Prenatal Exposure to Triclocarban Impairs ESR1 Signaling and Disrupts Epigenetic Status in Sex-Specific Ways as Well as Dysregulates the Expression of Neurogenesis- and Neurotransmitter-Related Genes in the Postnatal Mouse Brain
Abstract
:1. Introduction
2. Results
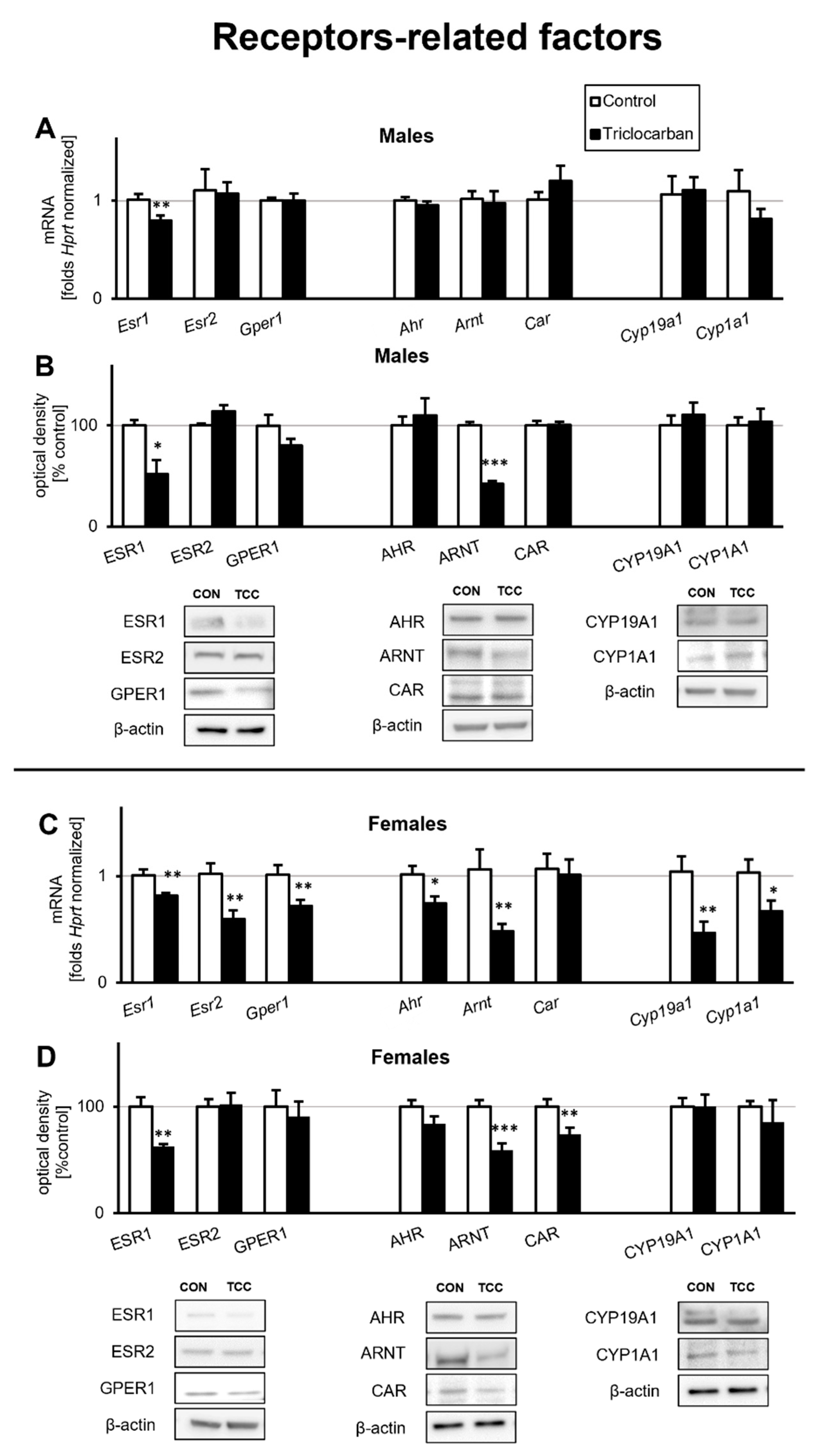
2.1. Triclocarban Downregulated Receptor Signaling in One-Month-Old Mice That Were Prenatally Exposed to Triclocarban
2.1.1. Triclocarban Inhibited Esr1 mRNA Expression and Decreased the Protein Levels of ESR1 and ARNT in One-Month-Old Male Mice That Were Prenatally Exposed to the Chemical
2.1.2. Triclocarban Inhibited the mRNA Expression of Esr1, Esr2, Gper1, Ahr, Arnt, Cyp19a1, and Cyp1a1, and Decreased ESR1, ARNT, and CAR Protein Levels in One-Month-Old Female Mice That Were Prenatally Exposed to the Chemical
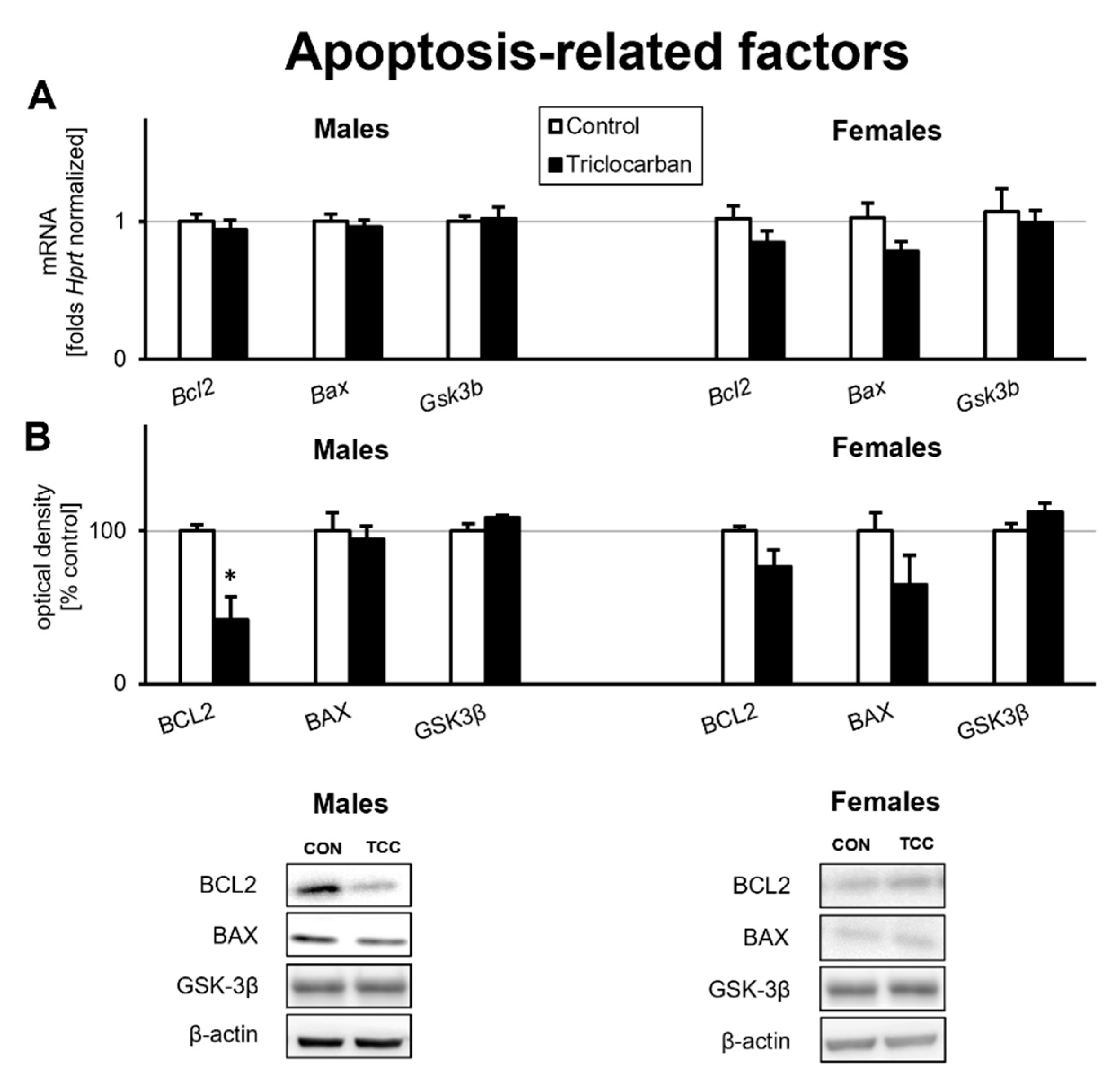
2.2. Prenatal Exposure to Triclocarban Did Not Affect the mRNA or Protein Expression of Apoptosis-Related Factors Other Than BCL2
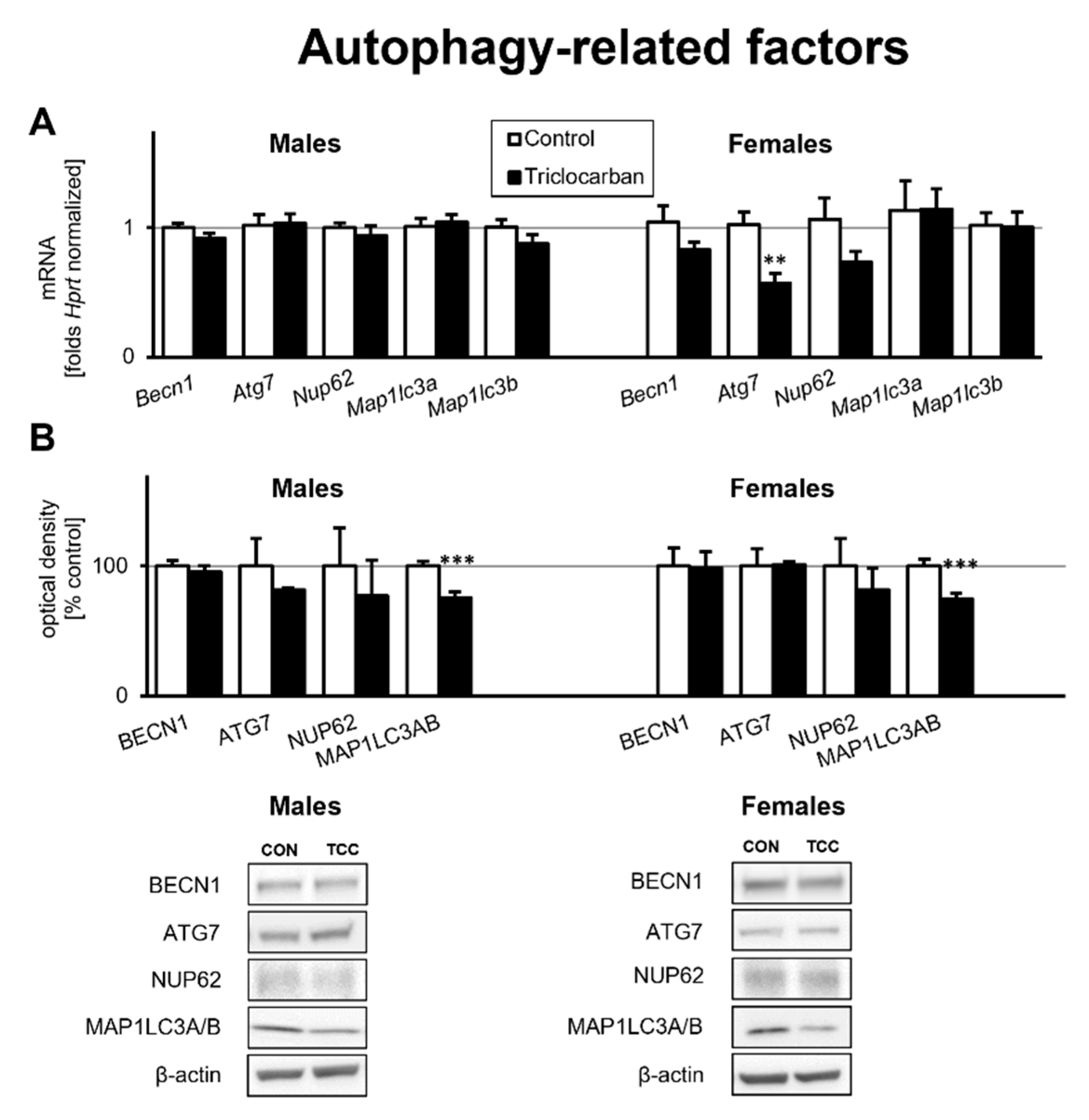
2.3. Prenatal Exposure to Triclocarban Did Not Affect the mRNA or Protein Expression of Autophagy-Related Factors Other Than Atg7 mRNA and MAP1LC3AB
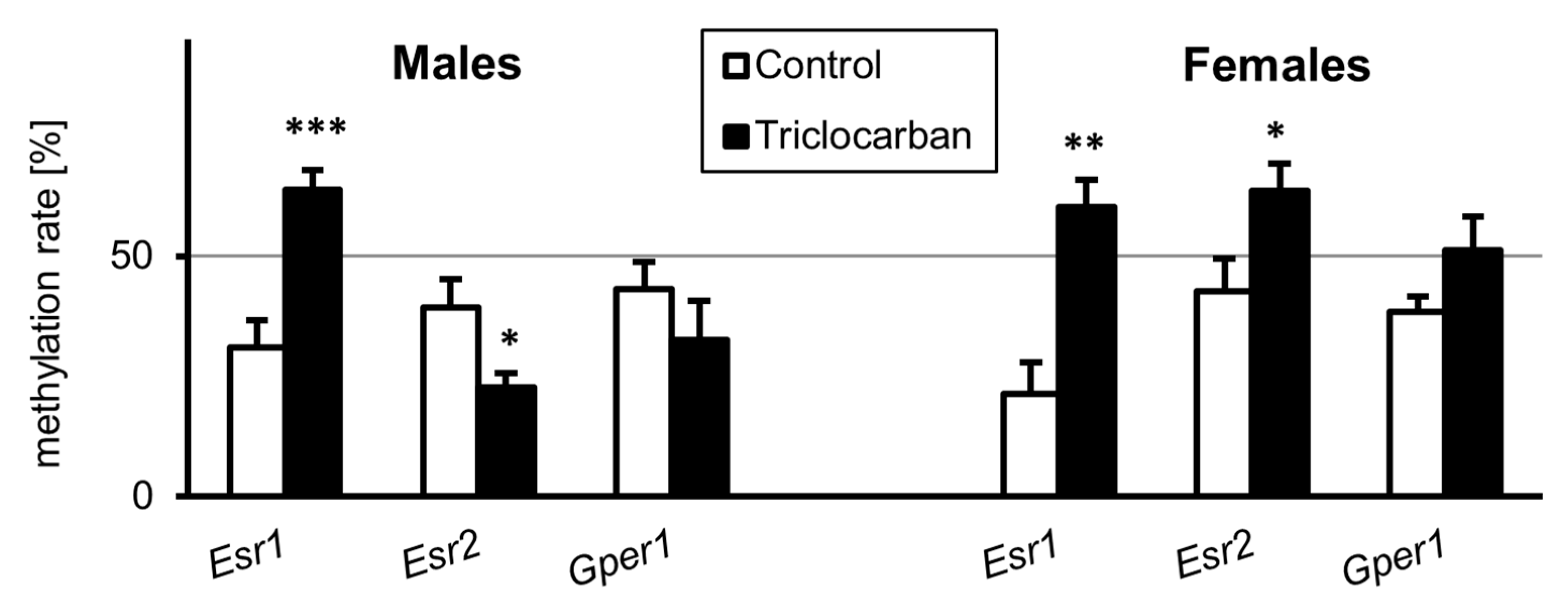
2.4. Prenatal Exposure to Triclocarban Caused Sex-Specific Disruption of the Methylation Statuses of Estrogen Receptor Genes
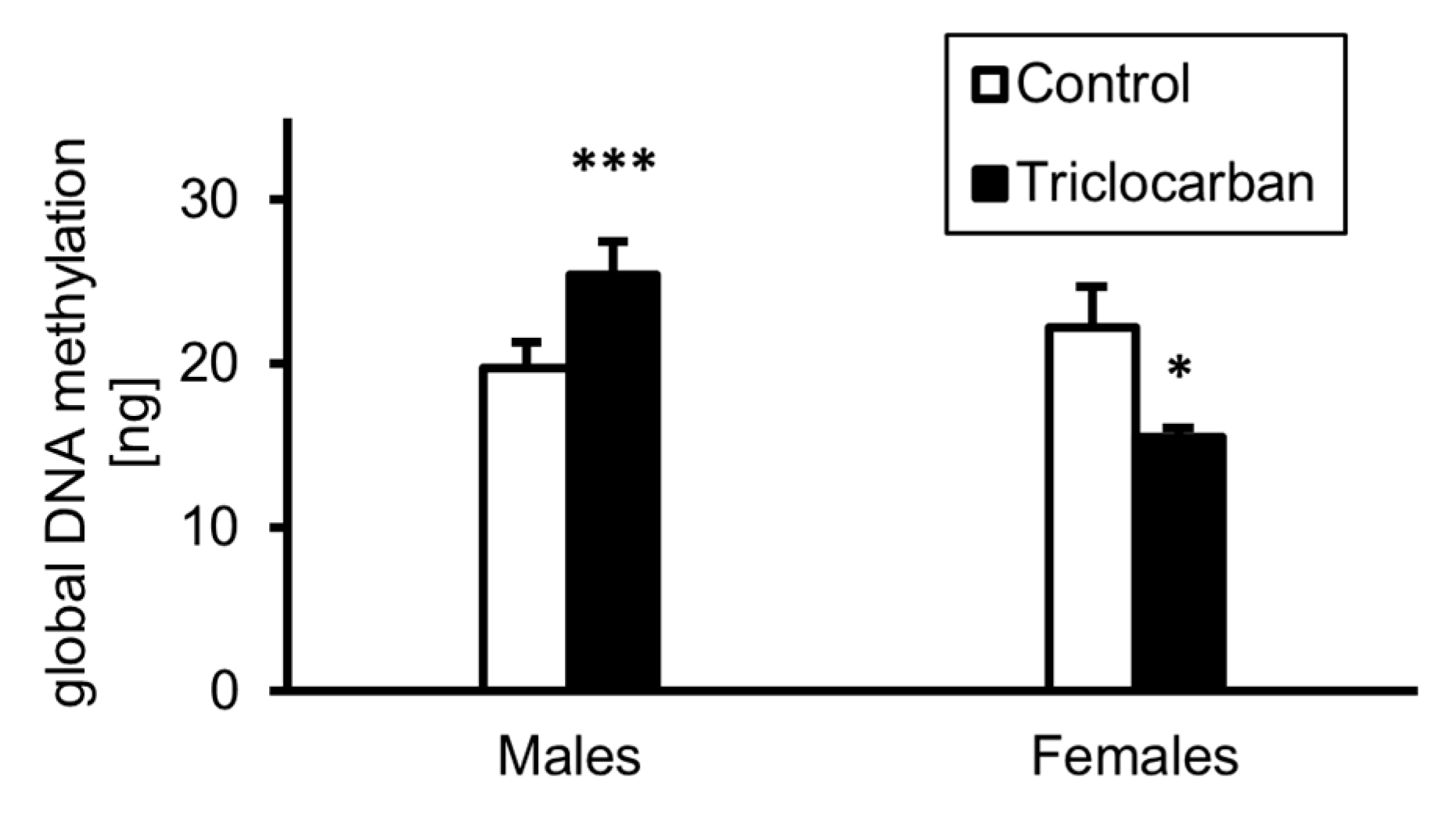
2.5. Prenatal Exposure to Triclocarban Caused Sex-Dependent Alterations in Global DNA Methylation
2.6. Triclocarban Dysregulated the Expression of Neurogenesis- and Neurotransmitter-Related Genes in One-Month-Old Mice That Were Prenatally Exposed to Triclocarban
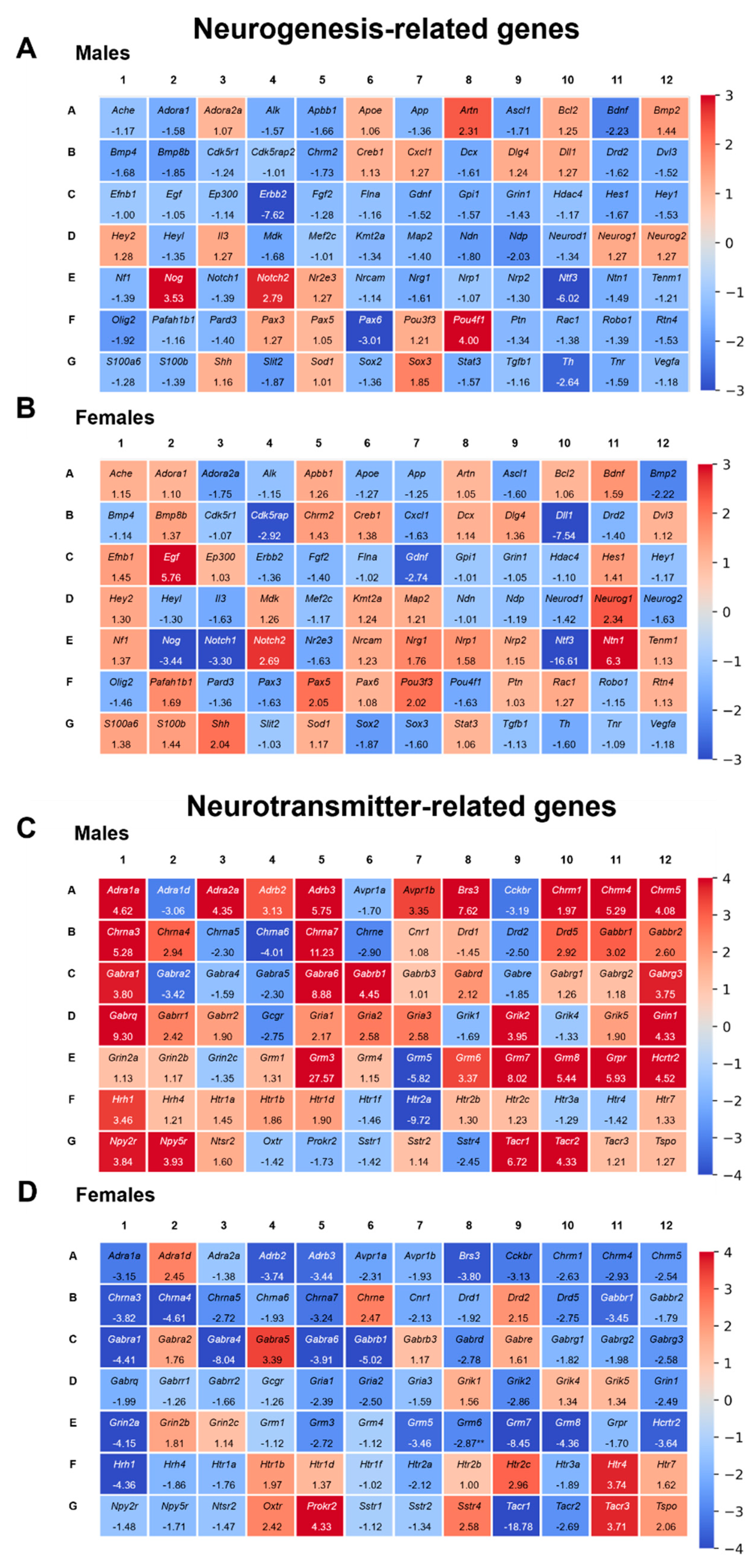
2.6.1. Triclocarban Dysregulated the Expression of Neurogenesis-Related Genes in One-Month-Old Male and Female Mice That Were Prenatally Exposed to the Chemical
2.6.2. Triclocarban Dysregulated the Expression of Neurotransmitter-Related Genes in One-Month-Old Male and Female Mice That Were Prenatally Exposed to the Chemical
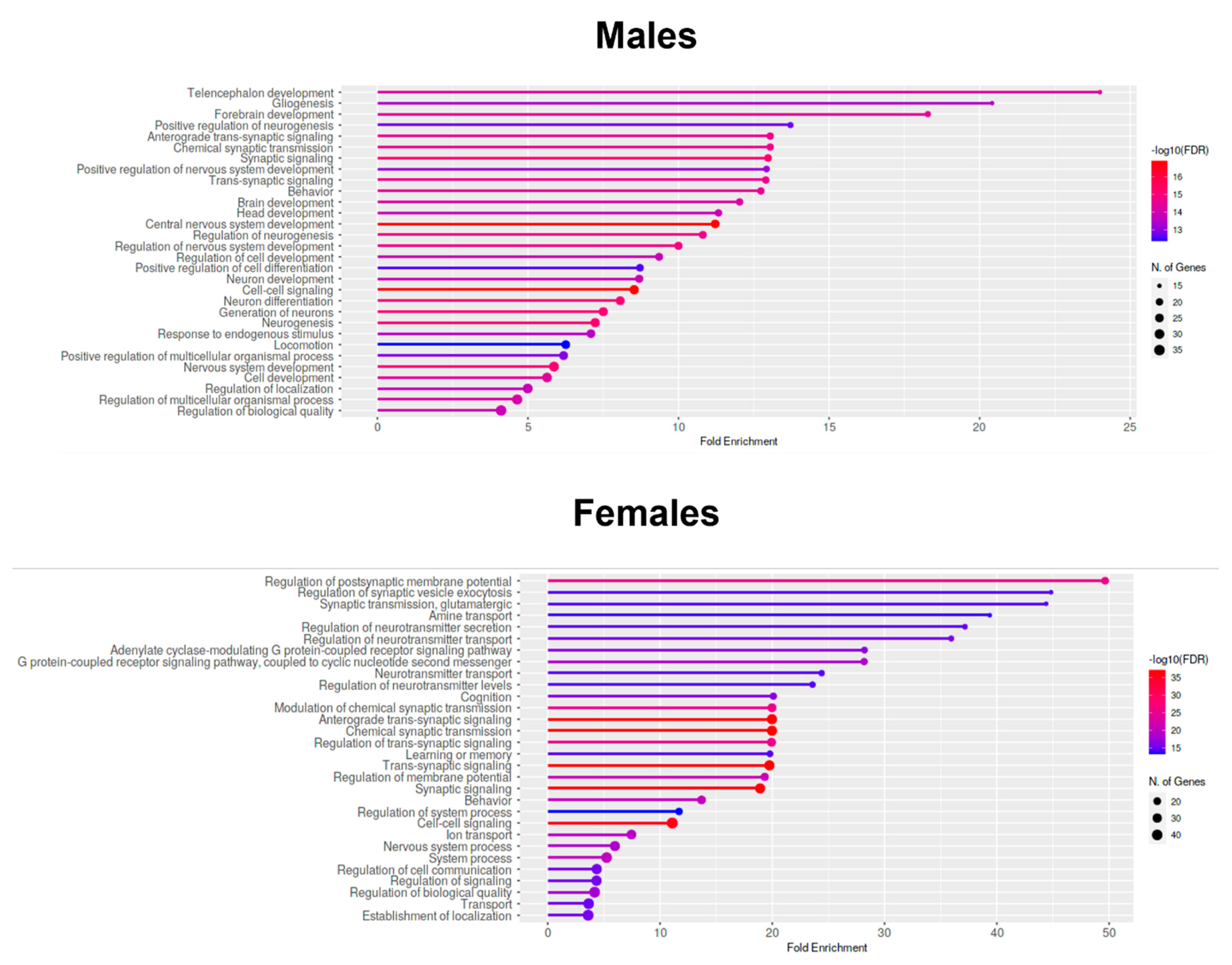
