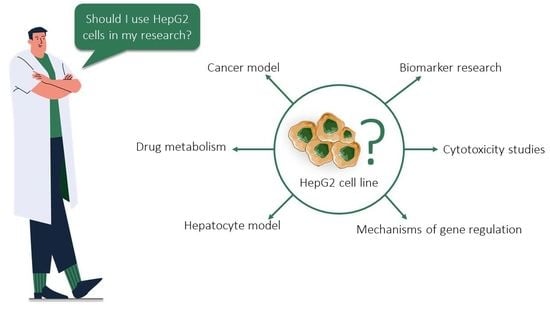
The Curious Case of the HepG2 Cell Line: 40 Years of Expertise
Abstract
1. Introduction
2. Historical Background
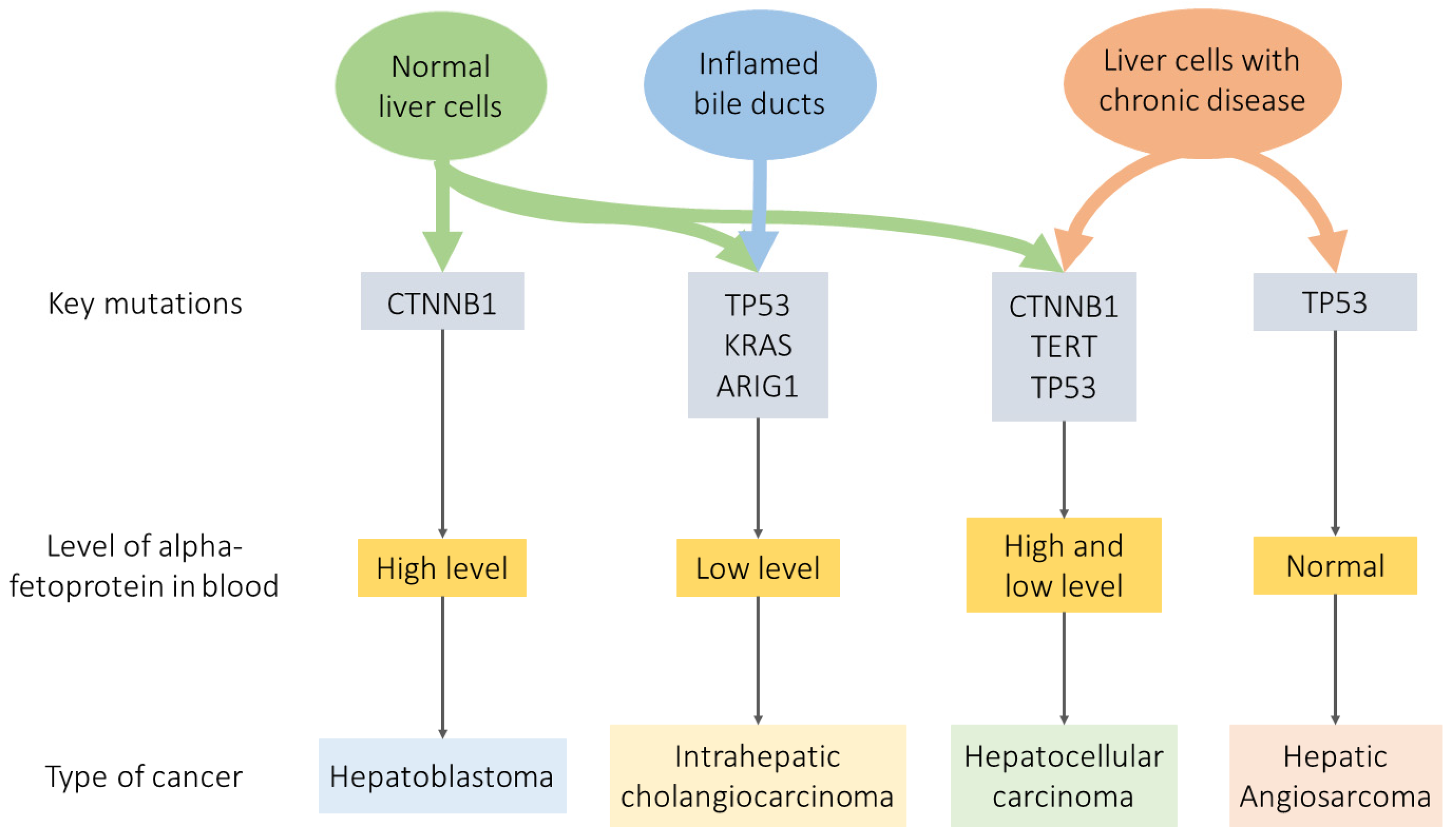
3. Comparison of Liver Cancer
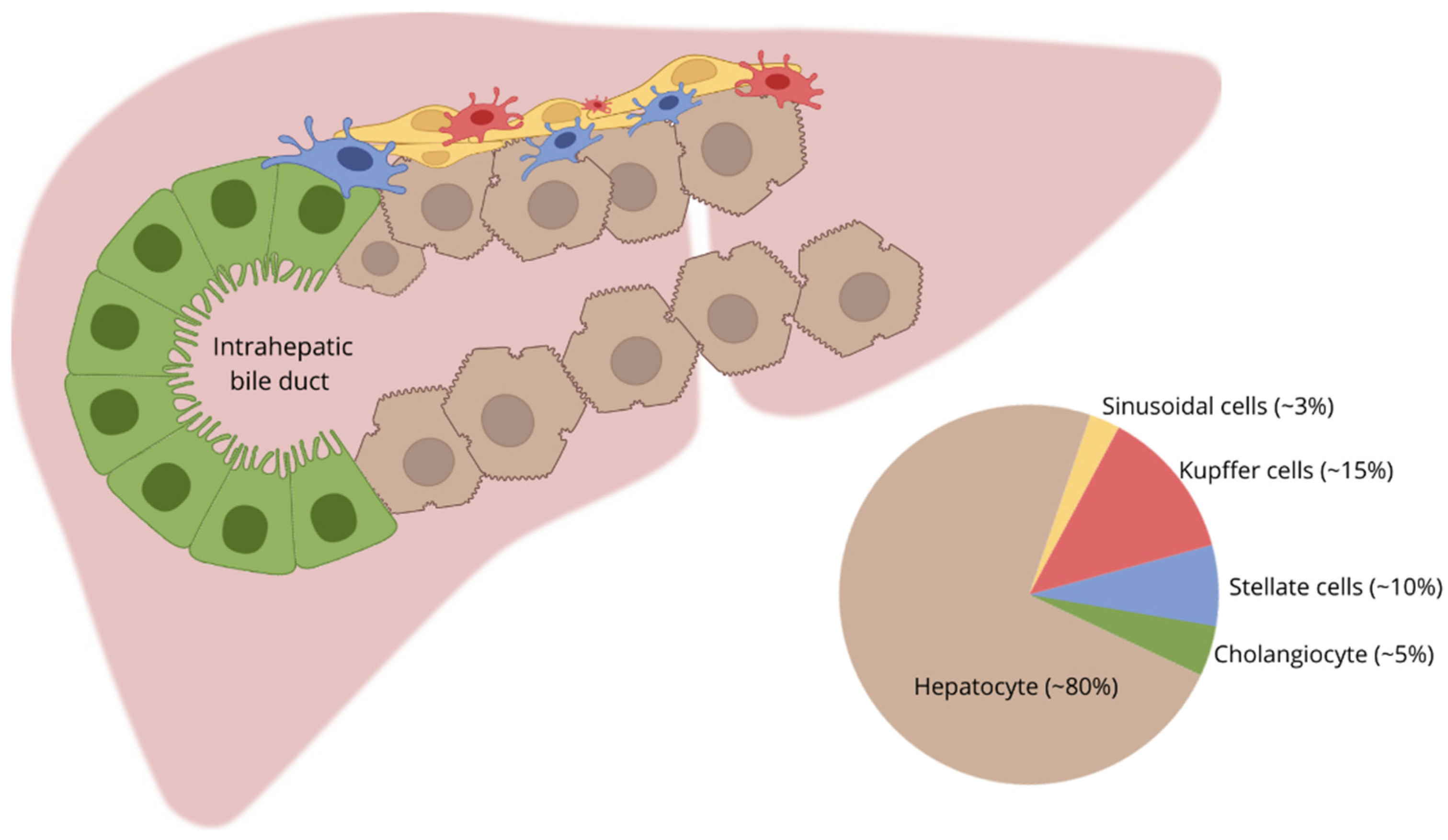
4. Comparison of HepG2, Normal Hepatocyte, HB, and HCC
4.1. Genome
4.2. Transcriptome
4.3. Proteome
4.4. Metabolome
4.5. Signalome
4.6. HepG2 for Biomedical Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kaur, G.; Dufour, J.M. Cell Lines. Spermatogenesis 2012, 2, 1–5. [Google Scholar] [CrossRef]
- Wang, L.; Cao, D.; Wei, C.; Meng, X.-J.; Jiang, X.; Tan, M. A Dual Vaccine Candidate against Norovirus and Hepatitis E Virus. Vaccine 2014, 32, 445–452. [Google Scholar] [CrossRef]
- Yang, Y.-J.; Pang, X.; Wang, B.; Yang, J.; Chen, X.-J.; Sun, X.-G.; Li, Q.; Zhang, J.; Guo, B.-L.; Ma, B.-P. Steroidal Saponins from Trillium Tschonoskii Rhizomes and Their Cytotoxicity against HepG2 Cells. Steroids 2020, 156, 108587. [Google Scholar] [CrossRef]
- Khanal, T.; Kim, H.G.; Hwang, Y.P.; Kong, M.J.; Kang, M.J.; Yeo, H.K.; Kim, D.H.; Jeong, T.C.; Jeong, H.G. Role of Metabolism by the Human Intestinal Microflora in Arbutin-Induced Cytotoxicity in HepG2 Cell Cultures. Biochem. Biophys. Res. Commun. 2011, 413, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Steinbrecht, S.; König, R.; Schmidtke, K.-U.; Herzog, N.; Scheibner, K.; Krüger-Genge, A.; Jung, F.; Kammerer, S.; Küpper, J.-H. Metabolic Activity Testing Can Underestimate Acute Drug Cytotoxicity as Revealed by HepG2 Cell Clones Overexpressing Cytochrome P450 2C19 and 3A4. Toxicology 2019, 412, 37–47. [Google Scholar] [CrossRef]
- Yokoyama, Y.; Sasaki, Y.; Terasaki, N.; Kawataki, T.; Takekawa, K.; Iwase, Y.; Shimizu, T.; Sanoh, S.; Ohta, S. Comparison of Drug Metabolism and Its Related Hepatotoxic Effects in HepaRG, Cryopreserved Human Hepatocytes, and HepG2 Cell Cultures. Biol. Pharm. Bull. 2018, 41, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-Y.; Li, J.; Ma, J.; Duan, F.; Zhong, P. Potential Role of Novel Hepatocellular Carcinoma-Associated Gene IDD01 in Promoting Tumorigenesis of HepG2 Cell Line. Chin. Med. J. 2006, 119, 1709–1714. [Google Scholar] [CrossRef] [PubMed]
- Wendt, H.; Hillmer, A.; Reimers, K.; Kuhbier, J.W.; Schäfer-Nolte, F.; Allmeling, C.; Kasper, C.; Vogt, P.M. Artificial Skin—Culturing of Different Skin Cell Lines for Generating an Artificial Skin Substitute on Cross-Weaved Spider Silk Fibres. PLoS ONE 2011, 6, e21833. [Google Scholar] [CrossRef]
- Joseph, S.C.; Blackman, B.A.; Kelly, M.L.; Phillips, M.; Beaury, M.W.; Martinez, I.; Parronchi, C.J.; Bitsaktsis, C.; Blake, A.D.; Sabatino, D. Synthesis, Characterization, and Biological Activity of Poly(Arginine)-Derived Cancer-Targeting Peptides in HepG2 Liver Cancer Cells. J. Pept. Sci. 2014, 20, 736–745. [Google Scholar] [CrossRef]
- Frevert, U.; Galinski, M.R.; Hügel, F.U.; Allon, N.; Schreier, H.; Smulevitch, S.; Shakibaei, M.; Clavijo, P. Malaria Circumsporozoite Protein Inhibits Protein Synthesis in Mammalian Cells. EMBO J. 1998, 17, 3816–3826. [Google Scholar] [CrossRef] [PubMed]
- Liver Regeneration: Biological and Pathological Mechanisms and Implications. Nature Reviews Gastroenterology & Hepatology. Available online: https://www.nature.com/articles/s41575-020-0342-4 (accessed on 24 November 2021).
- Wiśniewski, J.R.; Vildhede, A.; Norén, A.; Artursson, P. In-Depth Quantitative Analysis and Comparison of the Human Hepatocyte and Hepatoma Cell Line HepG2 Proteomes. J. Proteom. 2016, 136, 234–247. [Google Scholar] [CrossRef]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The Liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef] [PubMed]
- Genetic Liver Disease in Adults. Early Recognition of the Three Most Common Causes. PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/10689414/ (accessed on 24 November 2021).
- Castelli, G.; Pelosi, E.; Testa, U. Liver Cancer: Molecular Characterization, Clonal Evolution and Cancer Stem Cells. Cancers 2017, 9, 127. [Google Scholar] [CrossRef]
- Sharma, A.; Nagalli, S. Chronic Liver Disease. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Settivari, R.S.; Rowlands, J.C.; Wilson, D.M.; Arnold, S.M.; Spencer, P.J. Chapter 32—Application of Evolving Computational and Biological Platforms for Chemical Safety Assessment. In A Comprehensive Guide to Toxicology in Nonclinical Drug Development, 2nd ed.; Faqi, A.S., Ed.; Academic Press: Boston, MA, USA, 2017; pp. 843–873. ISBN 978-0-12-803620-4. [Google Scholar]
