Protocadherin-7 Regulates Osteoclast Differentiation through Intracellular SET-Binding Domain-Mediated RhoA and Rac1 Activation
Abstract
1. Introduction
2. Results
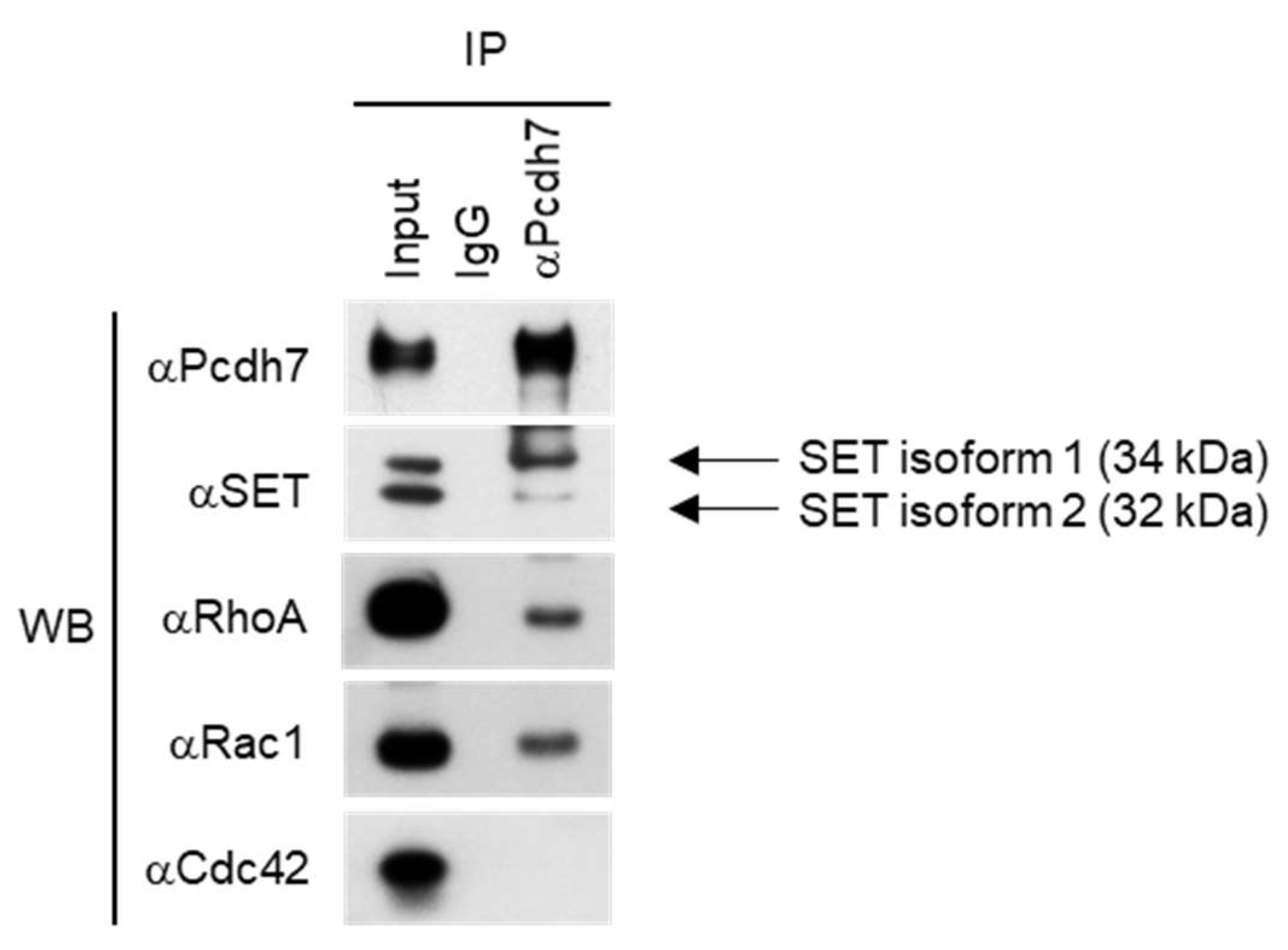
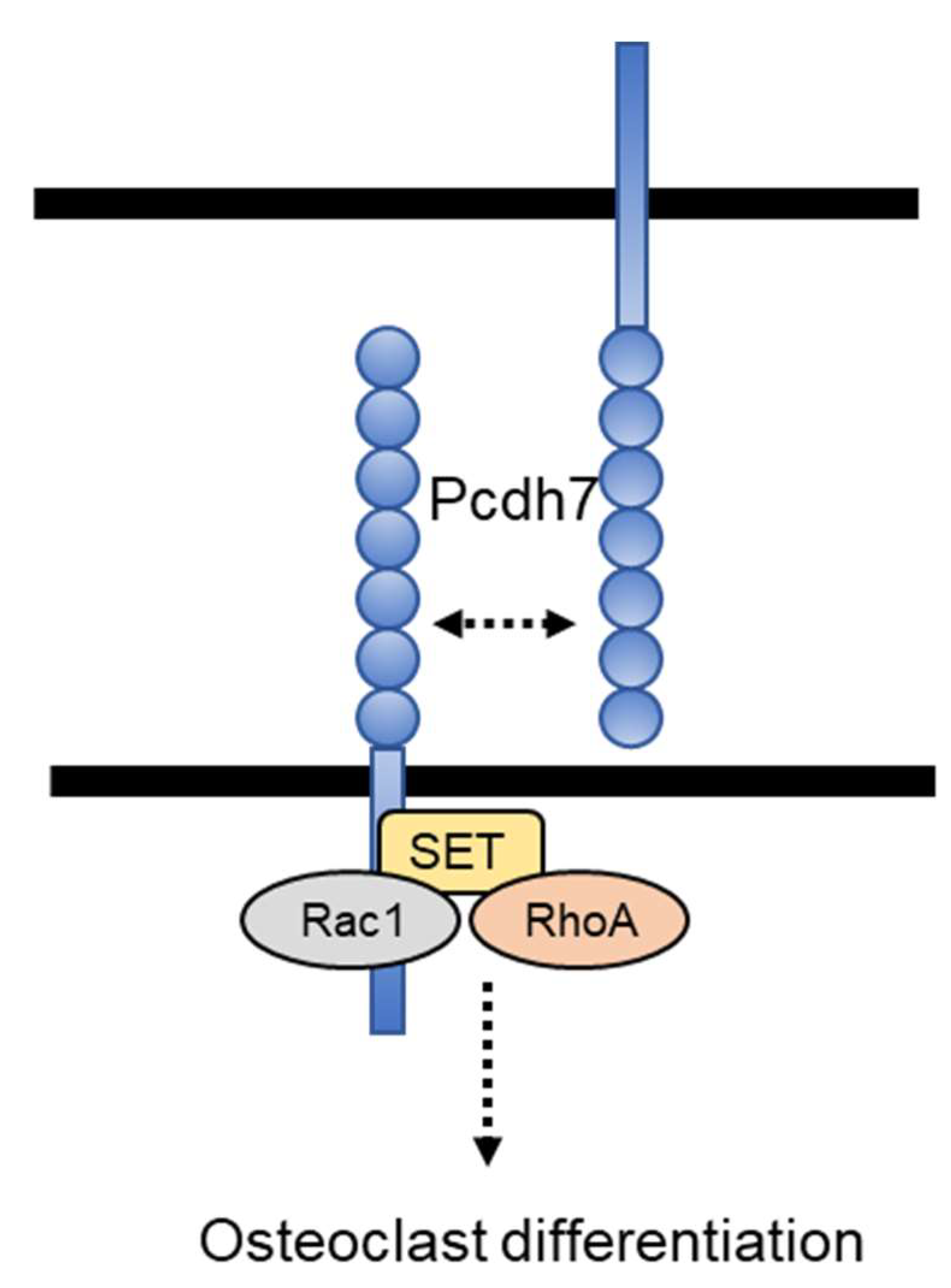
2.1. Pcdh7 Associates with SET and Small GTPases