2.7. Enrichment Analyses—GO Biological Process Analysis of Downregulated Genes in the Prefrontal Cortices of One-Month-Old Males and Females That Were Prenatally Exposed to Triclocarban
2.8. Triclocarban Crossed the Blood–Brain Barrier In Vitro
3. Discussion
4. Materials and Methods
4.1. Material
4.2. Animals
4.3. Treatment
4.4. Sampling
4.5. qPCR Analyses
4.6. Measurement of DNA Methylation of Specific Genes
4.7. Measurement of Global DNA Methylation
4.8. Profiling Neurogenesis-Related Genes and Neurotransmitter-Related Genes Using Microarray Assays
4.9. Western Blot Analysis
4.10. Measurement of Blood–Brain Barrier Permeability
4.11. Data Analysis
4.12. Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADHD | Attention-deficit hyperactivity disorder |
| Ahr/AHR | Aryl hydrocarbon receptor |
| Arnt/ARNT | Aryl hydrocarbon receptor nuclear translocator |
| ASD | Autism spectrum disorder |
| Atg7/ATG7 | Autophagy-related 7 |
| BBB | Blood–brain barrier |
| Car/CAR | Constitutive androstane receptor |
| ERs | Estrogen receptors |
| Esr1/ESR1 | Estrogen receptor 1; estrogen receptor alpha (ERα) |
| Esr2/ESR2 | Estrogen receptor 2; estrogen receptor beta (ERβ) |
| Gper1/GPER1 | G-protein-coupled estrogen receptor 1; G-protein-coupled receptor 30 (GPR30) |
| Map1lc3a/b/MAP1LC3AB | Microtubule associated protein 1 light chain 3 alpha/beta |
| TCC | Triclocarban (3,4,4′-trichlorocarbanilide) |
References
- Yun, H.; Liang, B.; Kong, D.; Li, X.; Wang, A. Fate, risk and removal of triclocarban: A critical review. J. Hazard Mater. 2020, 387, 121944. [Google Scholar] [CrossRef]
- Iacopetta, D.; Catalano, A.; Ceramella, J.; Saturnino, C.; Salvagno, L.; Ielo, I.; Drommi, D.; Scali, E.; Plutino, M.R.; Rosace, G.; et al. The Different Facets of Triclocarban: A Review. Molecules 2021, 26, 2811. [Google Scholar] [CrossRef]
- Bai, X.; Zhang, B.; He, Y.; Hong, D.; Song, S.; Huang, Y.; Zhang, T. Triclosan and triclocarbon in maternal-fetal serum, urine, and amniotic fluid samples and their implication for prenatal exposure. Environ. Pollut. 2020, 266, 115117. [Google Scholar] [CrossRef]