- Olsavsky Goyak, K.M.; Laurenzana, E.M.; Omiecinski, C.J. Hepatocyte Differentiation. Methods Mol. Biol. 2010, 640, 115–138. [Google Scholar] [CrossRef] [PubMed]
- Tanimizu, N.; Mitaka, T. Morphogenesis of Liver Epithelial Cells. Hepatol. Res. 2016, 46, 964–976. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, M.; Sustersic, T.; Filipovic, N. In Vitro Models and On-Chip Systems: Biomaterial Interaction Studies With Tissues Generated Using Lung Epithelial and Liver Metabolic Cell Lines. Front. Bioeng. Biotechnol. 2018, 6, 120. [Google Scholar] [CrossRef]
- Perugorria, M.J.; Olaizola, P.; Banales, J.M. Cholangiocyte-to-Hepatocyte Differentiation: A Context-Dependent Process and an Opportunity for Regenerative Medicine. Hepatology 2019, 69, 480–483. [Google Scholar] [CrossRef]
- Pfeifer, A.M.; Cole, K.E.; Smoot, D.T.; Weston, A.; Groopman, J.D.; Shields, P.G.; Vignaud, J.M.; Juillerat, M.; Lipsky, M.M.; Trump, B.F. Simian Virus 40 Large Tumor Antigen-Immortalized Normal Human Liver Epithelial Cells Express Hepatocyte Characteristics and Metabolize Chemical Carcinogens. Proc. Natl. Acad. Sci. USA 1993, 90, 5123–5127. [Google Scholar] [CrossRef] [PubMed]
- Godoy, P.; Hewitt, N.J.; Albrecht, U.; Andersen, M.E.; Ansari, N.; Bhattacharya, S.; Bode, J.G.; Bolleyn, J.; Borner, C.; Böttger, J.; et al. Recent Advances in 2D and 3D in Vitro Systems Using Primary Hepatocytes, Alternative Hepatocyte Sources and Non-Parenchymal Liver Cells and Their Use in Investigating Mechanisms of Hepatotoxicity, Cell Signaling and ADME. Arch. Toxicol. 2013, 87, 1315–1530. [Google Scholar] [CrossRef]
- Verrier, E.R.; Colpitts, C.C.; Schuster, C.; Zeisel, M.B.; Baumert, T.F. Cell Culture Models for the Investigation of Hepatitis B and D Virus Infection. Viruses 2016, 8, 261. [Google Scholar] [CrossRef] [PubMed]
- Verrier, E.R.; Colpitts, C.C.; Bach, C.; Heydmann, L.; Weiss, A.; Renaud, M.; Durand, S.C.; Habersetzer, F.; Durantel, D.; Abou-Jaoudé, G.; et al. A Targeted Functional RNA Interference Screen Uncovers Glypican 5 as an Entry Factor for Hepatitis B and D Viruses. Hepatology 2016, 63, 35–48. [Google Scholar] [CrossRef]
- Gripon, P.; Rumin, S.; Urban, S.; Le Seyec, J.; Glaise, D.; Cannie, I.; Guyomard, C.; Lucas, J.; Trepo, C.; Guguen-Guillouzo, C. Infection of a Human Hepatoma Cell Line by Hepatitis B Virus. Proc. Natl. Acad. Sci. USA 2002, 99, 15655–15660. [Google Scholar] [CrossRef]
- Monteil, M.; Migianu-Griffoni, E.; Sainte-Catherine, O.; Di Benedetto, M.; Lecouvey, M. Bisphosphonate Prodrugs: Synthesis and Biological Evaluation in HuH7 Hepatocarcinoma Cells. Eur. J. Med. Chem. 2014, 77, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Cavalloni, G.; Peraldo-Neia, C.; Varamo, C.; Casorzo, L.; Dell’Aglio, C.; Bernabei, P.; Chiorino, G.; Aglietta, M.; Leone, F. Establishment and Characterization of a Human Intrahepatic Cholangiocarcinoma Cell Line Derived from an Italian Patient. Tumour. Biol. 2016, 37, 4041–4052. [Google Scholar] [CrossRef] [PubMed]
- López-Terrada, D.; Cheung, S.W.; Finegold, M.J.; Knowles, B.B. Hep G2 Is a Hepatoblastoma-Derived Cell Line. Hum. Pathol. 2009, 40, 1512–1515. [Google Scholar] [CrossRef]
- Zhou, B.; Ho, S.S.; Greer, S.U.; Spies, N.; Bell, J.M.; Zhang, X.; Zhu, X.; Arthur, J.G.; Byeon, S.; Pattni, R.; et al. Haplotype-Resolved and Integrated Genome Analysis of the Cancer Cell Line HepG2. Nucleic Acids Res. 2019, 47, 3846–3861. [Google Scholar] [CrossRef] [PubMed]
- Saravanakumar, A. An Omics Based Approach for the Identification of Biomarkers of Non-Alcoholic Fatty Liver Using in Vitro Models of Hepatic Steatosis. Ph.D Thesis, University of Rhode Island, Kingston, RI, USA, 2019. pp. 1–184. [Google Scholar]
- Buick, J.K.; Williams, A.; Meier, M.J.; Swartz, C.D.; Recio, L.; Gagné, R.; Ferguson, S.S.; Engelward, B.P.; Yauk, C.L. A Modern Genotoxicity Testing Paradigm: Integration of the High-Throughput CometChip® and the TGx-DDI Transcriptomic Biomarker in Human HepaRGTM Cell Cultures. Front. Public Health 2021, 9, 1144. [Google Scholar] [CrossRef]
- HepaRGTM Cells. Cryopreserved. Available online: https://www.thermofisher.com/order/catalog/product/HPRGC10 (accessed on 28 November 2021).
