Caulerpin Mitigates Helicobacter pylori-Induced Inflammation via Formyl Peptide Receptors
Abstract
:1. Introduction
2. Results
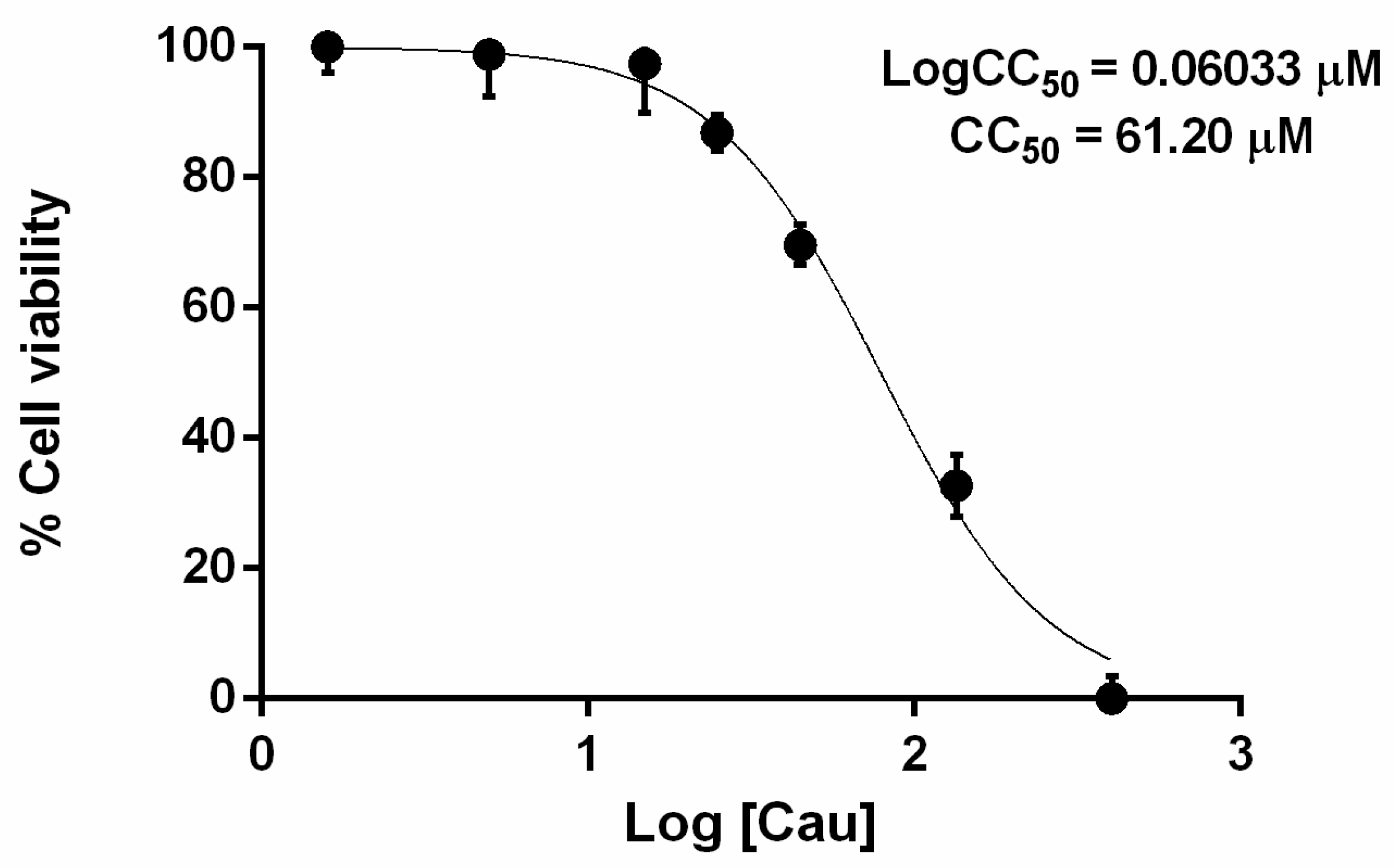
2.1. The Effect of Cau on Cell Viability
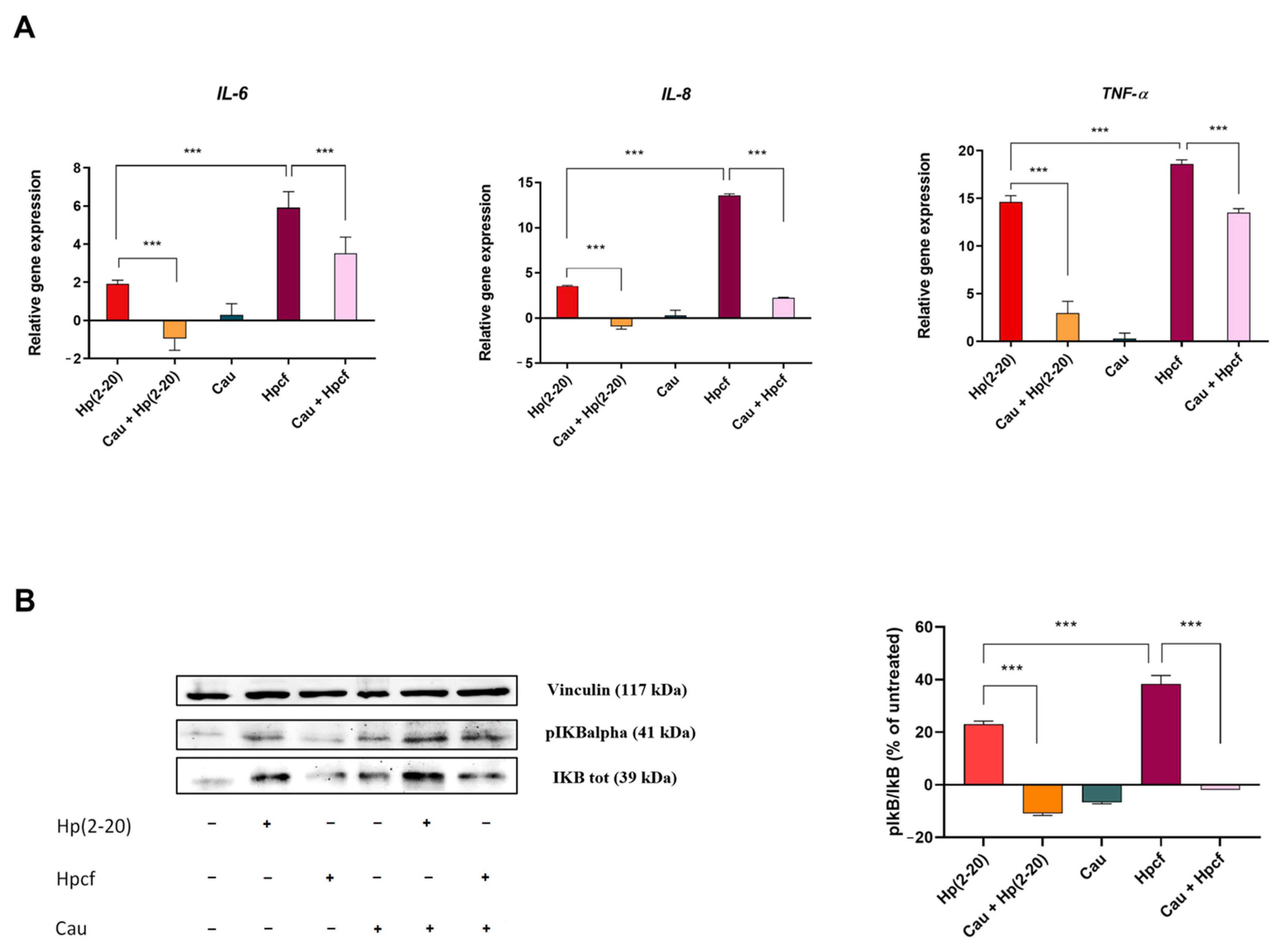
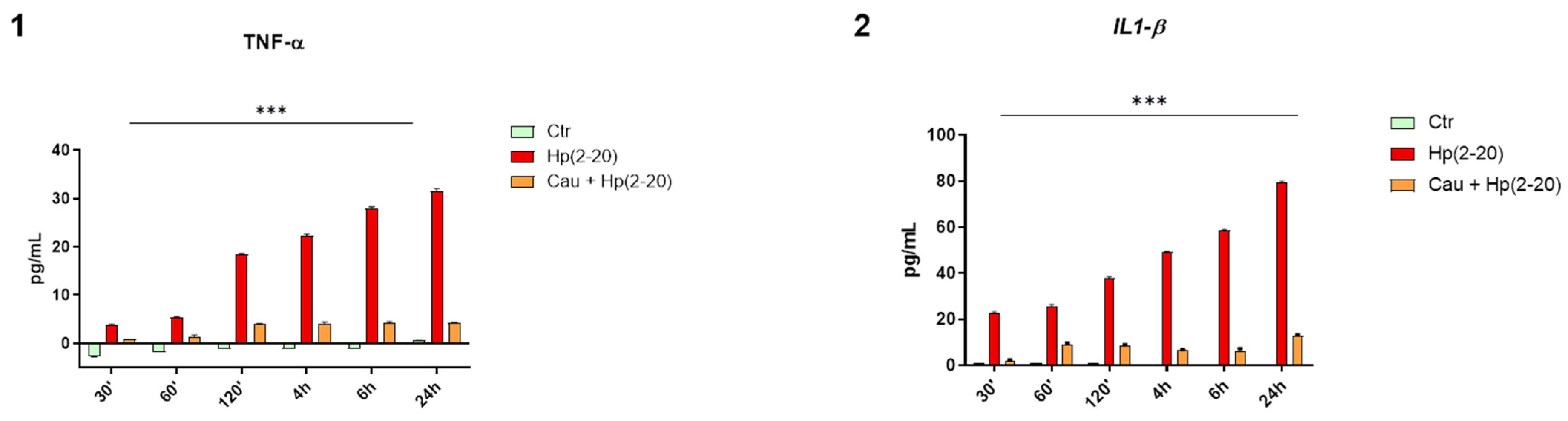
2.2. Cau Reverses the Pro-Inflammatory Action of Hp(2–20)
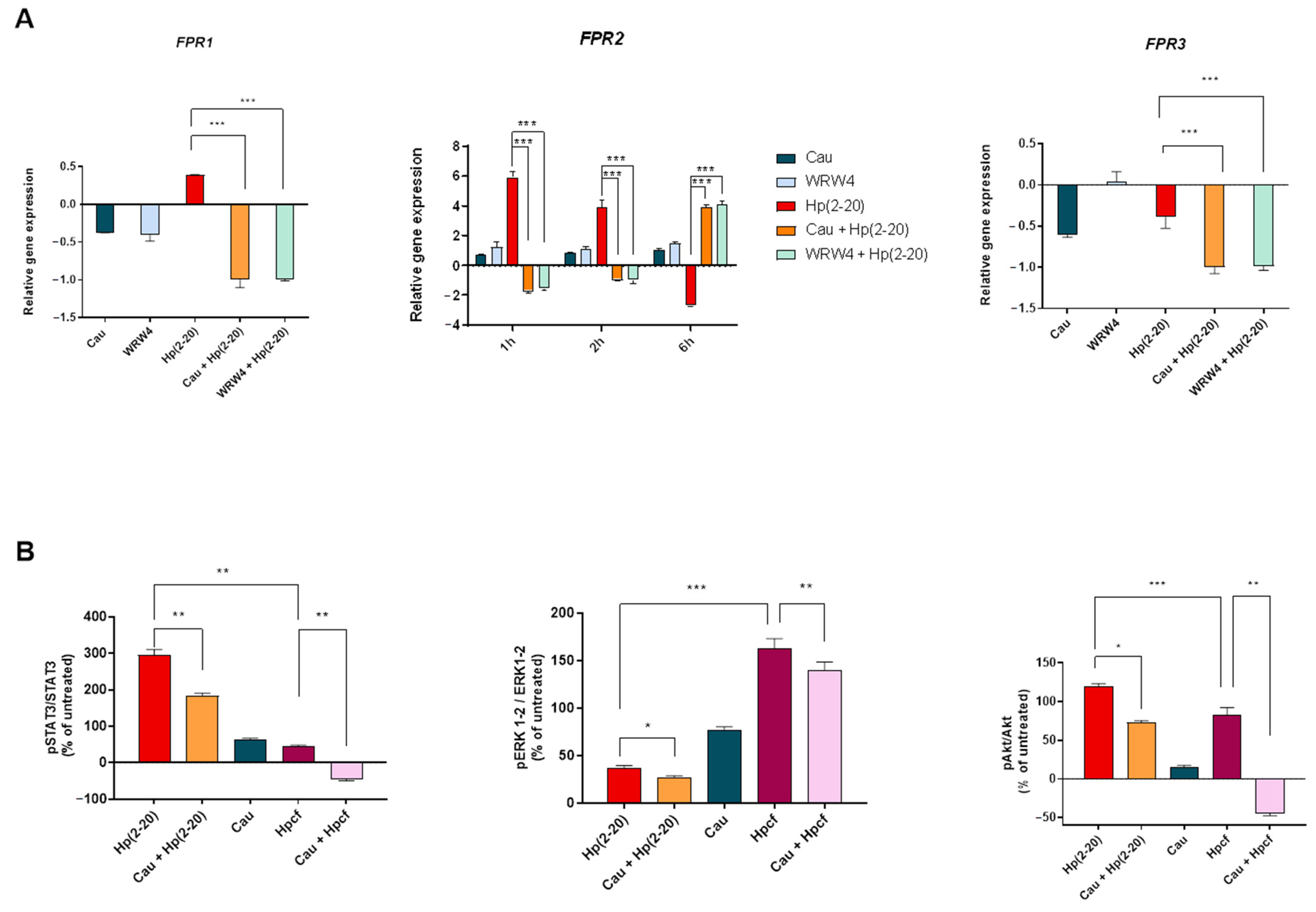
2.3. The Effect of Cau on Hp(2–20)-Induced Chemotactic Signals
2.4. Cau Prevents Hp(2–20)-Induced Inflammatory Response in Macrophages
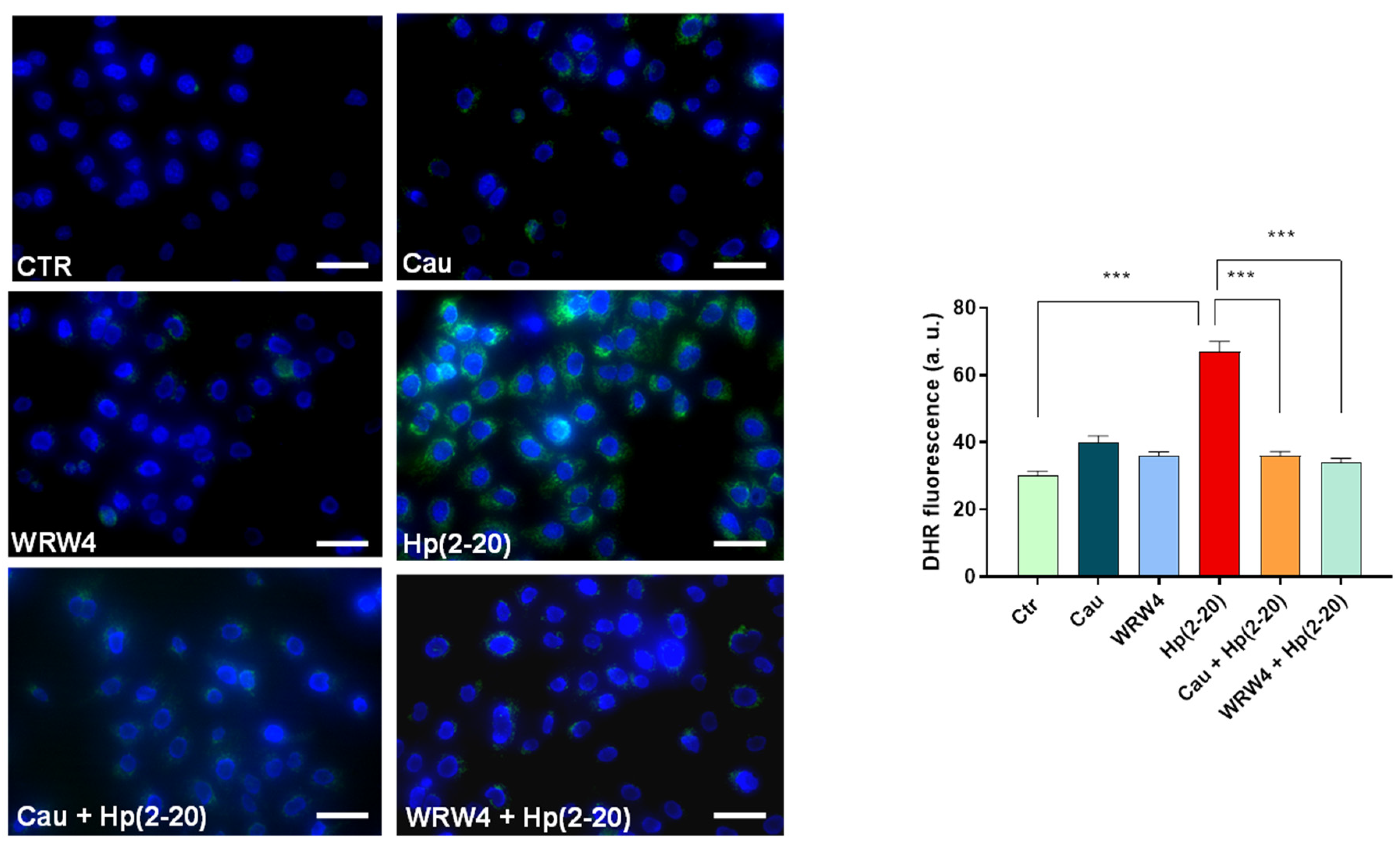
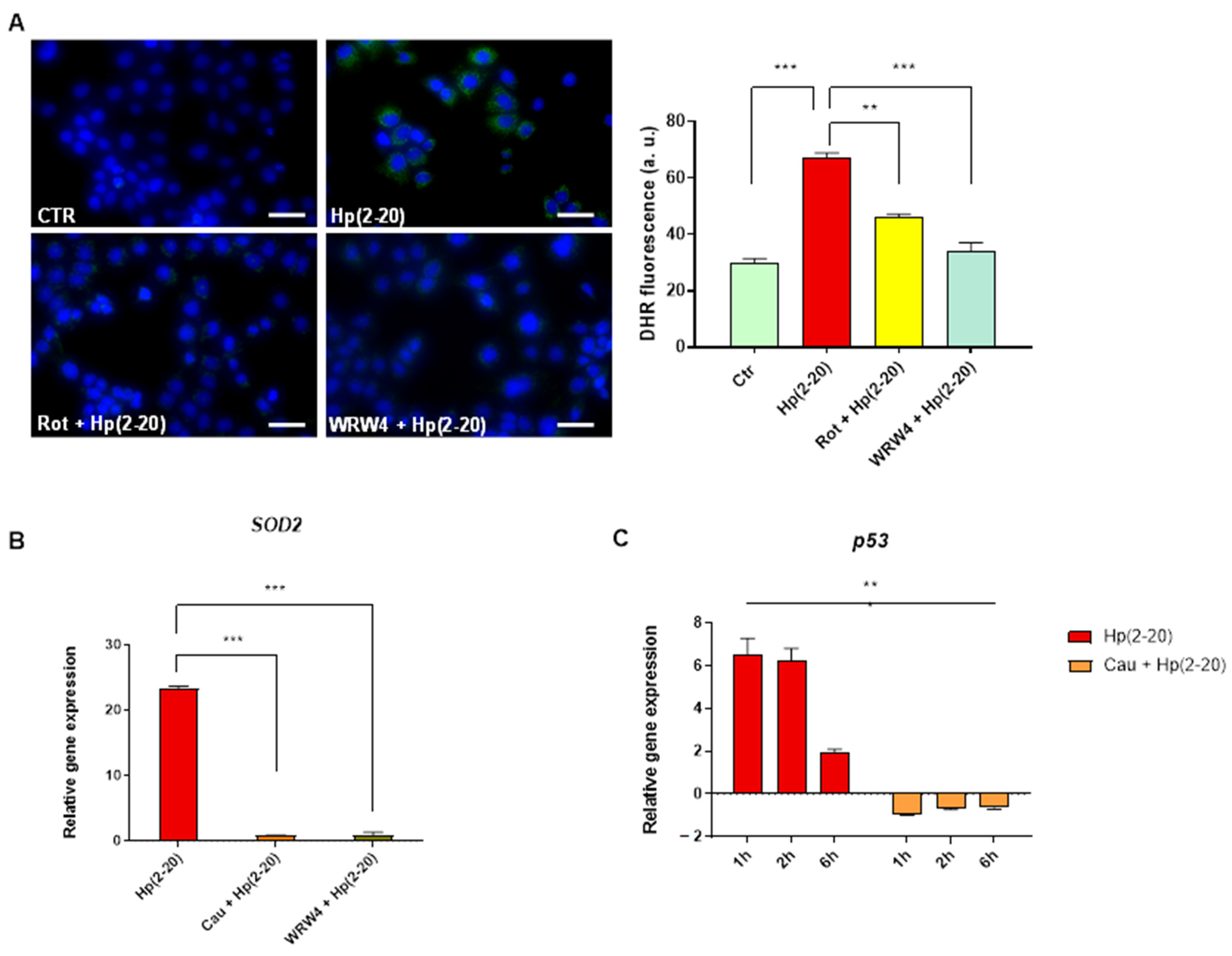
2.5. Cau Affects the Hp(2–20)-Induced ROS Production via Mitochondrial and NADPH Oxidase-Dependent Mechanisms
2.6. Cau Acts as FPR2 Inhibitor
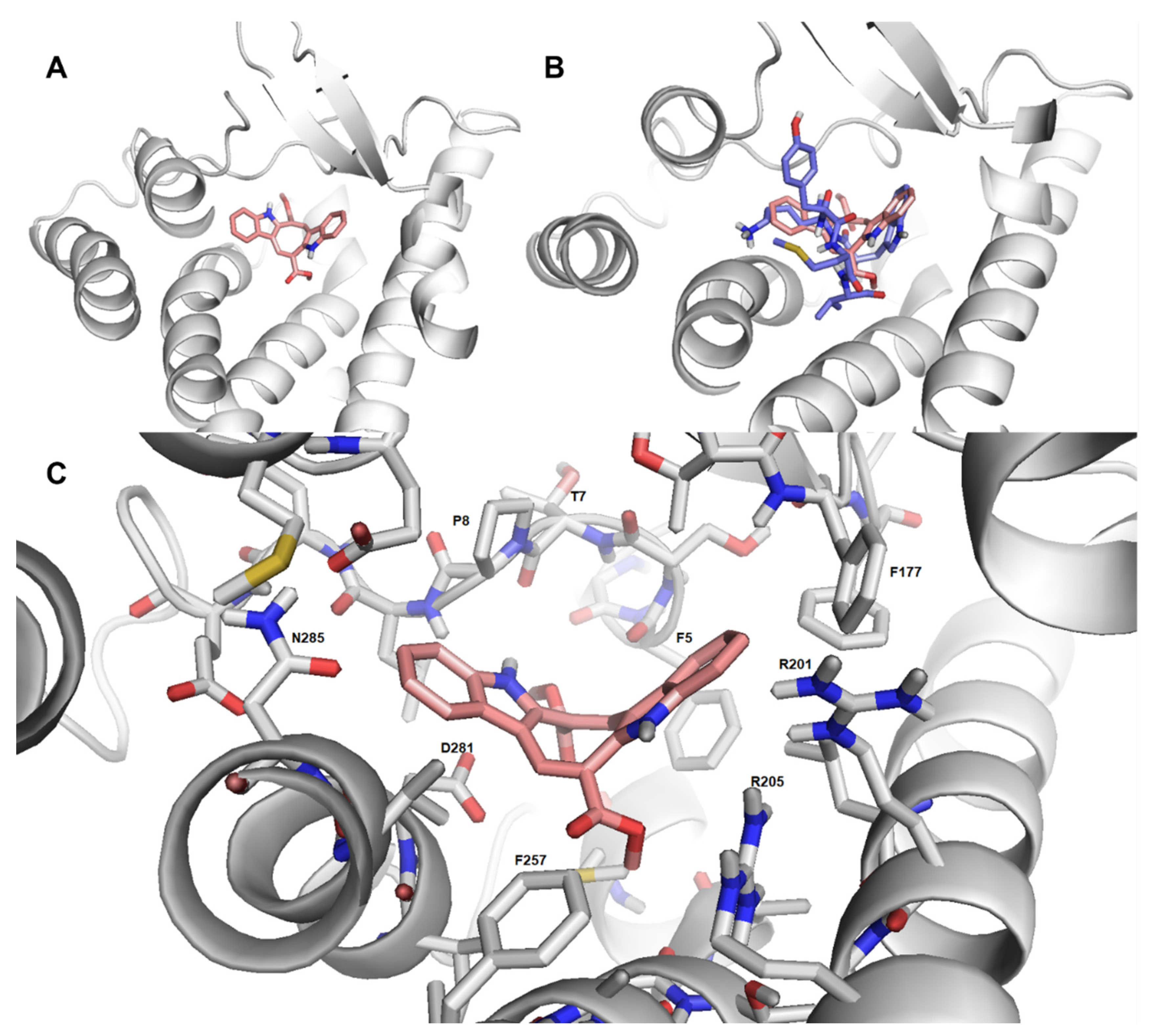
2.7. Cau-FPR2 Interaction: Predictive Computational Studies
2.8. Cau: A More Potent Anti-Inflammatory Molecule than Indomethacin
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Helicobacter Pylori Culture Filtrate Production
4.3. Cau Extraction and Purification
4.4. Cell Viability Assay
4.5. In Vitro Scratch Assay
4.6. RNA Extraction and Quantitative Real-Time PCR
4.7. Measurement of Cytokines Production in Macrophages
4.8. Intracellular ROS Measurement
4.9. Western Blotting for Protein Studies
4.10. Cytokines Assay
4.11. Molecular Modeling
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tshibangu-Kabamba, E.; Yamaoka, Y. Helicobacter pylori infection and antibiotic resistance—From biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 613–629. [Google Scholar] [CrossRef]
- Mentis, A.F.A.; Boziki, M.; Grigoriadis, N.; Papavassiliou, A.G. Helicobacter pylori infection and gastric cancer biology: Tempering a double-edged sword. Cell. Mol. Life Sci. 2019, 76, 2477–2486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, J.R.; Winter, J.A.; Robinson, K. Differential inflammatory response to Helicobacter pylori infection: Etiology and clinical outcomes. J. Inflamm. Res. 2015, 8, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Charitos, I.A.; D’agostino, D.; Topi, S.; Bottalico, L.; Li, C.-F.; Yang, C.-C.; Chiang, N.-J. 40 Years of Helicobacter pylori: A Revolution in Biomedical Thought. Gastroenterol. Insights 2021, 12, 111–135. [Google Scholar] [CrossRef]
- Durazzo, M.; Adriani, A.; Fagoonee, S.; Saracco, G.M.; Pellicano, R. Helicobacter pylori and Respiratory Diseases: 2021 Update. Microorganisms 2021, 9, 2033. [Google Scholar] [CrossRef]