- Kennedy, R.C.; Menn, F.M.; Healy, L.; Fecteau, K.A.; Hu, P.; Bae, J.; Gee, N.A.; Lasley, B.L.; Zhao, L.; Chen, J. Early life triclocarban exposure during lactation affects neonate rat survival. Reprod. Sci. 2015, 22, 75–89. [Google Scholar] [CrossRef] [Green Version]
- Aker, A.; McConnell, R.E.R.; Loch-Caruso, R.; Park, S.K.; Mukherjee, B.; Rosario, Z.Y.; Vélez-Vega, C.M.; Huerta-Montanez, G.; Alshawabkeh, A.N.; Cordero, J.F.; et al. Interactions between chemicals and non-chemical stressors: The modifying effect of life events on the association between triclocarban, phenols and parabens with gestational length in a Puerto Rican cohort. Sci. Total Environ. 2020, 708, 134719. [Google Scholar] [CrossRef] [PubMed]
- Pycke, B.F.; Geer, L.A.; Dalloul, M.; Abulafia, O.; Jenck, A.M.; Halden, R.U. Human fetal exposure to triclosan and triclocarban in an urban population from Brooklyn, New York. Environ. Sci. Technol. 2014, 48, 8831–8838. [Google Scholar] [CrossRef]
- Van Der Meer, T.P.; Artacho-Cordón, F.; Swaab, D.F.; Struik, D.; Makris, K.C.; Wolffenbuttel, B.H.R.; Frederiksen, H.; Van Vliet-Ostaptchouk, J.V. Distribution of Non-Persistent Endocrine Disruptors in Two Different Regions of the Human Brain. Int. J. Environ. Res. Public Health 2017, 14, 1059. [Google Scholar] [CrossRef] [Green Version]
- Kajta, M.; Wnuk, A.; Rzemieniec, J.; Lason, W.; Mackowiak, M.; Chwastek, E.; Staniszewska, M.; Nehring, I.; Wojtowicz, A.K. Triclocarban Disrupts the Epigenetic Status of Neuronal Cells and Induces AHR/CAR-Mediated Apoptosis. Mol. Neurobiol. 2019, 56, 3113–3131. [Google Scholar] [CrossRef] [PubMed]
- Kajta, M.; Rzemieniec, J.; Wnuk, A.; Lasoń, W. Triclocarban impairs autophagy in neuronal cells and disrupts estrogen receptor signaling via hypermethylation of specific genes. Sci. Total Environ. 2020, 701, 134818. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Zhu, J.; Han, W.; Wang, S.; Yan, Z.; Ma, D.; Goh, E.L.K.; Chen, T. Maternal methamphetamine exposure causes cognitive impairment and alteration of neurodevelopment-related genes in adult offspring mice. Neuropharmacology 2018, 140, 25–34. [Google Scholar] [CrossRef]