RhoA and Rac1 in Osteoclasts
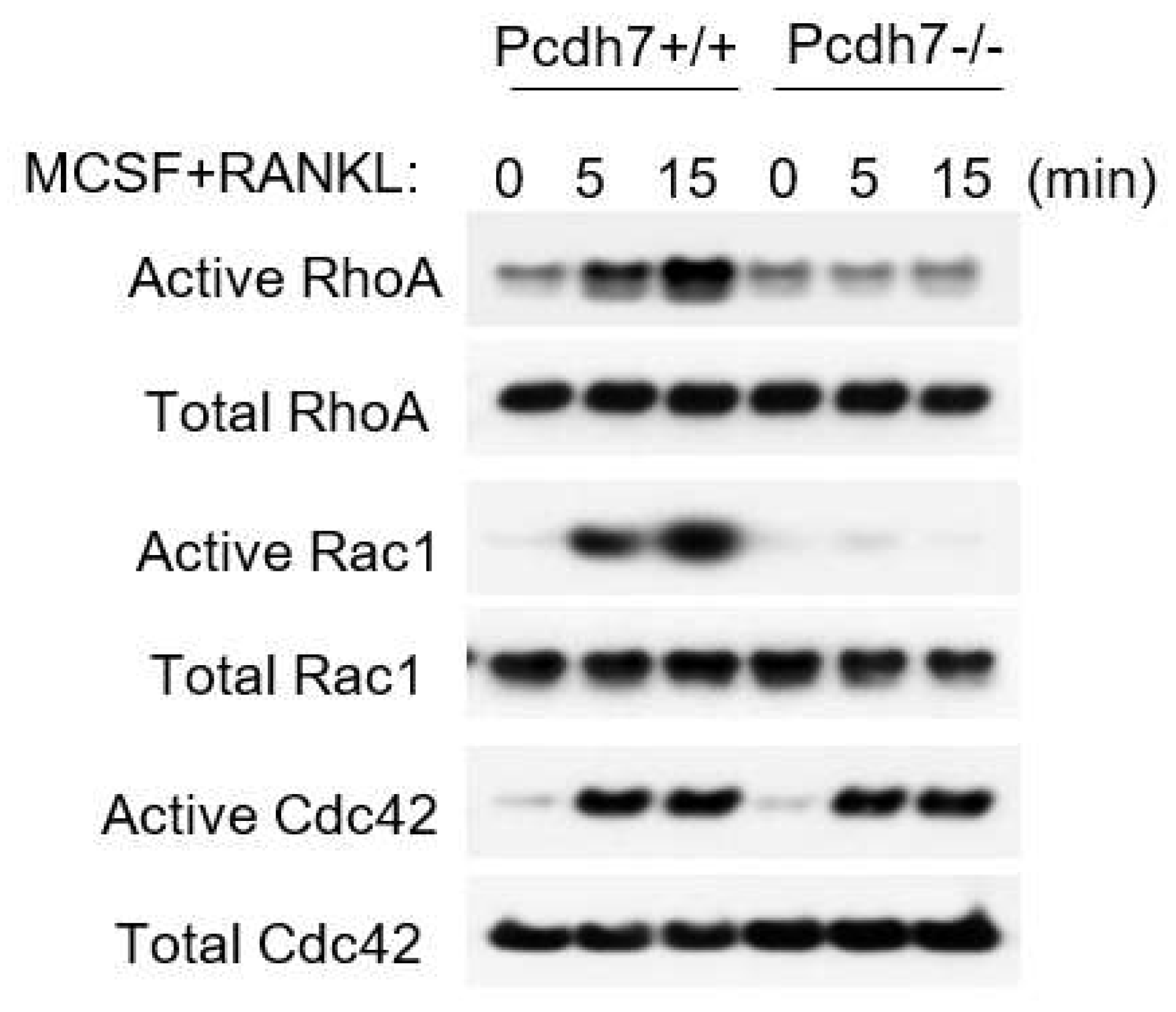
2.2. Pcdh7 Regulates Activation of Small GTPases RhoA and Rac1