- Mladenova, I. Helicobacter pylori and cardiovascular disease: Update 2019. Minerva Cardioangiol. 2019, 67, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Li, J.Y.; Wu, T.F.; Xu, J.H.; Huang, C.Z.; Cheng, D.; Chen, Q.K.; Yu, T. Helicobacter pylori Infection Is Associated with Type 2 Diabetes, Not Type 1 Diabetes: An Updated Meta-Analysis. Gastroenterol. Res. Pract. 2017, 2017, 5715403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mégraud, F.; Bessède, E.; Varon, C. Helicobacter pylori infection and gastric carcinoma. Clin. Microbiol. Infect. 2015, 21, 984–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wroblewski, L.E.; Peek, R.M., Jr.; Wilson, K.T. Helicobacter pylori and Gastric Cancer: Factors That Modulate Disease Risk. Clin. Microbiol. Rev. 2010, 23, 713–739. [Google Scholar] [CrossRef] [Green Version]
- Cuomo, P.; Papaianni, M.; Sansone, C.; Iannelli, A.; Iannelli, D.; Medaglia, C.; Paris, D.; Motta, A.; Capparelli, R. An In Vitro Model to Investigate the Role of Helicobacter pylori in Type 2 Diabetes, Obesity, Alzheimer’s Disease and Cardiometabolic Disease. Int. J. Mol. Sci. 2020, 21, 8369. [Google Scholar] [CrossRef]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2017, 9, 7204–7218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, G.M. A calculated response: Control of inflammation by the innate immune system. J. Clin. Investig. 2008, 118, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C. Points of control in inflammation. Nature 2002, 420, 846–852. [Google Scholar] [CrossRef]
- Van der Heijden, C.D.C.C.; Noz, M.P.; Joosten, L.A.B.; Netea, M.G.; Riksen, N.P.; Keating, S.T. Epigenetics and Trained Immunity. Antioxid. Redox Signal. 2018, 29, 1023–1040. [Google Scholar] [CrossRef] [PubMed]
- Paludan, S.R.; Pradeu, T.; Masters, S.L.; Mogensen, T.H. Constitutive immune mechanisms: Mediators of host defence and immune regulation. Nat. Rev. Immunol. 2021, 21, 137–150. [Google Scholar] [CrossRef]
- Raabe, C.A.; Gröper, J.; Rescher, U. Biased perspectives on formyl peptide receptors. Biochim. Biophys. Acta-Mol. Cell Res. 2019, 1866, 305–316. [Google Scholar] [CrossRef]
- Cattaneo, F.; Russo, R.; Castaldo, M.; Chambery, A.; Zollo, C.; Esposito, G.; Pedone, P.V.; Ammendola, R. Phosphoproteomic analysis sheds light on intracellular signaling cascades triggered by Formyl-Peptide Receptor 2. Sci. Rep. 2019, 9, 17894. [Google Scholar] [CrossRef]
- Muto, Y.; Guindon, S.; Umemura, T.; Kőhidai, L.; Ueda, H. Adaptive evolution of formyl peptide receptors in mammals. J. Mol. Evol. 2015, 80, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Boillat, M.; Carleton, A.; Rodriguez, I. From immune to olfactory expression: Neofunctionalization of formyl peptide receptors. Cell Tissue Res. 2021, 383, 387–393. [Google Scholar] [CrossRef]