- La Barbera, L.; Vedele, F.; Nobili, A.; D’Amelio, M.; Krashia, P. Neurodevelopmental Disorders: Functional Role of Ambra1 in Autism and Schizophrenia. Mol. Neurobiol. 2019, 56, 6716–6724. [Google Scholar] [CrossRef]
- Fleming, A.; Rubinsztein, D.C. Autophagy in Neuronal Development and Plasticity. Trends Neurosci. 2020, 43, 767–779. [Google Scholar] [CrossRef]
- Hess, J.L.; Radonjić, N.V.; Patak, J.; Glatt, S.J.; Faraone, S.V. Autophagy, apoptosis, and neurodevelopmental genes might underlie selective brain region vulnerability in attention-deficit/hyperactivity disorder. Mol. Psychiatry 2020. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.J.; Lee, T.Y.; Kim, N.S.; Kwon, J.S. The Role of Estrogen Receptors and Their Signaling across Psychiatric Disorders. Int. J. Mol. Sci. 2020, 22, 373. [Google Scholar] [CrossRef] [PubMed]
- Juricek, L.; Coumoul, X. The Aryl Hydrocarbon Receptor and the Nervous System. Int. J. Mol. Sci. 2018, 19, 2504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliviero, F.; Lukowicz, C.; Boussadia, B.; Forner-Piquer, I.; Pascussi, J.M.; Marchi, N.; Mselli-Lakhal, L. Constitutive Androstane Receptor: A Peripheral and a Neurovascular Stress or Environmental Sensor. Cells 2020, 9, 2426. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.J.; Vargas-Maya, N.; Pang, A.P.S.; Lum-Jones, A.; Li, D.; Khadka, V.; Sultana, R.; Blanchard, D.C.; Maunakea, A.K. Epigenetic Delay in the Neurodevelopmental Trajectory of DNA Methylation States in Autism Spectrum Disorders. Front. Genet. 2019, 10, 907. [Google Scholar] [CrossRef] [Green Version]
- Aref-Eshghi, E.; Kerkhof, J.; Pedro, V.P.; Groupe, D.I.F.; Barat-Houari, M.; Ruiz-Pallares, N.; Andrau, J.C.; Lacombe, D.; Van-Gils, J.; Fergelot, P.; et al. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020, 106, 356–370. [Google Scholar] [CrossRef]
- Huang, H.; Du, G.; Zhang, W.; Hu, J.; Wu, D.; Song, L.; Xia, Y.; Wang, X. The in vitro estrogenic activities of triclosan and triclocarban. J. Appl. Toxicol. 2014, 34, 1060–1067. [Google Scholar] [CrossRef]