- Yu, K.; Chen, B.; Aran, D.; Charalel, J.; Yau, C.; Wolf, D.M.; van ’t Veer, L.J.; Butte, A.J.; Goldstein, T.; Sirota, M. Comprehensive Transcriptomic Analysis of Cell Lines as Models of Primary Tumors across 22 Tumor Types. Nat. Commun. 2019, 10, 3574. [Google Scholar] [CrossRef]
- Qiu, Z.; Li, H.; Zhang, Z.; Zhu, Z.; He, S.; Wang, X.; Wang, P.; Qin, J.; Zhuang, L.; Wang, W.; et al. A Pharmacogenomic Landscape in Human Liver Cancers. Cancer Cell 2019, 36, 179–193. [Google Scholar] [CrossRef]
- Kasai, F.; Hirayama, N.; Ozawa, M.; Satoh, M.; Kohara, A. HuH-7 Reference Genome Profile: Complex Karyotype Composed of Massive Loss of Heterozygosity. Hum. Cell 2018, 31, 261–267. [Google Scholar] [CrossRef]
- Kawamoto, M.; Yamaji, T.; Saito, K.; Shirasago, Y.; Satomura, K.; Endo, T.; Fukasawa, M.; Hanada, K.; Osada, N. Identification of Characteristic Genomic Markers in Human Hepatoma HuH-7 and Huh7.5.1-8 Cell Lines. Front. Genet. 2020, 11, 546106. [Google Scholar] [CrossRef]
- Yu, M.; Selvaraj, S.K.; Liang-Chu, M.M.Y.; Aghajani, S.; Busse, M.; Yuan, J.; Lee, G.; Peale, F.; Klijn, C.; Bourgon, R.; et al. A Resource for Cell Line Authentication, Annotation and Quality Control. Nature 2015, 520, 307–311. [Google Scholar] [CrossRef]
- Heffelfinger, S.C.; Hawkins, H.H.; Barrish, J.; Taylor, L.; Darlington, G.J. SK HEP-1: A Human Cell Line of Endothelial Origin. Vitr. Cell Dev. Biol. 1992, 28, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF Gene in Human Cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Aden, D.P.; Fogel, A.; Plotkin, S.; Damjanov, I.; Knowles, B.B. Controlled Synthesis of HBsAg in a Differentiated Human Liver Carcinoma-Derived Cell Line. Nature 1979, 282, 615–616. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.; Gao, J.-H.; Zhao, C.; Tong, H.; Zheng, S.-P.; Huang, Z.-Y.; Liu, R.; Tang, C.-W.; Li, J. SK-Hep1: Not Hepatocellular Carcinoma Cells but a Cell Model for Liver Sinusoidal Endothelial Cells. Int. J. Clin. Exp. Pathol. 2018, 11, 2931–2938. [Google Scholar] [PubMed]
- Tomlinson, G.E.; Douglass, E.C.; Pollock, B.H.; Finegold, M.J.; Schneider, N.R. Cytogenetic Evaluation of a Large Series of Hepatoblastomas: Numerical Abnormalities with Recurring Aberrations Involving 1q12-Q21. Genes Chromosomes Cancer 2005, 44, 177–184. [Google Scholar] [CrossRef]
- Adesina, A.M.; Lopez-Terrada, D.; Wong, K.K.; Gunaratne, P.; Nguyen, Y.; Pulliam, J.; Margolin, J.; Finegold, M.J. Gene Expression Profiling Reveals Signatures Characterizing Histologic Subtypes of Hepatoblastoma and Global Deregulation in Cell Growth and Survival Pathways. Hum. Pathol. 2009, 40, 843–853. [Google Scholar] [CrossRef]
- Guengerich, F.P. Cytochrome P450 Research and The Journal of Biological Chemistry. J. Biol. Chem. 2019, 294, 1671–1680. [Google Scholar] [CrossRef]
- Pareek, A.; Godavarthi, A.; Issarani, R.; Nagori, B.P. Antioxidant and Hepatoprotective Activity of Fagonia Schweinfurthii (Hadidi) Hadidi Extract in Carbon Tetrachloride Induced Hepatotoxicity in HepG2 Cell Line and Rats. J. Ethnopharmacol. 2013, 150, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Ringelhan, M.; Pfister, D.; O’Connor, T.; Pikarsky, E.; Heikenwalder, M. The Immunology of Hepatocellular Carcinoma. Nat. Immunol. 2018, 19, 222–232. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.; Gogineni, V.; Saeian, K. Epidemiology of Primary and Secondary Liver Cancers. Semin. Interv. Radiol. 2006, 23, 47–63. [Google Scholar] [CrossRef]
- Angelico, R.; Grimaldi, C.; Saffioti, M.C.; Castellano, A.; Spada, M. Hepatocellular Carcinoma in Children: Hepatic Resection and Liver Transplantation. Transl. Gastroenterol. Hepatol. 2018, 3, 59. [Google Scholar] [CrossRef] [PubMed]
- Eichenmüller, M.; Trippel, F.; Kreuder, M.; Beck, A.; Schwarzmayr, T.; Häberle, B.; Cairo, S.; Leuschner, I.; Von Schweinitz, D.; Strom, T.M.; et al. The Genomic Landscape of Hepatoblastoma and Their Progenies with HCC-like Features. J Hepatol. 2014, 61, 1312–1320. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, T.F.M.; Rivas, M.P.; Costa, S.; Maschietto, M.; Rodrigues, T.; Sobral de Barros, J.; Barbosa, A.C.; Valieris, R.; Fernandes, G.R.; Bertola, D.R.; et al. Insights Into the Somatic Mutation Burden of Hepatoblastomas From Brazilian Patients. Front. Oncol. 2020, 10, 1–16. [Google Scholar] [CrossRef]
- Armengol, C.; Cairo, S.; Fabre, M.; Buendia, M.A. Wnt Signaling and Hepatocarcinogenesis: The Hepatoblastoma Model. Int. J. Biochem. Cell Biol. 2011, 43, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, M.L.; Buonaguro, L.; Tatangelo, F.; Botti, G.; Izzo, F.; Buonaguro, F.M. Mutations in TP53, CTNNB1 and PIK3CA Genes in Hepatocellular Carcinoma Associated with Hepatitis B and Hepatitis C Virus Infections. Genomics 2013, 102, 74–83. [Google Scholar] [CrossRef]
- Javanmard, D.; Najafi, M.; Babaei, M.R.; Karbalaie Niya, M.H.; Esghaei, M.; Panahi, M.; Safarnezhad Tameshkel, F.; Tavakoli, A.; Jazayeri, S.M.; Ghaffari, H.; et al. Investigation of CTNNB1 Gene Mutations and Expression in Hepatocellular Carcinoma and Cirrhosis in Association with Hepatitis B Virus Infection. Infect Agent Cancer 2020, 15, 37. [Google Scholar] [CrossRef]
- Tarnow, G.; McLachlan, A. β-Catenin Signaling Regulates the In Vivo Distribution of Hepatitis B Virus Biosynthesis across the Liver Lobule. J. Virol. 2021, 95, e0078021. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.-S.; Wang, Z.; He, X.-J.; Diplas, B.H.; Yang, R.; Killela, P.J.; Liang, J.; Meng, Q.; Ye, Z.-Y.; Wang, W.; et al. Recurrent TERT Promoter Mutations Identified in a Large-Scale Study of Multiple Tumor Types Are Associated with Increased TERT Expression and Telomerase Activation. Eur. J. Cancer 2015, 51, 969–976. [Google Scholar] [CrossRef]
- Nault, J.-C.; Ningarhari, M.; Rebouissou, S.; Zucman-Rossi, J. The Role of Telomeres and Telomerase in Cirrhosis and Liver Cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 544–558. [Google Scholar] [CrossRef]
- Patel, B.; Taiwo, R.; Kim, A.H.; Dunn, G.P. TERT, a Promoter of CNS Malignancies. Neurooncol. Adv. 2020, 2, vdaa025. [Google Scholar] [CrossRef] [PubMed]
- Nault, J.C.; Mallet, M.; Pilati, C.; Calderaro, J.; Bioulac-Sage, P.; Laurent, C.; Laurent, A.; Cherqui, D.; Balabaud, C.; Zucman-Rossi, J. High Frequency of Telomerase Reverse-Transcriptase Promoter Somatic Mutations in Hepatocellular Carcinoma and Preneoplastic Lesions. Nat. Commun. 2013, 4, 2218. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, D.; Saitta, C.; Giosa, D.; Di Tocco, F.C.; Musolino, C.; Caminiti, G.; Chines, V.; Franzè, M.S.; Alibrandi, A.; Navarra, G.; et al. Frequency of Somatic Mutations in TERT Promoter, TP53 and CTNNB1 Genes in Patients with Hepatocellular Carcinoma from Southern Italy. Oncol. Lett. 2020, 19, 2368–2374. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.J.; Vousden, K.H. P53 Mutations in Cancer. Nat. Cell Biol. 2013, 15, 2–8. [Google Scholar] [CrossRef]
- Bossi, G.; Lapi, E.; Strano, S.; Rinaldo, C.; Blandino, G.; Sacchi, A. Mutant P53 Gain of Function : Reduction of Tumor Malignancy of Human Cancer Cell Lines through Abrogation of Mutant P53 Expression. Oncogene 2006, 25, 304–309. [Google Scholar] [CrossRef]