- Wang, H.; Peng, X.; Ge, Y.; Zhang, S.; Wang, Z.; Fan, Y.; Huang, W.; Qiu, M.; Ye, R.D. A Ganoderma- Derived Compound Exerts Inhibitory Effect through Formyl Peptide Receptor 2. Front. Pharmacol. 2020, 11, 337. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.; Chen, K.; Gong, W.; Yoshimura, T.; Le, Y.; Wang, Y.; Wang, J.M. The Contribution of Chemoattractant GPCRs, Formylpeptide Receptors, to Inflammation and Cancer. Front. Endocrinol. 2020, 11, 17. [Google Scholar] [CrossRef] [Green Version]
- Cattaneo, F.; Parisi, M.; Ammendola, R. Distinct Signaling Cascades Elicited by Different Formyl Peptide Receptor 2 (FPR2) Agonists. Int. J. Mol. Sci. 2013, 14, 7193–7230. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-C.; Kim, M.-H.; Hossain, M.A.; Shin, S.Y.; Kim, Y.; Stella, L.; Wade, J.D.; Park, Y.; Hahm, K.-S. Amphipathic α-helical peptide, HP (2–20), and its analogues derived from Helicobacter pylori: Pore formation mechanism in various lipid compositions. Biochim. Biophys. Acta (BBA)-Biomembr. 2007, 1778, 229–241. [Google Scholar] [CrossRef]
- Bylund, J.; Christophe, T.; Boulay, F.; Nyström, T.; Karlsson, A.; Dahlgren, C. Proinflammatory Activity of a Cecropin-Like Antibacterial Peptide from Helicobacter pylori. Antimicrob. Agents Chemother. 2001, 45, 1700–1704. [Google Scholar] [CrossRef] [Green Version]
- Cuomo, P.; Papaianni, M.; Capparelli, R.; Medaglia, C. The Role of Formyl Peptide Receptors in Permanent and Low-Grade Inflammation: Helicobacter pylori Infection as a Model. Int. J. Mol. Sci. 2021, 22, 3706. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.-L.; Ji, C.-D.; Tang, J.; Wang, Y.-X.; Xiang, D.-F.; Li, H.-Q.; Liu, W.-W.; Wang, J.-X.; Yan, H.-Z.; Wang, Y.; et al. FPR2 promotes invasion and metastasis of gastric cancer cells and predicts the prognosis of patients. Sci. Rep. 2017, 7, 3153. [Google Scholar] [CrossRef] [PubMed]
- Vitale, R.M.; D’Aniello, E.; Gorbi, S.; Martella, A.; Silvestri, C.; Giuliani, M.E.; Fellous, T.; Gentile, A.; Carbone, M.; Cutignano, A.; et al. Fishing for Targets of Alien Metabolites: A Novel Peroxisome Proliferator-Activated Receptor (PPAR) Agonist from a Marine Pest. Mar. Drugs 2018, 16, 431. [Google Scholar] [CrossRef] [Green Version]