- Li, H.; Zhao, Y.; Chen, L.; Su, Y.; Li, X.; Jin, L.; Ge, R.S. Triclocarban and Triclosan Inhibit Human Aromatase via Different Mechanisms. Biomed. Res. Int. 2017, 2017, 8284097. [Google Scholar] [CrossRef] [Green Version]
- Chung, E.; Genco, M.C.; Megrelis, L.; Ruderman, J.V. Effects of bisphenol A and triclocarban on brain-specific expression of aromatase in early zebrafish embryos. Proc. Natl. Acad. Sci. USA 2011, 108, 17732–17737. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.L.; Ueki, K.; Kumagai, K.; Araki, R.; Otsuki, Y. Regulation of bcl-2 transcription by estrogen receptor-α and c-Jun in human endometrium. Med. Mol. Morphol. 2014, 47, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Mandour, D.A.; Aidaros, A.A.; Mohamed, S. Potential long-term developmental toxicity of in utero and lactational exposure to Triclocarban (TCC) in hampering ovarian folliculogenesis in rat offspring. Acta Histochem. 2021, 123, 151772. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Xie, W.; Zheng, W.; Yin, D.; Luo, R.; Guo, F. Targeted binding of estradiol with ESR1 promotes proliferation of human chondrocytes in vitro by inhibiting activation of ERK signaling pathway. Nan Fang Yi Ke Da Xue Xue Bao 2019, 39, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Yueh, M.F.; Li, T.; Evans, R.M.; Hammock, B.; Tukey, R.H. Triclocarban mediates induction of xenobiotic metabolism through activation of the constitutive androstane receptor and the estrogen receptor alpha. PLoS ONE 2012, 7, e37705. [Google Scholar] [CrossRef]
- Kajta, M.; Wójtowicz, A.K.; Maćkowiak, M.; Lasoń, W. Aryl hydrocarbon receptor-mediated apoptosis of neuronal cells: A possible interaction with estrogen receptor signaling. Neuroscience 2009, 158, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Tarnow, P.; Tralau, T.; Hunecke, D.; Luch, A. Effects of triclocarban on the transcription of estrogen, androgen and aryl hydrocarbon receptor responsive genes in human breast cancer cells. Toxicol. Vitr. 2013, 27, 1467–1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Qian, Q.; Xia, M.; Wang, X.; Wang, H. Trichlorocarban induces developmental and immune toxicity to zebrafish (Danio rerio) by targeting TLR4/MyD88/NF-κB signaling pathway. Environ. Pollut. 2021, 273, 116479. [Google Scholar] [CrossRef]