- Villanueva, A.; Hoshida, Y. Depicting the Role of TP53 in Hepatocellular Carcinoma Progression. J. Hepatol. 2011, 55, 724–725. [Google Scholar] [CrossRef] [PubMed]
- Simbolo, M.; Vicentini, C.; Ruzzenente, A.; Brunelli, M.; Conci, S.; Fassan, M.; Mafficini, A.; Rusev, B.; Corbo, V.; Capelli, P.; et al. Genetic Alterations Analysis in Prognostic Stratified Groups Identified TP53 and ARID1A as Poor Clinical Performance Markers in Intrahepatic Cholangiocarcinoma. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Ruzzenente, A.; Fassan, M.; Conci, S.; Simbolo, M.; Lawlor, R.T.; Pedrazzani, C.; Capelli, P.; D’Onofrio, M.; Iacono, C.; Scarpa, A.; et al. Cholangiocarcinoma Heterogeneity Revealed by Multigene Mutational Profiling: Clinical and Prognostic Relevance in Surgically Resected Patients. Ann. Surg. Oncol. 2016, 23, 1699–1707. [Google Scholar] [CrossRef]
- Dakkak, W. AACR Project GENIE: Powering Precision M. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, H.; Zhao, Y.; Pan, Q.; Mao, A.; Zhu, W.; Zhang, N.; Lin, Z.; Zhou, J.; Wang, Y.; et al. Comprehensive Molecular Profiling of Intrahepatic Cholangiocarcinoma in the Chinese Population and Therapeutic Experience. J. Transl. Med. 2020, 18, 1–11. [Google Scholar] [CrossRef]
- Buettner, S.; Van Vugt, J.L.A.; Ijzermans, J.N.M.; Koerkamp, B.G. Intrahepatic Cholangiocarcinoma: Current Perspectives. OncoTargets Ther. 2017, 10, 1131–1142. [Google Scholar] [CrossRef]
- Perugorria, M.J.; Olaizola, P.; Labiano, I.; Esparza-Baquer, A.; Marzioni, M.; Marin, J.J.G.; Bujanda, L.; Banales, J.M. Wnt-β-Catenin Signalling in Liver Development, Health and Disease. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 121–136. [Google Scholar] [CrossRef]
- Wang, W.; Smits, R.; Hao, H.; He, C. Wnt/β-Catenin Signaling in Liver Cancers. Cancers 2019, 11, E926. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Q. Alpha-Fetoprotein and Hepatocellular Carcinoma Immunity. Can. J. Gastroenterol. Hepatol. 2018, 2018, 9049252. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J.; Cao, D.; Yang, J.; Shen, K.; Huang, H.; Shi, X. Alpha-Fetoprotein (AFP)-Producing Epithelial Ovarian Carcinoma (EOC): A Retrospective Study of 27 Cases. Arch. Gynecol. Obs. 2021, 304, 1043–1053. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Lu, Y.; Li, W.; Guo, J.; Dong, X.; Lin, B.; Chen, Y.; Xie, X.; Li, M. Hepatitis B Virus X Protein Driven Alpha Fetoprotein Expression to Promote Malignant Behaviors of Normal Liver Cells and Hepatoma Cells. J. Cancer 2016, 7, 935–946. [Google Scholar] [CrossRef]
- Houwelingen, L.V.; Sandoval, J.A. Alpha-Fetoprotein in Malignant Pediatric Conditions; IntechOpen: London, UK, 2016; ISBN 9789535125815. [Google Scholar]
- Toyoda, H.; Bregerie, O.; Vallet, A.; Nalpas, B.; Pivert, G.; Brechot, C.; Desdouets, C. Changes to Hepatocyte Ploidy and Binuclearity Profiles during Human Chronic Viral Hepatitis. Gut 2005, 54, 297–302. [Google Scholar] [CrossRef]
- The Ploidy Conveyor of Mature Hepatocytes as a Source of Genetic Variation. Nature 2010, 467, 707–710. Available online: https://www.nature.com/articles/nature09414 (accessed on 2 December 2021). [CrossRef]
- Li, H.; Feng, Z.; Tsang, T.C.; Tang, T.; Jia, X.; He, X.; Pennington, M.E.; Badowski, M.S.; Liu, A.K.M.; Chen, D.; et al. Fusion of HepG2 Cells with Mesenchymal Stem Cells Increases Cancer-Associated and Malignant Properties: An in Vivo Metastasis Model. Oncol. Rep. 2014, 32, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Hui, L. Progress in Human Liver Organoids. J. Mol. Cell Biol. 2020, 12, 607–617. [Google Scholar] [CrossRef]
- Weir, E.G.; Ali, S.Z. Hepatoblastoma: Cytomorphologic Characteristics in Serous Cavity Fluids. Cancer 2002, 96, 267–274. [Google Scholar] [CrossRef] [PubMed]
- De Sánchez, V.C.; Chávez, E.; Velasco-Loyden, G.; Lozano-Rosas, M.G.; Aparicio-Cadena, A.R. Interaction of Mitochondrial and Epigenetic Regulation in Hepatocellular Carcinoma. In Liver Cancer; IntechOpen: London, UK, 2016; p. 13. [Google Scholar] [CrossRef]
- Wei, J.; Fang, D. Endoplasmic Reticulum Stress Signaling and the Pathogenesis of Hepatocarcinoma. Int. J. Mol. Sci. 2021, 22, 1799. [Google Scholar] [CrossRef]
- Chen, H.L.; Chiu, T.S.; Chen, P.J.; Chen, D.S. Cytogenetic Studies on Human Liver Cancer Cell Lines. Cancer Genet. Cytogenet. 1993, 65, 161–166. [Google Scholar] [CrossRef]
- Wang, X.; Li, T.; Su, X.; Li, J.; Li, W.; Gan, J.; Wu, T.; Kong, L.; Zhang, T.; Tang, M.; et al. Genotoxic Effects of Silver Nanoparticles with/without Coating in Human Liver HepG2 Cells and in Mice. J. Appl. Toxicol. 2019, 39, 908–918. [Google Scholar] [CrossRef]
- Lee, C.; Ling, Q.; Chang, M. Characterization of De Novo Genomic Alterations in HBV-Producing Characterization of De Novo Genomic Alterations in HBV-Producing Hepatoma Cells Using Array Comparative Genomic Hybridization. J. Cancer Mol. 2014, 1, 93–95. [Google Scholar]
- Simon, D.; Aden, D.P.; Knowles, B.B. Chromosomes of Human Hepatoma Cell Lines. Int. J. Cancer 1982, 30, 27–33. [Google Scholar] [CrossRef]
- Wong, N.; Lai, P.; Pang, E.; Wai-Tong Leun, T.; Wan-Yee Lau, J.; James Johnson, P. A Comprehensive Karyotypic Study on Human Hepatocellular Carcinoma by Spectral Karyotyping. Hepatology 2000, 32, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Luquet, I.; Laı, J.L. Hyperdiploid Karyotypes in Acute Myeloid Leukemia Define a Novel Entity : A Study of 38 Patients from the Groupe Francophone de Cytogenetique Hematologique (GFCH). Leukemia 2008, 22, 132–137. [Google Scholar] [CrossRef]
- Heim, S.; Alimena, G.; Billstrm, R.; Diverio, D.; Kristoffersson, U.; Mandahl, N.; Nanni, M.; Mitelman, F. Tetraploid Karyotype (92,XXYY) in Two Patients with Acute Lymphoblastic Leukemia Sverre. Cancer Genet. Cytogenet. 1987, 133, 129–133. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Hein, M.Y.; Cox, J.; Mann, M. A “Proteomic Ruler” for Protein Copy Number and Concentration Estimation without Spike-in Standards. Mol. Cell. Proteom. 2014, 13, 3497–3506. [Google Scholar] [CrossRef]
- Yang, T.; Wen, Y.; Li, J.; Tan, T.; Yang, J.; Pan, J.; Hu, C.; Yao, Y.; Zhang, J.; Xin, Y.; et al. NRAS and KRAS Polymorphisms Are Not Associated with Hepatoblastoma Susceptibility in Chinese Children. Exp. Hematol. Oncol. 2019, 8, 1–7. [Google Scholar] [CrossRef]
- Yang, M.H.; Yen, C.H.; Chen, Y.F.; Fang, C.C.; Li, C.H.; Lee, K.J.; Lin, Y.H.; Weng, C.H.; Liu, T.T.; Huang, S.F.; et al. Somatic Mutations of PREX2 Gene in Patients with Hepatocellular Carcinoma. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bévant, K.; Coulouarn, C. Landscape of Genomic Alterations in Hepatocellular Carcinoma: Current Knowledge and Perspectives for Targeted Therapies. Hepato. Biliary Surg. Nutr. 2017, 6, 404–407. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Halpert, G.; Lerner, M.G.; Hu, H.; Dimitrion, P.; Weiss, M.J.; He, J.; Philosophe, B.; Burkhart, R.; Burns, W.R.; et al. Protein Synthesis Inhibitor Omacetaxine Is Effective against Hepatocellular Carcinoma. JCI Insight 2021, 6, e138197. [Google Scholar] [CrossRef]