- Máximo, P.; Ferreira, L.M.; Branco, P.; Lima, P.; Lourenço, A. Secondary Metabolites and Biological Activity of Invasive Macroalgae of Southern Europe. Mar. Drugs 2018, 8, 265. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Morgan, J.B.; Coothankandaswamy, V.; Liu, R.; Jekabsons, M.B.; Mahdi, F.; Nagle, D.G.; Zhou, Y.-D. The Caulerpa Pigment Caulerpin Inhibits HIF-1 Activation and Mitochondrial Respiration. J. Nat. Prod. 2009, 72, 2104–2109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souza, C.R.M.; Bezerra, W.P.; Souto, J.T. Marine Alkaloids with Anti-Inflammatory Activity: Current Knowledge and Future Perspectives. Mar. Drugs 2020, 18, 147. [Google Scholar] [CrossRef] [Green Version]
- De Sá Alves, F.R.; Barreiro, E.J.; Fraga, C.A. From nature to drug discovery: The indole scaffold as a “privileged structure”. Mini Rev. Med. Chem. 2009, 9, 782–793. [Google Scholar] [CrossRef]
- Guerra, A.S.; Malta, D.J.; Laranjeira, L.P.; Maia, M.B.; Colaço, N.C.; de Lima, M.C.; Galdino, S.L.; da Rocha Pitta, I.; Gonçalves-Silva, T. Anti-inflammatory and antinociceptive activities of indole-imidazolidine derivatives. Int. Immunopharmacol. 2011, 11, 1816–1822. [Google Scholar] [CrossRef] [Green Version]
- Moore, B.B.; Kunkel, S.L. Attracting Attention: Discovery of IL-8/CXCL8 and the Birth of the Chemokine Field. J. Immunol. 2019, 202, 3–4. [Google Scholar] [CrossRef] [Green Version]
- De Paulis, A.; Prevete, N.; Fiorentino, I.; Walls, A.F.; Curto, M.; Petraroli, A.; Castaldo, V.; Ceppa, P.; Fiocca, R.; Marone, G. Basophils Infiltrate Human Gastric Mucosa at Sites of Helicobacter pylori Infection, and Exhibit Chemotaxis in Response to H. pylori-derived Peptide Hp(2–20). J. Immunol. 2004, 172, 7734–7743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louwe, P.A.; Badiola Gomez, L.; Webster, H.; Perona-Wright, G.; Bain, C.C.; Forbes, S.J.; Jenkins, S.J. Recruited macrophages that colonize the post-inflammatory peritoneal niche convert into functionally divergent resident cells. Nat. Commun. 2021, 12, 1770. [Google Scholar] [CrossRef] [PubMed]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Ruytinx, P.; Proost, P.; Van Damme, J.; Struyf, S. Chemokine-Induced Macrophage Polarization in Inflammatory Conditions. Front. Immunol. 2018, 9, 1930. [Google Scholar] [CrossRef] [Green Version]
- Gregory, J.L.; Morand, E.F.; McKeown, S.J.; Ralph, J.A.; Hall, P.; Yang, Y.H.; McColl, S.R.; Hickey, M.J. Macrophage Migration Inhibitory Factor Induces Macrophage Recruitment via CC Chemokine Ligand 2. J. Immunol. 2006, 177, 8072–8079. [Google Scholar] [CrossRef] [Green Version]
- Ruytinx, P.; Proost, P.; Van Damme, J.; Struyf, S. Pro-Resolving FPR2 Agonists Regulate NADPH Oxidase-Dependent Phosphorylation of HSP27, OSR1, and MARCKS and Activation of the Respective Upstream Kinases. Antioxidants 2021, 10, 134. [Google Scholar] [CrossRef]
