces an autophagy-related process and sensitizes normal human keratinocytes to cell death in response to growth factor deprivation. Virology 2009, 385, 192–197. [Google Scholar] [CrossRef]
- Wang, N.; Zhan, T.; Ke, T.; Huang, X.; Ke, D.; Wang, Q.; Li, H. Increased expression of RRM2 by human papillomavirus E7 oncoprotein promotes angiogenesis in cervical cancer. Br. J. Cancer 2014, 110, 1034–1044. [Google Scholar] [CrossRef]
- Shankar, S.; Prasad, D.; Sanawar, R.; Das, A.V.; Pillai, M.R. TALEN based HPV-E7 editing triggers necrotic cell death in cervical cancer cells. Sci. Rep. 2017, 7, 5500. [Google Scholar] [CrossRef]
- Phelps, W.C.; Münger, K.; Yee, C.L.; Barnes, J.A.; Howley, P.M. Structure-function analysis of the human papillomavirus type 16 E7 oncoprotein. J. Virol. 1992, 66, 2418–2427. [Google Scholar] [CrossRef]
- Ohlenschläger, O.; Seiboth, T.; Zengerling, H.; Briese, L.; Marchanka, A.; Ramachandran, R.; Baum, M.; Korbas, M.; Meyer-Klaucke, W.; Dürst, M.; et al. Solution structure of the partially folded high-risk human papilloma virus 45 oncoprotein E7. Oncogene 2006, 25, 5953–5959. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Bromberg-White, J.L.; Meyers, C. The role of the human papillomavirus type 18 E7 oncoprotein during the complete viral life cycle. Virology 2005, 338, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Clements, A.; Zhao, K.; Marmorstein, R. Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor. J. Biol. Chem. 2006, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Jansma, A.L.; Martinez-Yamout, M.A.; Liao, R.; Sun, P.; Dyson, H.J.; Wright, P.E. The high-risk HPV16 E7 oncoprotein mediates interaction between the transcriptional coactivator CBP and the retinoblastoma protein pRb. J. Mol. Biol. 2014, 426, 4030–4048. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.S.; Edmonds, C.; Fisher, C.; Schiller, J.T.; Lowy, D.R.; Vousden, K.H. The region of the HPV E7 oncoprotein homologous to adenovirus E1a and Sv40 large T antigen contains separate domains for Rb binding and casein kinase II phosphorylation. EMBO J. 1990, 9, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chen, W.; Roman, A. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation. Proc. Natl. Acad. Sci. USA 2006, 103, 437. [Google Scholar] [CrossRef]
- Chellappan, S.; Kraus, V.B.; Kroger, B.; Munger, K.; Howley, P.M.; Phelps, W.C.; Nevins, J.R. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 1992, 89, 4549. [Google Scholar] [CrossRef]
- McIntyre, M.C.; Frattini, M.G.; Grossman, S.R.; Laimins, L.A. Human papillomavirus type 18 E7 protein requires intact Cys-X-X-Cys motifs for zinc binding, dimerization, and transformation but not for Rb binding. J. Virol. 1993, 67, 3142. [Google Scholar] [CrossRef]
- Balsitis, S.; Dick, F.; Lee, D.; Farrell, L.; Hyde, R.K.; Griep, A.E.; Dyson, N.; Lambert, P.F. Examination of the pRb-Dependent and pRb-Independent Functions of E7 In Vivo. J. Virol. 2005, 79, 11392. [Google Scholar] [CrossRef]
- Todorovic, B.; Massimi, P.; Hung, K.; Shaw, G.S.; Banks, L.; Mymryk, J.S. Systematic analysis of the amino acid residues of human papillomavirus type 16 E7 conserved region 3 involved in dimerization and transformation. J. Virol. 2011, 85, 10048–10057. [Google Scholar] [CrossRef][Green Version]
- Berezutskaya, E.; Yu, B.; Morozov, A.; Raychaudhuri, P.; Bagchi, S. Differential regulation of the pocket domains of the retinoblastoma family proteins by the HPV16 E7 oncoprotein. Cell Growth Differ. 1997, 8, 1277–1286. [Google Scholar]
- Pang, C.L.; Toh, S.Y.; He, P.; Teissier, S.; Ben Khalifa, Y.; Xue, Y.; Thierry, F. A functional interaction of E7 with B-Myb-MuvB complex promotes acute cooperative transcriptional activation of both S- and M-phase genes. (129 c). Oncogene 2014, 33, 4039–4049. [Google Scholar] [CrossRef] [PubMed]
- Hatterschide, J.; Bohidar, A.E.; Grace, M.; Nulton, T.J.; Kim, H.W.; Windle, B.; Morgan, I.M.; Munger, K.; White, E.A. PTPN14 degradation by high-risk human papillomavirus E7 limits keratinocyte differentiation and contributes to HPV-mediated oncogenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 7033. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.S.; Kee, S.H.; Kim, J.W.; Park, N.H.; Kang, S.B.; Chang, W.H.; Lee, H.P. Major Sequence Variants in E7 Gene of Human Papillomavirus Type 16 from Cervical Cancerous and Noncancerous Lesions of Korean Women. Gynecol. Oncol. 1997, 66, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Pillai, M.R.; Hariharan, R.; Babu, J.M.; Lakshmi, S.; Chiplunkar, S.V.; Patkar, M.; Tongaonkar, H.; Dinshaw, K.; Jayshree, R.S.; Reddy, B.K.M.; et al. Molecular variants of HPV-16 associated with cervical cancer in Indian population. Int. J. Cancer 2009, 125, 91–103. [Google Scholar] [CrossRef]