- Kajta, M.; Wnuk, A.; Rzemieniec, J.; Litwa, E.; Lason, W.; Zelek-Molik, A.; Nalepa, I.; Rogóż, Z.; Grochowalski, A.; Wojtowicz, A.K. Depressive-like effect of prenatal exposure to DDT involves global DNA hypomethylation and impairment of GPER1/ESR1 protein levels but not ESR2 and AHR/ARNT signaling. J. Steroid. Biochem. Mol. Biol. 2017, 171, 94–109. [Google Scholar] [CrossRef] [PubMed]
- Schebb, N.H.; Flores, I.; Kurobe, T.; Franze, B.; Ranganathan, A.; Hammock, B.D.; The, S.J. Bioconcentration, metabolism and excretion of triclocarban in larval Qurt medaka (Oryzias latipes). Aquat. Toxicol. 2011, 105, 448–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, J.; Wei, L.; Shi, Y.; Zhang, J.; Wu, Q.; Shao, B. Chinese population exposure to triclosan and triclocarban as measured via human urine and nails. Environ. Geochem. Health 2016, 38, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Geer, L.A.; Pycke, B.F.G.; Waxenbaum, J.; Sherer, D.M.; Abulafia, O.; Halden, R.U. Association of birth outcomes with fetal exposure to parabens, triclosan and triclocarban in an immigrant population in Brooklyn, New York. J. Hazard Mater. 2017, 323, 177–183. [Google Scholar] [CrossRef] [Green Version]
- Schebb, N.H.; Inceoglu, B.; Ahn, K.C.; Morisseau, C.; Gee, S.J.; Hammock, B.D. Investigation of human exposure to triclocarban after showering and preliminary evaluation of its biological effects. Environ. Sci. Technol. 2011, 45, 3109–3115. [Google Scholar] [CrossRef] [Green Version]
- Wnuk, A.; Rzemieniec, J.; Staroń, J.; Litwa, E.; Lasoń, W.; Bojarski, A.; Kajta, M. Prenatal Exposure to Benzophenone-3 Impairs Autophagy, Disrupts RXRs/PPARγ Signaling, and Alters Epigenetic and Post-Translational Statuses in Brain Neurons. Mol. Neurobiol. 2019, 56, 4820–4837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rzemieniec, J.; Bratek, E.; Wnuk, A.; Przepiórska, K.; Salińska, E.; Kajta, M. Neuroprotective effect of 3,3′-Diindolylmethane against perinatal asphyxia involves inhibition of the AhR and NMDA signaling and hypermethylation of specific genes. Apoptosis 2020, 25, 747–762. [Google Scholar] [CrossRef]