- Heinz, S.; Freyberger, A.; Lawrenz, B.; Schladt, L.; Schmuck, G.; Ellinger-Ziegelbauer, H. Mechanistic Investigations of the Mitochondrial Complex I Inhibitor Rotenone in the Context of Pharmacological and Safety Evaluation. Sci. Rep. 2017, 7, 45465. [Google Scholar] [CrossRef] [Green Version]
- Berkers, C.R.; Maddocks, O.D.; Cheung, E.C.; Mor, I.; Vousden, K.H. Metabolic regulation by p53 family members. Cell Metab. 2013, 18, 617–633. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Chen, Y.; Clair, D.K.S. ROS and p53: Versatile partnership. Free Radic. Biol. Med. 2008, 44, 1529–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas, S.Y.; Nogueira, M.I.; Azmitia, E.C. Antagonist-induced increase in 5-HT1A-receptor expression in adult rat hippocampus and cortex. Synapse 2007, 61, 531–539. [Google Scholar] [CrossRef]
- Toth, M.; Shenk, T. Antagonist-mediated down-regulation of 5-hydroxytryptamine type 2 receptor gene expression: Modulation of transcription. Mol. Pharmacol. 1994, 45, 1095–1100. [Google Scholar] [PubMed]
- Annunziata, M.C.; Parisi, M.; Esposito, G.; Fabbrocini, G.; Ammendola, R.; Cattaneo, F. Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling. Int. J. Mol. Sci. 2020, 21, 3818. [Google Scholar] [CrossRef] [PubMed]
- Estevão, M.S.; Carvalho, L.C.; Ferreira, L.M.; Fernandes, E.; Manuel, M.; Marques, B. Analysis of the antioxidant activity of an indole library: Cyclic voltammetry versus ROS scavenging activity. Tetrahedron Lett. 2011, 52, 101–106. [Google Scholar] [CrossRef]
- Schepetkin, I.A.; Khlebnikov, A.I.; Giovannoni, M.P.; Kirpotina, L.N.; Cilibrizzi, A.; Quinn, M.T. Development of small molecule non-peptide formyl peptide receptor (FPR) ligands and molecular modeling of their recognition. Curr. Med. Chem. 2014, 21, 1478–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perianin, A.; Gaudry, M.; Marquetty, C.; Giroud, J.-P.; Hakim, J. Protective effect of indomethacin against chemotactic deactivation of human neutrophils induced by formylated peptide. Biochem. Pharmacol. 1988, 37, 1693–1698. [Google Scholar] [CrossRef]
- Rajindrajith, S.; Devanarayana, N.M.; de Silva, H.J. Helicobacter pylori infection in children. Saudi J. Gastroenterol. 2009, 15, 86–94. [Google Scholar] [CrossRef]
- Kumari, A.; Singh, R.K. Medicinal chemistry of indole derivatives: Current to future therapeutic prospectives. Bioorg. Chem. 2019, 89, 103021. [Google Scholar] [CrossRef]
- Kumar, S. A brief review of the biological potential of indole derivatives. Future J. Pharm. Sci. 2020, 6, 121. [Google Scholar] [CrossRef]
- Kaushik, N.K.; Kaushik, N.; Attri, P.; Kumar, N.; Kim, C.H.; Verma, A.K.; Choi, E.H. Biomedical Importance of Indoles. Molecules 2013, 18, 6620–6662. [Google Scholar] [CrossRef]