- Park, J.S.; Shin, S.; Kim, E.-C.; Kim, J.E.; Kim, Y.B.; Oh, S.; Roh, E.Y.; Yoon, J.H. Association of human papillomavirus type 16 and its genetic variants with cervical lesion in Korea. APMIS 2016, 124, 950–957. [Google Scholar] [CrossRef]
- Ramas, V.; Mirazo, S.; Bonilla, S.; Ruchansky, D.; Arbiza, J. Analysis of human papillomavirus 16 E6, E7 genes and Long Control Region in cervical samples from Uruguayan women. Gene 2018, 654, 103–109. [Google Scholar] [CrossRef]
- Hoffmann, M.; Lohrey, C.; Hunziker, A.; Kahn, T.; Schwarz, E. Human papillomavirus type 16 E6 and E7 genotypes in head-and-neck carcinomas. Oral Oncol. 2004, 40, 520–524. [Google Scholar] [CrossRef]
- Mirabello, L.; Yeager, M.; Yu, K.; Clifford, G.M.; Xiao, Y.; Zhu, B.; Cullen, M.; Boland, J.F.; Wentzensen, N.; Nelson, C.W.; et al. HPV16 E7 Genetic Conservation Is Critical to Carcinogenesis. Cell 2017, 170, 1164–1174.e6. [Google Scholar] [CrossRef]
- Zine El Abidine, A.; Tomaić, V.; Bel Haj Rhouma, R.; Massimi, P.; Guizani, I.; Boubaker, S.; Ennaifer, E.; Banks, L. A naturally occurring variant of HPV-16 E7 exerts increased transforming activity through acquisition of an additional phospho-acceptor site. Virology 2017, 500, 218–225. [Google Scholar] [CrossRef]
- Chemes, L.B.; Glavina, J.; Alonso, L.G.; Marino-Buslje, C.; de Prat-Gay, G.; Sánchez, I.E. Sequence Evolution of the Intrinsically Disordered and Globular Domains of a Model Viral Oncoprotein. PLoS ONE 2012, 7, e47661. [Google Scholar] [CrossRef]
- Noval, M.G.; Gallo, M.; Perrone, S.n.; Salvay, A.G.; Chemes, L.B.; de Prat-Gay, G. Conformational Dissection of a Viral Intrinsically Disordered Domain Involved in Cellular Transformation. PLoS ONE 2013, 8, e72760. [Google Scholar] [CrossRef] [PubMed]
- Giarrè, M.; Caldeira, S.; Malanchi, I.; Ciccolini, F.; Leão, M.J.; Tommasino, M. Induction of pRb degradation by the human papillomavirus type 16 E7 protein is essential to efficiently overcome p16INK4a-imposed G1 cell cycle Arrest. J. Virol. 2001, 75, 4705–4712. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.D.R.; Jeanmougin, F.O.; Higgins, D.G. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices11Edited by G. Von Heijne. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2016, 14, 71–73. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Glykos, N.M. Software news and updates carma: A molecular dynamics analysis program. J. Comput. Chem. 2006, 27, 1765–1768. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Zhang, Y. Protein-protein complex structure predictions by multimeric threading and template recombination. Structure 2011, 19, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Valdovinos-Torres, H.; Orozco-Morales, M.; Pedroza-Saavedra, A.; Padilla-Noriega, L.; Esquivel-Guadarrama, F.; Gutierrez-Xicotencatl, L. Different Isoforms of HPV-16 E7 Protein are Present in Cytoplasm and Nucleus. Open Virol. J. 2008, 2, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, M.O.; Hošeka, T.; Calçadaa, E.O.; Castiglia, F.; Massimi, P.; Banks, L.; Felli, I.C.; Pierattelli, R. Monitoring HPV-16 E7 phosphorylation events. Virology 2017, 503, 70–75. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bello-Rios, C.; Montaño, S.; Garibay-Cerdenares, O.L.; Araujo-Arcos, L.E.; Leyva-Vázquez, M.A.; Illades-Aguiar, B. Modeling and Molecular Dynamics of the 3D Structure of the HPV16 E7 Protein and Its Variants. Int. J. Mol. Sci. 2021, 22, 1400. https://doi.org/10.3390/ijms22031400
Bello-Rios C, Montaño S, Garibay-Cerdenares OL, Araujo-Arcos LE, Leyva-Vázquez MA, Illades-Aguiar B. Modeling and Molecular Dynamics of the 3D Structure of the HPV16 E7 Protein and Its Variants. International Journal of Molecular Sciences. 2021; 22(3):1400. https://doi.org/10.3390/ijms22031400
Chicago/Turabian StyleBello-Rios, Ciresthel, Sarita Montaño, Olga Lilia Garibay-Cerdenares, Lilian Esmeralda Araujo-Arcos, Marco Antonio Leyva-Vázquez, and Berenice Illades-Aguiar. 2021. "Modeling and Molecular Dynamics of the 3D Structure of the HPV16 E7 Protein and Its Variants" International Journal of Molecular Sciences 22, no. 3: 1400. https://doi.org/10.3390/ijms22031400
APA StyleBello-Rios, C., Montaño, S., Garibay-Cerdenares, O. L., Araujo-Arcos, L. E., Leyva-Vázquez, M. A., & Illades-Aguiar, B. (2021). Modeling and Molecular Dynamics of the 3D Structure of the HPV16 E7 Protein and Its Variants. International Journal of Molecular Sciences, 22(3), 1400. https://doi.org/10.3390/ijms22031400