- Ally, A.; Balasundaram, M.; Carlsen, R.; Chuah, E.; Clarke, A.; Dhalla, N.; Holt, R.A.; Jones, S.J.M.; Lee, D.; Ma, Y.; et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341. [Google Scholar] [CrossRef] [PubMed]
- Cevik, D. Common Telomerase Reverse Transcriptase Promoter Mutations in Hepatocellular Carcinomas from Different Geographical Locations. World J. Gastroenterol. 2015, 21, 311. [Google Scholar] [CrossRef]
- Sumazin, P.; Chen, Y.; Treviño, L.R.; Sarabia, S.F.; Hampton, O.A.; Patel, K.; Mistretta, T.-A.; Zorman, B.; Thompson, P.; Heczey, A.; et al. Genomic Analysis of Hepatoblastoma Identifies Distinct Molecular and Prognostic Subgroups. Hepatology 2017, 65, 104–121. [Google Scholar] [CrossRef]
- Cagatay, T.; Ozturk, M. P53 Mutation as a Source of Aberrant Beta-Catenin Accumulation in Cancer Cells. Oncogene 2002, 21, 7971–7980. [Google Scholar] [CrossRef]
- Woodfield, S.E.; Shi, Y.; Patel, R.H.; Chen, Z.; Shah, A.P.; Srivastava, R.K.; Whitlock, R.S.; Ibarra, A.M.; Larson, S.R.; Sarabia, S.F.; et al. MDM4 Inhibition: A Novel Therapeutic Strategy to Reactivate P53 in Hepatoblastoma. Sci. Rep. 2021, 11, 2967. [Google Scholar] [CrossRef]
- Hussain, S.P.; Schwank, J.; Staib, F.; Wang, X.W.; Harris, C.C. TP53 Mutations and Hepatocellular Carcinoma: Insights into the Etiology and Pathogenesis of Liver Cancer. Oncogene 2007, 26, 2166–2176. [Google Scholar] [CrossRef] [PubMed]
- Pyatnitskiy, M.A.; Arzumanian, V.A.; Radko, S.P.; Ptitsyn, K.G.; Vakhrushev, I.V.; Poverennaya, E.V.; Ponomarenko, E.A. Oxford Nanopore MinION Direct RNA-Seq for Systems Biology. Biology 2021, 10, 1131. [Google Scholar] [CrossRef]
- Tyakht, A.V.; Ilina, E.N.; Alexeev, D.G.; Ischenko, D.S.; Gorbachev, A.Y.; Semashko, T.A.; Larin, A.K.; Selezneva, O.V.; Kostryukova, E.S.; Karalkin, P.A.; et al. RNA-Seq Gene Expression Profiling of HepG2 Cells: The Influence of Experimental Factors and Comparison with Liver Tissue. BMC Genom. 2014, 15, 1–9. [Google Scholar] [CrossRef]
- Nasution, A.A.; Adnindya, M.R.; Indriyani; Liem, I.K. Roles of DLK1 in Liver Development and Oncogenesis. OnLine J. Biol. Sci. 2017, 17, 309–315. [Google Scholar] [CrossRef]
- Xu, X.; Liu, R.F.; Zhang, X.; Huang, L.Y.; Chen, F.; Fei, Q.L.; Han, Z.G. DLK1 as a Potential Target against Cancer Stem/Progenitor Cells of Hepatocellular Carcinoma. Mol. Cancer Ther. 2012, 11, 629–638. [Google Scholar] [CrossRef]
- Luo, J.; Ren, B.; Keryanov, S.; Tseng, G.C.; Rao, U.N.M.; Monga, S.P.; Strom, S.; Demetris, A.J.; Nalesnik, M.; Yan, P.; et al. Transcriptomic and Genomic Analysis of Human Hepatocellular Carcinomas and Hepatoblastomas. Hepatology 2007, 44, 1012–1024. [Google Scholar] [CrossRef]
- Pivonello, C.; Negri, M.; De Martino, M.C.; Napolitano, M.; de Angelis, C.; Provvisiero, D.P.; Cuomo, G.; Auriemma, R.S.; Simeoli, C.; Izzo, F.; et al. The Dual Targeting of Insulin and Insulin-like Growth Factor 1 Receptor Enhances the MTOR Inhibitor-Mediated Antitumor Efficacy in Hepatocellular Carcinoma. Oncotarget 2016, 7, 9718–9731. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Desai, V.; Wang, J.; Epstein, D.M.; Miglarese, M.; Buck, E. Epithelial—Mesenchymal Transition Predicts Sensitivity to the Dual IGF-1R / IR Inhibitor OSI-906 in Hepatocellular Carcinoma Cell Lines. Mol. Cancer Ther. 2012, 11, 503–514. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Martinez-Quetglas, I.; Pinyol, R.; Dauch, D.; Torrecilla, S.; Tovar, V.; Moeini, A.; Alsinet, C.; Portela, A.; Rodriguez-Carunchio, L.; Solé, M.; et al. IGF2 Is Up-Regulated by Epigenetic Mechanisms in Hepatocellular Carcinomas and Is an Actionable Oncogene Product in Experimental Models. Gastroenterology 2016, 151, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H. Liver Cancer: IGF2—An Epigenetic Oncodriver in HCC. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 625. [Google Scholar] [CrossRef]
- Honda, S.; Arai, Y.; Haruta, M.; Sasaki, F.; Ohira, M.; Yamaoka, H.; Horie, H.; Nakagawara, A.; Hiyama, E.; Todo, S.; et al. Loss of Imprinting of IGF2 Correlates with Hypermethylation of the H19 Differentially Methylated Region in Hepatoblastoma. Br. J. Cancer 2008, 99, 1891–1899. [Google Scholar] [CrossRef]
- Ekström, T.J. Altered Expression of Members of the IGF-Axis in Hepatoblastoma. Br. J. Cancer 2000, 82, 1561–1567. [Google Scholar]
- Weiss, J.B.W.; Wagner, A.E.; Eberherr, C.; Haberle, B.; Vokuhl, C.; Von Schweinitz, D.; Kappler, R. High Expression of IGF2-Derived Intronic MiR-483 Predicts Outcome in Hepatoblastoma. Cancer Biomark. 2020, 28, 321–328. [Google Scholar] [CrossRef]
- Shen, G.; Shen, H.; Zhang, J.; Yan, Q.; Liu, H. DNA Methylation in Hepatoblastoma-a Literature Review. Ital. J. Pediatr. 2020, 46, 1–9. [Google Scholar] [CrossRef]
- Nussbaum, T.; Samarin, J.; Ehemann, V.; Bissinger, M.; Ryschich, E.; Khamidjanov, A.; Yu, X.; Gretz, N.; Schirmacher, P.; Breuhahn, K. Autocrine Insulin-like Growth Factor-II Stimulation of Tumor Cell Migration Is a Progression Step in Human Hepatocarcinogenesis. Hepatology 2008, 48, 146–156. [Google Scholar] [CrossRef]
- Waly, A.A.; El-Ekiaby, N.; Assal, R.A.; Abdelrahman, M.M.; Hosny, K.A.; El Tayebi, H.M.; Esmat, G.; Breuhahn, K.; Abdelaziz, A.I. Methylation in MIRLET7A3 Gene Induces the Expression of IGF-II and Its MRNA Binding Proteins IGF2BP-2 and 3 in Hepatocellular Carcinoma. Front. Physiol. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Breuhahn, K.; Longerich, T.; Schirmacher, P. Dysregulation of Growth Factor Signaling in Human Hepatocellular Carcinoma. Oncogene 2006, 25, 3787–3800. [Google Scholar] [CrossRef]
- Chen, L.; Li, M.; Li, Q.; Wang, C.; Xie, S. DKK1 Promotes Hepatocellular Carcinoma Cell Migration and Invasion through β-Catenin/MMP7 Signaling Pathway. Mol. Cancer 2013, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, N.; Chen, Y.; Sun, L.; Li, L.; Liu, H. Elevated GPC3 Level Promotes Cell Proliferation in Liver Cancer. Oncol. Lett. 2018, 16, 970–976. [Google Scholar] [CrossRef]
- Jia, H.L.; Ye, Q.H.; Qin, L.X.; Budhu, A.; Forgues, M.; Chen, Y.; Liu, Y.K.; Sun, H.C.; Wang, L.; Lu, H.Z.; et al. Gene Expression Profiling Reveals Potential Biomarkers of Human Hepatocellular Carcinoma. Clin. Cancer Res. 2007, 13, 1133–1139. [Google Scholar] [CrossRef]
- Maass, T.; Sfakianakis, I.; Staib, F.; Krupp, M.; Galle, R.P.; Teufel, A. Microarray-Based Gene Expression Analysis of Hepatocellular Carcinoma. Curr. Genom. 2010, 11, 261–268. [Google Scholar] [CrossRef][Green Version]
- Sun, C.K.; Chua, M.S.; He, J.; So, S.K. Suppression of Glypican 3 Inhibits Growth of Hepatocellular Carcinoma Cells through Up-Regulation of TGF-Β2. Neoplasia 2011, 13, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, M.; Seki, M.; Kawai, T.; Yoshida, K.; Yoshida, M.; Isobe, T.; Hoshino, N. Integrated Multiomics Analysis of Hepatoblastoma Unravels Its Heterogeneity and Provides Novel Druggable Targets. NPJ Precis. Oncol. 2020, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Vastrad, B.; Vastrad, C.; Kotturshetti, I. Identification of Potential Core Genes in Hepatoblastoma via Bioinformatics Analysis. Genet. Genom. Med. 2020, 120, 10069–10081. [Google Scholar] [CrossRef]