- Bae, Y.-S.; Lee, H.Y.; Jo, E.J.; Kim, J.I.; Kang, H.-K.; Ye, R.D.; Kwak, J.-Y.; Ryu, S.H. Identification of Peptides That Antagonize Formyl Peptide Receptor-Like 1-Mediated Signaling. J. Immunol. 2004, 173, 607–614. [Google Scholar] [CrossRef] [Green Version]
- Tak, P.P.; Firestein, G.S. NF-κB: A key role in inflammatory diseases. J. Clin. Investig. 2001, 107, 7–11. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniguchi, K.; Karin, M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Xu, H.; Chen, X.; Tang, G.; Gu, L.; Wang, Y. Helicobacter pylori cytotoxin-associated gene A activates tumor necrosis factor-α and interleukin-6 in gastric epithelial cells through P300/CBP-associated factor-mediated nuclear factor-κB p65 acetylation. Mol. Med. Rep. 2015, 12, 6337–6345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devi, S.; Ansari, S.A.; Vadivelu, J.; Mégraud, F.; Tenguria, S.; Ahmed, N. Helicobacter pylori Antigen HP0986 (TieA) Interacts with Cultured Gastric Epithelial Cells and Induces IL8 Secretion via NF-κB Mediated Pathway. Helicobacter 2014, 19, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Atri, C.; Guerfali, F.Z.; Laouini, D. Role of Human Macrophage Polarization in Inflammation during Infectious Diseases. Int. J. Mol. Sci. 2018, 19, 1801. [Google Scholar] [CrossRef] [Green Version]
- De Paulis, A.; Prevete, N.; Rossi, F.W.; Rivellese, F.; Salerno, F.; Delfino, G.; Liccardo, B.; Avilla, E.; Montuori, N.; Mascolo, M.; et al. Helicobacter pylori Hp(2–20) promotes migration and proliferation of gastric epithelial cells by interacting with formyl peptide receptors in vitro and accelerates gastric mucosal healing in vivo. J. Immunol. 2009, 183, 3761–3769. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.-H.; Yi, E.H.; Ye, S.-K. Signal transducer and activator of transcription 3 as a therapeutic target for cancer and the tumor microenvironment. Arch. Pharmacal Res. 2016, 39, 1085–1099. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Yang, M.; Ding, Y.; Yu, L.; Chen, J. Formyl peptide receptor 2 expression predicts poor prognosis and promotes invasion and metastasis in epithelial ovarian cancer. Oncol. Rep. 2017, 38, 3297–3308. [Google Scholar] [CrossRef] [Green Version]
- Jiang, N.; Dai, Q.; Su, X.; Fu, J.; Feng, X.; Peng, J. Role of PI3K/AKT pathway in cancer: The framework of malignant behavior. Mol. Biol. Rep. 2020, 47, 4587–4629. [Google Scholar] [CrossRef]