- Wnuk, A.; Rzemieniec, J.; Przepiórska, K.; Wesołowska, J.; Wójtowicz, A.K.; Kajta, M. Autophagy-related neurotoxicity is mediated via AHR and CAR in mouse neurons exposed to DDE. Sci. Total Environ. 2020, 742, 140599. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Wnuk, A.; Rzemieniec, J.; Litwa, E.; Lasoń, W.; Kajta, M. Prenatal exposure to benzophenone-3 (BP-3) induces apoptosis, disrupts estrogen receptor expression and alters the epigenetic status of mouse neurons. J. Steroid Biochem. Mol. Biol. 2018, 182, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Eads, C.A.; Danenberg, K.D.; Kawakami, K.; Saltz, L.B.; Blake, C.; Shibata, D.; Danenberg, P.V.; Laird, P.W. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000, 28, E32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wnuk, A.; Rzemieniec, J.; Lasoń, W.; Krzeptowski, W.; Kajta, M. Apoptosis Induced by the UV Filter Benzophenone-3 in Mouse Neuronal Cells Is Mediated via Attenuation of Erα/Pparγ and Stimulation of Erβ/Gpr30 Signaling. Mol. Neurobiol. 2018, 55, 2362–2383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rzemieniec, J.; Wnuk, A.; Lasoń, W.; Bilecki, W.; Kajta, M. The neuroprotective action of 3,3′-diindolylmethane against ischemia involves an inhibition of apoptosis and autophagy that depends on HDAC and AhR/CYP1A1 but not ERα/CYP19A1 signaling. Apoptosis 2019, 24, 435–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wnuk, A.; Przepiórska, K.; Rzemieniec, J.; Pietrzak, B.; Kajta, M. Selective Targeting of Non-nuclear Estrogen Receptors with PaPE-1 as a New Treatment Strategy for Alzheimer’s Disease. Neurotox. Res. 2020, 38, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Deli, M.A.; Nakao, S.; Honda, M.; Hayashi, K.; Nakaoke, R.; Kataoka, Y.; Niwa, M. Pericytes from brain microvessels strengthen the barrier integrity in primary cultures of rat brain endothelial cells. Cell. Mol. Neurobiol. 