- Xu, Y.; Zheng, Y.; Li, T. Modulation of IGF2BP1 by Long Non-Coding RNA HCG11 Suppresses Apoptosis of Hepatocellular Carcinoma Cells via MAPK Signaling Transduction. Int. J. Oncol. 2017, 51, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, H.; Guo, X.; Zhu, Z.; Cai, H.; Kong, X. Insulin-like Growth Factor 2 MRNA-Binding Protein 1 ( IGF2BP1 ) in Cancer. J. Hematol. Oncol. 2018, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Mu, Q.; Huang, H. Review Article The Roles of Insulin-Like Growth Factor 2 MRNA-Binding Protein 2 in Cancer and Cancer Stem Cells. Stem Cells Int. 2018, 2018, 4217259. [Google Scholar] [CrossRef] [PubMed]
- Gerets, H.H.J.; Tilmant, K.; Gerin, B.; Chanteux, H.; Depelchin, B.O.; Dhalluin, S.; Atienzar, F.A. Characterization of Primary Human Hepatocytes, HepG2 Cells, and HepaRG Cells at the MRNA Level and CYP Activity in Response to Inducers and Their Predictivity for the Detection of Human Hepatotoxins. Cell Biol. Toxicol. 2012, 28, 69–87. [Google Scholar] [CrossRef]
- Berger, B.; Donzelli, M.; Maseneni, S.; Boess, F. Comparison of Liver Cell Models Using the Basel Phenotyping Cocktail. Front. Pharmacol. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Shankar, K.; Sciences, M.; Rock, L. Cytochrome P450; Elsevier: Amsterdam, The Netherlands, 2014; Volume 1, pp. 1125–1127. [Google Scholar] [CrossRef]
- Choi, J.M.; Oh, S.J.; Lee, S.Y.; Im, J.H.; Oh, J.M.; Ryu, C.S.; Kwak, H.C.; Lee, J.Y.; Kang, K.W.; Kim, S.K. HepG2 Cells as an in Vitro Model for Evaluation of Cytochrome P450 Induction by Xenobiotics. Arch. Pharmacal. Res. 2015, 38, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Jiang, X.; Qi, X. Downregulation of CYP2A6 and CYP2C8 in Tumor Tissues Is Linked to Worse Overall Survival and Recurrence-Free Survival from Hepatocellular Carcinoma. BioMed Res. Int. 2018, 2018, 5859415. [Google Scholar] [CrossRef]
- Gupta, R.; Schrooders, Y.; Hauser, D.; van Herwijnen, M.; Albrecht, W.; ter Braak, B.; Brecklinghaus, T.; Castell, J.V.; Elenschneider, L.; Escher, S.; et al. Comparing in Vitro Human Liver Models to in Vivo Human Liver Using RNA-Seq. Arch. Toxicol. 2021, 95, 573–589. [Google Scholar] [CrossRef]
- Jennen, D.G.J.; Magkoufopoulou, C.; Ketelslegers, H.B.; van Herwijnen, M.H.M.; Kleinjans, J.C.S.; van Delft, J.H.M. Comparison of HepG2 and HepaRG by Whole-Genome Gene Expression Analysis for the Purpose of Chemical Hazard Identification. Toxicol. Sci. 2010, 115, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Koepsell, H.; Gizak, A.; Rakus, D. Absolute Protein Quantification Allows Differentiation of Cell-Specific Metabolic Routes and Functions. Proteomics 2015, 15, 1316–1325. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Turpeinen, M.; Klein, K.; Schwab, M. Functional Pharmacogenetics / Genomics of Human Cytochromes P450 Involved in Drug Biotransformation. Anal. Bioanal. Chem. 2008, 392, 1093–1108. [Google Scholar] [CrossRef]
- Vildhede, A.; Wiśniewski, J.R.; Norén, A.; Karlgren, M.; Artursson, P. Comparative Proteomic Analysis of Human Liver Tissue and Isolated Hepatocytes with a Focus on Proteins Determining Drug Exposure. J. Proteome Res. 2015, 14, 3305–3314. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Braeuning, A.; Ruck, P.; Seitz, G.; Armeanu-Ebinger, S.; Fuchs, J.; Warmann, S.W.; Schwarz, M. Differential Expression of Glutamine Synthetase and Cytochrome P450 Isoforms in Human Hepatoblastoma. Toxicology 2011, 281, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Chen, L.; Yang, Y.C.; Wang, X.M.; Wang, R.Y.; Li, L.; Wen, W.; Chang, Y.X.; Chen, C.Y.; Tang, J.; et al. CYP3A5 Functions as a Tumor Suppressor in Hepatocellular Carcinoma by Regulating MTORC2/Akt Signaling. Cancer Res. 2015, 75, 1470–1481. [Google Scholar] [CrossRef]
- Westerink, W.M.A.; Schoonen, W.G.E.J. Phasea II Enzyme Levels in HepG2 Cells and Cryopreserved Primary Human Hepatocytes and Their Induction in HepG2 Cells. Toxicol. In Vitro 2007, 21, 1592–1602. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhou, J.; Shi, J.; Peng, X.; Qi, X.; Wang, Y. Drug-Metabolizing Activity, Protein and Gene Expression of UDP-Glucuronosyltransferases Are Significantly Altered in Hepatocellular Carcinoma Patients. PLoS ONE 2015, 10, e0127524. [Google Scholar] [CrossRef]
- Xie, C.; Yan, T.; Chen, J.; Li, X.; Zou, J.; Zhu, L.; Lu, L. LC-MS / MS Quantification of Sulfotransferases Is Better than Conventional Immunogenic Methods in Determining Human Liver SULT Activities : Implication in Precision Medicine. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Huang, L.; Coughtrie, M.W.H.; Hsu, H. Down-Regulation of Dehydroepiandrosterone Sulfotransferase Gene in Human Hepatocellular Carcinoma. Mol. Cell. Endocrinol. 2005, 231, 87–94. [Google Scholar] [CrossRef]
- Cells, H.; Qu, X.; Zhang, Y.; Zhang, S.; Zhai, J.; Gao, H.; Tao, L. Dysregulation of BSEP and MRP2 May Play an Important Role in Isoniazid-Induced Liver Injury via the SIRT1/FXR Pathway in Rats And. Biol. Pharm. Bull. 2018, 41, 1211–1218. [Google Scholar]
- Lee, T.K.; Hammond, C.L.; Ballatori, N.; Lee, H.C.; Pharmacol, T.A. Intracellular Glutathione Regulates Taurocholate Transport in HepG2 Cells Intracellular Glutathione Regulates Taurocholate Transport In. Toxicol. Appl. Pharmacol. 2001, 215, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Knisely, A.S.; Strautnieks, S.S.; Meier, Y.; Stieger, B.; Byrne, J.A.; Portmann, B.C.; Moore, L.; Raftos, J.; Arnell, H.; Cielecka-kuszyk, J.; et al. Hepatocellular Carcinoma in Ten Children Under Five Years of Age With Bile Salt Export Pump Deficiency. Hepatology 2006, 44, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Knisely, A.S.; Portmann, B.C.; Hill, D. Letter to the Editor Deficiency of BSEP in PFIC with Hepatocellular Malignancy. Natl. Libr. Med. 2006, 10, 644–645. [Google Scholar] [CrossRef]
- Pérez-Pineda, S.I.; Baylón-Pacheco, L.; Espíritu-Gordillo, P.; Tsutsumi, V.; Rosales-Encina, J.L. Effect of Bile Acids on the Expression of MRP3 and MRP4: An In Vitro Study in HepG2 Cell Line. Ann. Hepatol. 2021, 24, 100325. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Z.; Feng, J.H.; Bin Chen, Y.; Liao, N.S.; Su, Y.; Zou, C.Y. Metabolic Profiling of Normal Hepatocyte and Hepatocellular Carcinoma Cells via 1H Nuclear Magnetic Resonance Spectroscopy. Cell Biol. Int. 2018, 42, 425–434. [Google Scholar] [CrossRef]
- Ye, F.; Chen, C.; Qin, J.; Liu, J.; Zheng, A.C. Genetic Profiling Reveals an Alarming Rate of Cross-Contamination among Human Cell Lines Used in China. FASEB J. 2015, 29, 4268–4272. [Google Scholar] [CrossRef]
- Bayet-Robert, M.; Loiseau, D.; Rio, P.; Demidem, A.; Barthomeuf, C.; Stepien, G.; Morvan, D. Quantitative Two-Dimensional HRMAS 1H-NMR Spectroscopy-Based Metabolite Profiling of Human Cancer Cell Lines and Response to Chemotherapy. Magn. Reson. Med. 2010, 63, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Halama, A.; Möller, G.; Adamski, J. Metabolic Signatures in Apoptotic Human Cancer Cell Lines. OMICS J. Integr. Biol. 2011, 15, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Sniderman, A.D.; Kalant, D.; Vu, H.; Monge, J.C.; Tao, Y.; Cianflone, K. The Role of Amino Acids in ApoB100 Synthesis and Catabolism in Human HepG2 Cells. J. Biol. Chem. 1993, 268, 26920–26926. [Google Scholar] [CrossRef]