- Wu, D. Signaling mechanisms for regulation of chemotaxis. Cell Res. 2005, 15, 52–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coffelt, S.B.; Tomchuck, S.L.; Zwezdaryk, K.J.; Danka, E.S.; Scandurro, A.B. Leucine Leucine-37 Uses Formyl Peptide Receptor–Like 1 to Activate Signal Transduction Pathways, Stimulate Oncogenic Gene Expression, and Enhance the Invasiveness of Ovarian Cancer Cells. Mol. Cancer Res. 2009, 7, 907–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leite, M.; Marques, M.S.; Melo, J.; Pinto, M.T.; Cavadas, B.; Aroso, M.; Gomez-Lazaro, M.; Seruca, R.; Figueiredo, C. Helicobacter Pylori Targets the EPHA2 Receptor Tyrosine Kinase in Gastric Cells Modulating Key Cellular Functions. Cells 2020, 9, 513. [Google Scholar] [CrossRef] [Green Version]
- Page, T.H.; Smolinska, M.; Gillespie, J.; Urbaniak, A.M.; Foxwell, B.M. Tyrosine kinases and inflammatory signalling. Curr. Mol. Med. 2009, 9, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Gschwind, A.; Fischer, O.M.; Ullrich, A. The discovery of receptor tyrosine kinases: Targets for cancer therapy. Nat. Rev. Cancer 2004, 4, 361–370. [Google Scholar] [CrossRef]
- Paletta-Silva, R.; Rocco-Machado, N.; Meyer-Fernandes, J.R. NADPH oxidase biology and the regulation of tyrosine kinase receptor signaling and cancer drug cytotoxicity. Int. J. Mol. Sci. 2013, 14, 3683–3704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 2014, 20, 1126–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dikalov, S. Crosstalk between mitochondria and NADPH oxidases. Free Radic. Biol. Med. 2011, 51, 1289–1301. [Google Scholar] [CrossRef] [Green Version]
- Butcher, L.D.; den Hartog, G.; Ernst, P.B.; Crowe, S.E. Oxidative Stress Resulting From Helicobacter pylori Infection Contributes to Gastric Carcinogenesis. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 316–322. [Google Scholar] [CrossRef] [Green Version]
- Fukai, T.; Ushio-Fukai, M. Cross-Talk between NADPH Oxidase and Mitochondria: Role in ROS Signaling and Angiogenesis. Cells 2020, 9, 1849. [Google Scholar] [CrossRef] [PubMed]
- Magliozzi, L.; Maselli, V.; Almada, F.; Di Cosmo, A.; Mollo, E.; Polese, G. Effect of the algal alkaloid caulerpin on neuropeptide Y (NPY) expression in the central nervous system (CNS) of Diplodus sargus. J. Comp. Physiol. 2019, 205, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Chay, C.I.C.; Cansino, R.G.; Pinzón, C.I.E.; Torres-Ochoa, R.O.; Martínez, R. Synthesis and Anti-Tuberculosis Activity of the Marine Natural Product Caulerpin and Its Analogues. Mar. Drugs 2014, 12, 1757–1772. [Google Scholar] [CrossRef]