2007, 27, 687–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, S.; Deli, M.A.; Kawaguchi, H.; Shimizudani, T.; Shimono, T.; Kittel, A.; Tanaka, K.; Niwa, M. A new blood-brain barrier model using primary rat brain endothelial cells, pericytes and astrocytes. Neurochem. Int. 2009, 54, 253–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wnuk, A.; Rzemieniec, J.; Przepiórska, K.; Pietrzak, B.A.; Maćkowiak, M.; Kajta, M. Prenatal Exposure to Triclocarban Impairs ESR1 Signaling and Disrupts Epigenetic Status in Sex-Specific Ways as Well as Dysregulates the Expression of Neurogenesis- and Neurotransmitter-Related Genes in the Postnatal Mouse Brain. Int. J. Mol. Sci. 2021, 22, 13121. https://doi.org/10.3390/ijms222313121
Wnuk A, Rzemieniec J, Przepiórska K, Pietrzak BA, Maćkowiak M, Kajta M. Prenatal Exposure to Triclocarban Impairs ESR1 Signaling and Disrupts Epigenetic Status in Sex-Specific Ways as Well as Dysregulates the Expression of Neurogenesis- and Neurotransmitter-Related Genes in the Postnatal Mouse Brain. International Journal of Molecular Sciences. 2021; 22(23):13121. https://doi.org/10.3390/ijms222313121
Chicago/Turabian StyleWnuk, Agnieszka, Joanna Rzemieniec, Karolina Przepiórska, Bernadeta Angelika Pietrzak, Marzena Maćkowiak, and Małgorzata Kajta. 2021. "Prenatal Exposure to Triclocarban Impairs ESR1 Signaling and Disrupts Epigenetic Status in Sex-Specific Ways as Well as Dysregulates the Expression of Neurogenesis- and Neurotransmitter-Related Genes in the Postnatal Mouse Brain" International Journal of Molecular Sciences 22, no. 23: 13121. https://doi.org/10.3390/ijms222313121
APA StyleWnuk, A., Rzemieniec, J., Przepiórska, K., Pietrzak, B. A., Maćkowiak, M., & Kajta, M. (2021). Prenatal Exposure to Triclocarban Impairs ESR1 Signaling and Disrupts Epigenetic Status in Sex-Specific Ways as Well as Dysregulates the Expression of Neurogenesis- and Neurotransmitter-Related Genes in the Postnatal Mouse Brain. International Journal of Molecular Sciences, 22(23), 13121. https://doi.org/10.3390/ijms222313121