- Miyazaki, T.; Matsuzaki, Y. Taurine and Liver Diseases: A Focus on the Heterogeneous Protective Properties of Taurine. Amino Acids 2014, 46, 101–110. [Google Scholar] [CrossRef]
- Van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The Critical Role of Phosphatidylcholine and Phosphatidylethanolamine Metabolism in Health and Disease. Biochim. Biophys. Acta (BBA)—Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Harvey, A.J. Overview of Cell Signaling Pathways in Cancer. In Predictive Biomarkers in Oncology: Applications in Precision Medicine; Badve, S., Kumar, G.L., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 167–182. ISBN 9783319952284. [Google Scholar]
- Yip, H.Y.K.; Papa, A. Signaling Pathways in Cancer: Therapeutic Targets, Combinatorial Treatments, and New Developments. Cells 2021, 10, 659. [Google Scholar] [CrossRef]
- Balzarini, P.; Benetti, A.; Invernici, G.; Cristini, S.; Zicari, S.; Caruso, A.; Gatta, L.B.; Berenzi, A.; Imberti, L.; Zanotti, C.; et al. Transforming Growth Factor-Beta1 Induces Microvascular Abnormalities through a down-Modulation of Neural Cell Adhesion Molecule in Human Hepatocellular Carcinoma. Lab. Investig. 2012, 92, 1297–1309. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Monga, S.P. β-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology 2015, 148, 1294–1310. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Li, R.-P.; Liang, P.; Zhou, Y.-L.; Wang, G.-W. MiR-125a Inhibits the Migration and Invasion of Liver Cancer Cells via Suppression of the PI3K/AKT/MTOR Signaling Pathway. Oncol. Lett. 2015, 10, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Liu, B.R.; Qu, B.; Xing, H.; Gao, S.L.; Yin, J.M.; Wang, X.F.; Cheng, Y.Q. Silencing STAT3 May Inhibit Cell Growth through Regulating Signaling Pathway, Telomerase, Cell Cycle, Apoptosis and Angiogenesis in Hepatocellular Carcinoma: Potential Uses for Gene Therapy. Neoplasma 2011, 58, 158–171. [Google Scholar] [CrossRef]
- Wang, X.-H.; Chen, Z.-G.; Xu, R.-L.; Lv, C.-Q.; Liu, J.; Du, B. TGF-Β1 Signaling Pathway Serves a Role in HepG2 Cell Regulation by Affecting the Protein Expression of PCNA, Gankyrin, P115, XIAP and Survivin. Oncol. Lett. 2017, 13, 3239–3246. [Google Scholar] [CrossRef][Green Version]
- Sha, Y.-L.; Liu, S.; Yan, W.-W.; Dong, B. Wnt/β-Catenin Signaling as a Useful Therapeutic Target in Hepatoblastoma. Biosci. Rep. 2019, 39, BSR20192466. [Google Scholar] [CrossRef]
- Liu, J.; Li, G.; Liu, D.; Liu, J. FH535 Inhibits the Proliferation of HepG2 Cells via Downregulation of the Wnt/β-Catenin Signaling Pathway. Mol. Med. Rep. 2014, 9, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guo, S.; Zhao, R.; Liu, Y.; Yang, G. STAT3-Activated Long Non-Coding RNA Lung Cancer Associated Transcript 1 Drives Cell Proliferation, Migration, and Invasion in Hepatoblastoma Through Regulation of the MiR-301b/STAT3 Axis. Hum. Gene 2019, 30, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Lu, P.; Song, G.; Liu, Q.; Zhu, D.; Liu, X. Involvement of PI3K/Akt, ERK and P38 Signaling Pathways in Emodin-Mediated Extrinsic and Intrinsic Human Hepatoblastoma Cell Apoptosis. Food Chem. Toxicol. 2016, 92, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Ferrín, G.; Guerrero, M.; Amado, V.; Rodríguez-Perálvarez, M.; De la Mata, M. Activation of MTOR Signaling Pathway in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, E1266. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zaidi, S.; Rao, S.; Chen, J.-S.; Phan, L.; Farci, P.; Su, X.; Shetty, K.; White, J.; Zamboni, F.; et al. Analysis of Genomes and Transcriptomes of Hepatocellular Carcinomas Identifies Mutations and Gene Expression Changes in the Transforming Growth Factor-β Pathway. Gastroenterology 2018, 154, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Immunomodulatory TGF-β Signaling in Hepatocellular Carcinoma. PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/31353124/ (accessed on 29 November 2021).
- Archakov, A.; Aseev, A.; Bykov, V.; Grigoriev, A.; Govorun, V.; Ivanov, V.; Khlunov, A.; Lisitsa, A.; Mazurenko, S.; Makarov, A.A.; et al. Gene-Centric View on the Human Proteome Project: The Example of the Russian Roadmap for Chromosome 18. Proteomics 2011, 11, 1853–1856. [Google Scholar] [CrossRef]
- Orsburn, B.C. Evaluation of the Sensitivity of Proteomics Methods Using the Absolute Copy Number of Proteins in a Single Cell as a Metric. Proteomes 2021, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The Size of the Human Proteome: The Width and Depth. Int. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Dial, S.; Shi, L.; Branham, W.; Liu, J.; Fang, J.-L.; Green, B.; Deng, H.; Kaput, J.; Ning, B. Similarities and Differences in the Expression of Drug-Metabolizing Enzymes between Human Hepatic Cell Lines and Primary Human Hepatocytes. Drug. Metab. Dispos. 2011, 39, 528–538. [Google Scholar] [CrossRef]
- Ahlin, G.; Hilgendorf, C.; Karlsson, J.; Szigyarto, C.A.-K.; Uhlén, M.; Artursson, P. Endogenous Gene and Protein Expression of Drug-Transporting Proteins in Cell Lines Routinely Used in Drug Discovery Programs. Drug. Metab. Dispos. 2009, 37, 2275–2283. [Google Scholar] [CrossRef] [PubMed]
- Zhen, R.; Chen, S.; Ning, B.; Guo, L. Use of Liver-Derived Cell Lines for the Study of Drug-Induced Liver Injury; Humana Press: New York, NY, USA, 2018. [Google Scholar] [CrossRef]
- Upgrading HepG2 Cells with Adenoviral Vectors That Encode Drug-Metabolizing Enzymes: Application for Drug Hepatotoxicity Testing. PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/27671376/ (accessed on 29 November 2021).
- Takemura, A.; Gong, S.; Sato, T.; Kawaguchi, M.; Sekine, S.; Kazuki, Y.; Horie, T.; Ito, K. Evaluation of Parent- and Metabolite-Induced Mitochondrial Toxicities Using CYP-Introduced HepG2 Cells. J. Pharm. Sci. 2021, 110, 3306–3312. [Google Scholar] [CrossRef]
- López-Terrada, D.; Gunaratne, P.H.; Adesina, A.M.; Pulliam, J.; Hoang, D.M.; Nguyen, Y.; Mistretta, T.-A.; Margolin, J.; Finegold, M.J. Histologic Subtypes of Hepatoblastoma Are Characterized by Differential Canonical Wnt and Notch Pathway Activation in DLK+ Precursors. Hum. Pathol. 2009, 40, 783–794. [Google Scholar] [CrossRef]
- Matsumoto, S.; Yamamichi, T.; Shinzawa, K.; Kasahara, Y.; Nojima, S.; Kodama, T.; Obika, S.; Takehara, T.; Morii, E.; Okuyama, H.; et al. GREB1 Induced by Wnt Signaling Promotes Development of Hepatoblastoma by Suppressing TGFβ Signaling. Nat. Commun. 2019, 10, 3882. [Google Scholar] [CrossRef]
- Regel, I.; Eichenmüller, M.; Mahajan, U.M.; Hagl, B.; Benitz, S.; Häberle, B.; Vokuhl, C.; von Schweinitz, D.; Kappler, R. Downregulation of SFRP1 Is a Protumorigenic Event in Hepatoblastoma and Correlates with Beta-Catenin Mutations. J. Cancer Res. Clin. Oncol. 2020, 146, 1153–1167. [Google Scholar] [CrossRef]
- Epidermal Growth Factor Receptor/Heme Oxygenase-1 Axis Is Involved in Chemoresistance to Cisplatin and Pirarubicin in HepG2 Cell Lines and Hepatoblastoma Specimens. PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/31559456/ (accessed on 29 November 2021).