- Maiti, B.C.; Thomson, R.H.; Mahendran, M. ChemInform Abstract: The structure of caulerpin, a pigment from caulerpa algae. Chem. Informationsd. 1978, 9, 32. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Papaianni, M.; Paris, D.; Woo, S.L.; Fulgione, A.; Rigano, M.M.; Parrilli, E.; Tutino, M.L.; Marra, R.; Manganiello, G.; Casillo, A.; et al. Plant Dynamic Metabolic Response to Bacteriophage Treatment after Xanthomonas campestris pv. campestris Infection. Front. Microbiol. 2020, 11, 732. [Google Scholar] [CrossRef]
- Fulgione, A.; Papaianni, M.; Cuomo, P.; Paris, D.; Romano, M.; Tuccillo, C.; Palomba, L.; Medaglia, C.; De Seta, M.; Esposito, N.; et al. Interaction between MyD88, TIRAP and IL1RL1 against Helicobacter pylori infection. Sci. Rep. 2020, 10, 15831. [Google Scholar] [CrossRef]
- Cuomo, P.; Papaianni, M.; Fulgione, A.; Guerra, F.; Capparelli, R.; Medaglia, C. An Innovative Approach to Control, H. pylori-Induced Persistent Inflammation and Colonization. Microorganisms 2020, 8, 1214. [Google Scholar] [CrossRef] [PubMed]
- Iannelli, D.; D’Apice, L.; Cottone, C.; Viscardi, M.; Scala, F.; Zoina, A.; Del Sorbo, G.; Spigno, P.; Capparelli, R. Simultaneous detection of cucumber mosaic virus, tomato mosaic virus and potato virus Y by flow cytometry. J. Virol. Methods 1997, 69, 137–145. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuomo, P.; Medaglia, C.; Allocca, I.; Montone, A.M.I.; Guerra, F.; Cabaro, S.; Mollo, E.; Eletto, D.; Papaianni, M.; Capparelli, R. Caulerpin Mitigates Helicobacter pylori-Induced Inflammation via Formyl Peptide Receptors. Int. J. Mol. Sci. 2021, 22, 13154. https://doi.org/10.3390/ijms222313154
Cuomo P, Medaglia C, Allocca I, Montone AMI, Guerra F, Cabaro S, Mollo E, Eletto D, Papaianni M, Capparelli R. Caulerpin Mitigates Helicobacter pylori-Induced Inflammation via Formyl Peptide Receptors. International Journal of Molecular Sciences. 2021; 22(23):13154. https://doi.org/10.3390/ijms222313154
Chicago/Turabian StyleCuomo, Paola, Chiara Medaglia, Ivana Allocca, Angela Michela Immacolata Montone, Fabrizia Guerra, Serena Cabaro, Ernesto Mollo, Daniela Eletto, Marina Papaianni, and Rosanna Capparelli. 2021. "Caulerpin Mitigates Helicobacter pylori-Induced Inflammation via Formyl Peptide Receptors" International Journal of Molecular Sciences 22, no. 23: 13154. https://doi.org/10.3390/ijms222313154
APA StyleCuomo, P., Medaglia, C., Allocca, I., Montone, A. M. I., Guerra, F., Cabaro, S., Mollo, E., Eletto, D., Papaianni, M., & Capparelli, R. (2021). Caulerpin Mitigates Helicobacter pylori-Induced Inflammation via Formyl Peptide Receptors. International Journal of Molecular Sciences, 22(23), 13154. https://doi.org/10.3390/ijms222313154