- Romanelli, R.G.; Petrai, I.; Robino, G.; Efsen, E.; Novo, E.; Bonacchi, A.; Pagliai, G.; Grossi, A.; Parola, M.; Navari, N.; et al. Thrombopoietin Stimulates Migration and Activates Multiple Signaling Pathways in Hepatoblastoma Cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, G120–G128. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ye, M.; He, J.; Zhang, J.; Liu, B.; Liu, X.; Xie, L.; Wei, M.; Dong, R.; Li, K.; Ma, D.; et al. USP7 Promotes Hepatoblastoma Progression through Activation of PI3K/AKT Signaling Pathway. Cancer Biomark. 2021, 31, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Liu, B.; Liu, X.; Xie, L.; He, J.; Zhang, J.; Dong, R.; Ma, D.; Dong, K.; Ye, M. ARID1B/SUB1-Activated LncRNA HOXA-AS2 Drives the Malignant Behaviour of Hepatoblastoma through Regulation of HOXA3. J. Cell Mol. Med. 2021, 25, 3524–3536. [Google Scholar] [CrossRef]
- Rauch, C.; Jennings, P.; Wilmes, A. Vitro Toxicology Systems; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar] [CrossRef]
- Perea, L.; Coll, M.; Sancho-Bru, P. Assessment of Liver Fibrotic Insults In Vitro; Humana Press: New York, NY, USA, 2015; Volume 1250, ISBN 9781493920747. [Google Scholar]
- Bouma, M.E.; Rogier, E.; Verthier, N.; Labarre, C.; Feldmann, G. Further Cellular Investigation of the Human Hepatoblastoma-Derived Cell Line HepG2: Morphology and Immunocytochemical Studies of Hepatic-Secreted Proteins. Vitr. Cell. Dev. Biol. J. Tissue Cult. Assoc. 1989, 25, 267–275. [Google Scholar] [CrossRef]
- Krithika, R.; Mohankumar, R.; Verma, R.J.; Shrivastav, P.S.; Mohamad, I.L.; Gunasekaran, P.; Narasimhan, S. Isolation, Characterization and Antioxidative Effect of Phyllanthin against CCl4-Induced Toxicity in HepG2 Cell Line. Chem. Biol. Interact. 2009, 181, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Xuan, J.; Chen, S.; Ning, B.; Tolleson, W.H.; Guo, L. Development of HepG2-Derived Cells Expressing Cytochrome P450s for Assessing Metabolism-Associated Drug-Induced Liver Toxicity. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Ooka, M.; Lynch, C.; Xia, M. Application of in Vitro Metabolism Activation in High-Throughput Screening. Int. J. Mol. Sci. 2020, 21, 8182. [Google Scholar] [CrossRef] [PubMed]
- Anton, D.; Burckel, H.; Josset, E.; Noel, G. Three-Dimensional Cell Culture: A Breakthrough in Vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef] [PubMed]
- Shah, U.K.; de Mallia, J.O.; Singh, N.; Chapman, K.E.; Doak, S.H.; Jenkins, G.J.S. A Three-Dimensional in Vitro HepG2 Cells Liver Spheroid Model for Genotoxicity Studies. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 2018, 825, 51–58. [Google Scholar] [CrossRef] [PubMed]
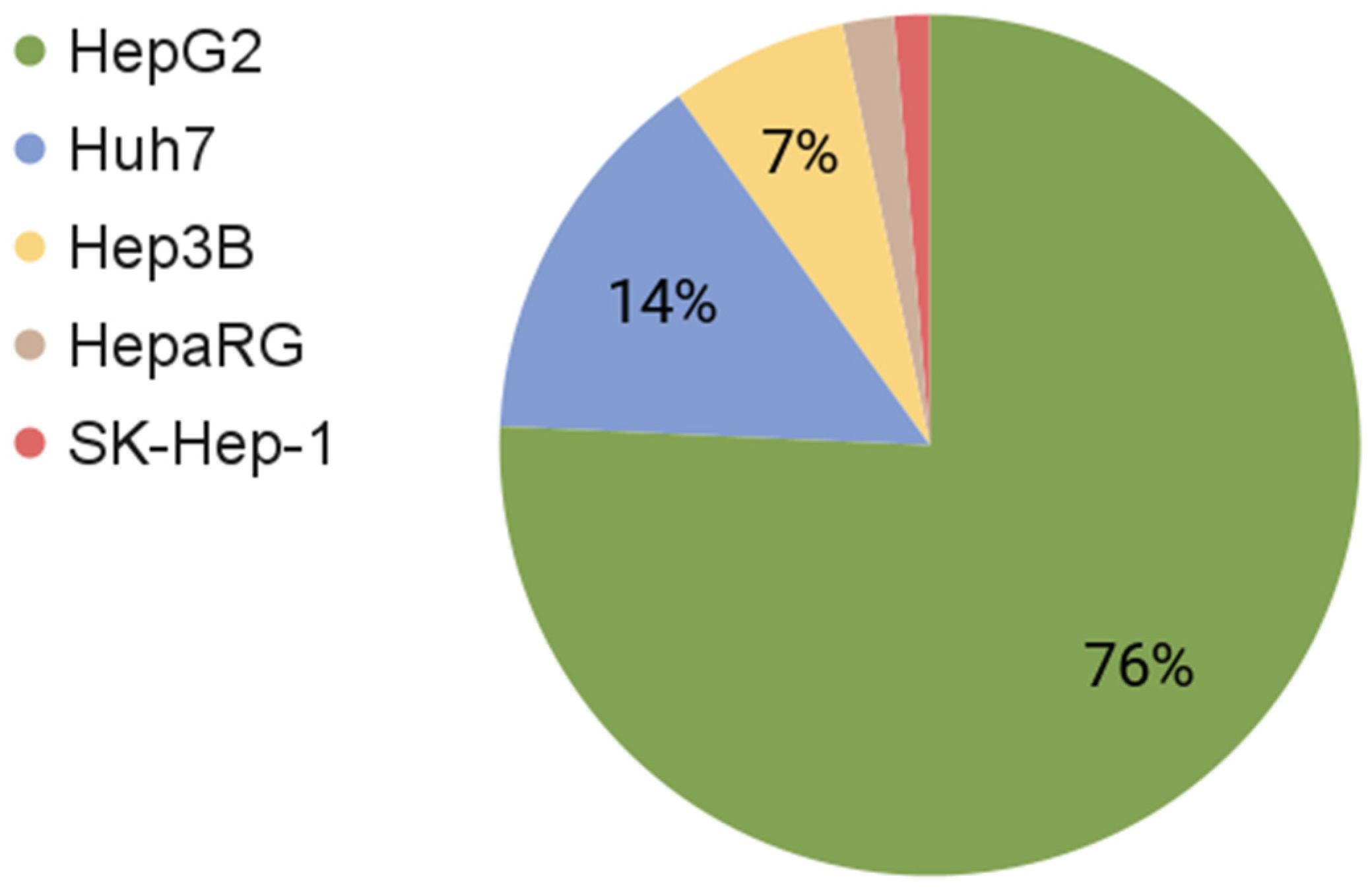
| Cell Type | Mutated Genes | Number of Chromosomes | Number of Articles (PubMed) | Reference | |
|---|---|---|---|---|---|
| HepG2 | HB | CTNNB1 | 50–60 | 34,021 | [12,30] |
| HepaRG | HCC | PLIN2, ANXA1, H2AFY, SNX1, GCHFR, APO | 46 | 880 | [26,31,32,33] |
| Huh7 | HCC | KDR, POLD3, TERT, TP53 | 55–63 | 6463 | [34,35,36,37] |
| Hep3B | HCC | AXIN1, RB1 | ≈60 | 2994 | [34,38] |
| SK-Hep-1 | Adenocarcinoma | CDKN2A, BRAF | 56–64 | 602 | [39,40] |
| HepG2 Cells | Hepatocyte Cells | Cells with HB | Cells with HCC | |
|---|---|---|---|---|
| Cell size and shape | 12–19 µm, polygonal | 15 µm, cube | 10–20 µm, round or angulated | >10 µm, spindle-shaped and show bizarre anaplastic figures |
| Subcellular components | Large nuclei, 3–7 nucleoli, low mitochondrial content, and poorly developed SER 1 | Two or more nuclei occupy 5–7% of the cell volume; high SER1 and mitochondria | Small, round, inconspicuous nucleoli; low mitochondrial and RER 2 content | The numbers of mitochondria and ER 3 is reduced, and have an abnormal structure, characteristic of stressful conditions |
| Number of chromosomes | 50–60 | Polyploidy | Aneuploidy, >46 | Aneuploidy, >46 |
| Genome stability, DNA content | 7.5 pg genomic DNA, genome unstable | ~6 pg genomic DNA, stable genome | Genome unstable | Genome unstable |
| Type of Research | Advantages | Disadvantages | Recommendations | Valid References 1 |
|---|---|---|---|---|
| Toxicity tests |
| According to the article (Ren et al.), the HepG2 cell line may not be a suitable model in investigating metabolism-mediated toxicity without additional modification due to it is lack of metabolic capability [174,175]. For example, the cell line can be used in studies of CYP inducers [176]. | No valid experiment. | |
| Drug metabolism | No valid experiment. | |||
| HB model | The correct use of HepG2 is an HB model, as it has many of the characteristics of HB. According to the article (Lopez-Terrada D et al.), “the correct attribution of the tumor of origin of this cell line is of crucial interest for investigators studying the biology of hepatocellular neoplasms, particularly those engaged in novel biology-based classifications, clinical stratification, and therapeutic interventions for pediatric and adult patient” [29]. | [178,179,180,181,182,183] | ||
| HCC model | Partially similar genome and transcriptome profiles. | At the genomic and transcriptomic levels has been shown to be similar to hepatoblastoma. | According to meta-analysis, the cell line has some similar mutations to HCC. However, there are a large number of cell lines derived from HCC cells, such as Huh-7, HepaRG, etc. Additionally, their use is relevant, as there are more overlaps in the genetic and transcriptome profiles. | Questionable. |
| Hepatocyte model | No advantages. |
| There are non-tumor cell lines such as THLE-2 and THLE-3 that have characteristics similar to hepatocytes [22]. | No valid experiment. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arzumanian, V.A.; Kiseleva, O.I.; Poverennaya, E.V. The Curious Case of the HepG2 Cell Line: 40 Years of Expertise. Int. J. Mol. Sci. 2021, 22, 13135. https://doi.org/10.3390/ijms222313135
Arzumanian VA, Kiseleva OI, Poverennaya EV. The Curious Case of the HepG2 Cell Line: 40 Years of Expertise. International Journal of Molecular Sciences. 2021; 22(23):13135. https://doi.org/10.3390/ijms222313135
Chicago/Turabian StyleArzumanian, Viktoriia A., Olga I. Kiseleva, and Ekaterina V. Poverennaya. 2021. "The Curious Case of the HepG2 Cell Line: 40 Years of Expertise" International Journal of Molecular Sciences 22, no. 23: 13135. https://doi.org/10.3390/ijms222313135
APA StyleArzumanian, V. A., Kiseleva, O. I., & Poverennaya, E. V. (2021). The Curious Case of the HepG2 Cell Line: 40 Years of Expertise. International Journal of Molecular Sciences, 22(23), 13135. https://doi.org/10.3390/ijms222313135