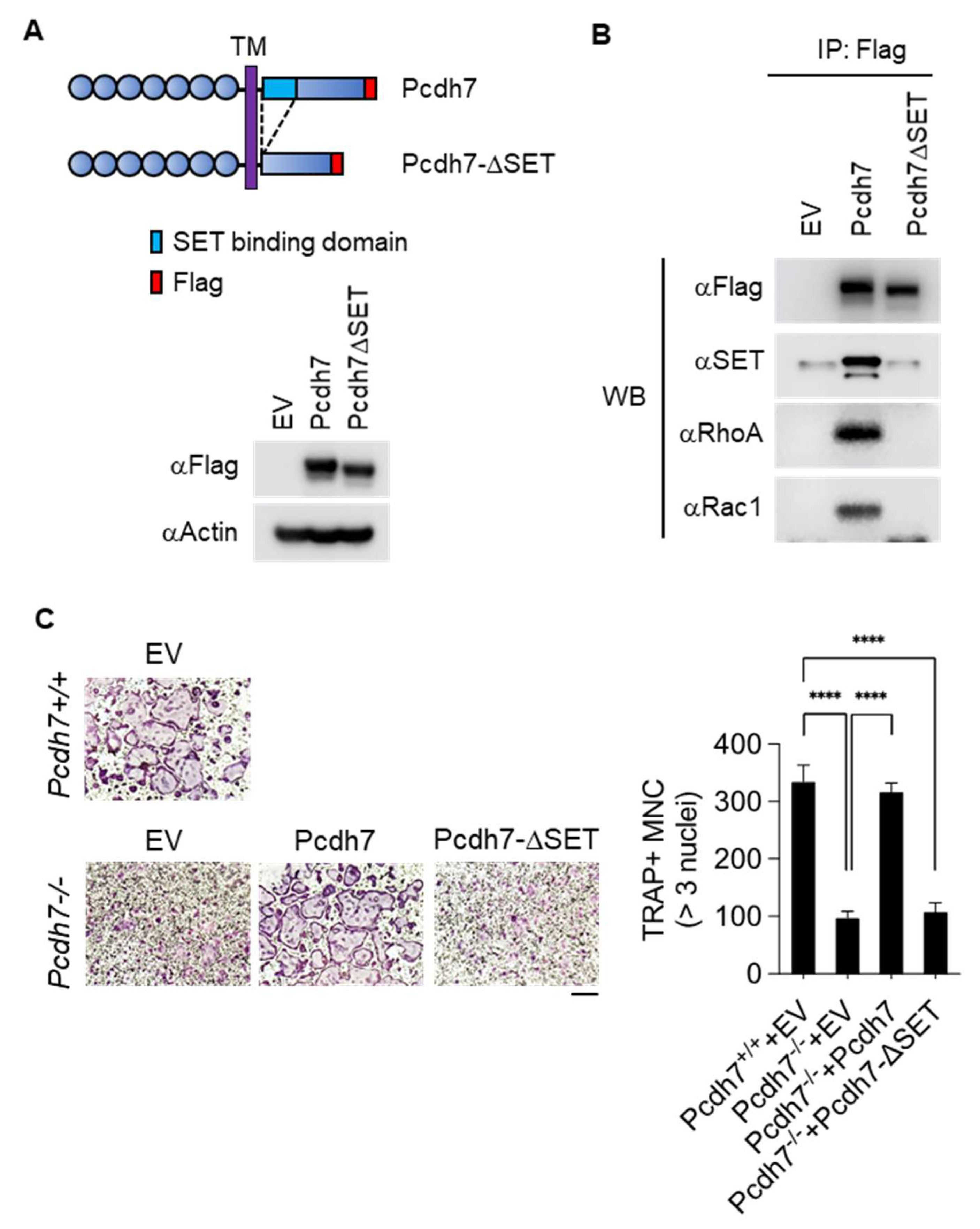
2.3. SET-Binding Domain in Pcdh7 Intracellular Region Is Required for Pcdh7-Mediated Regulation of Osteoclast Differentiation
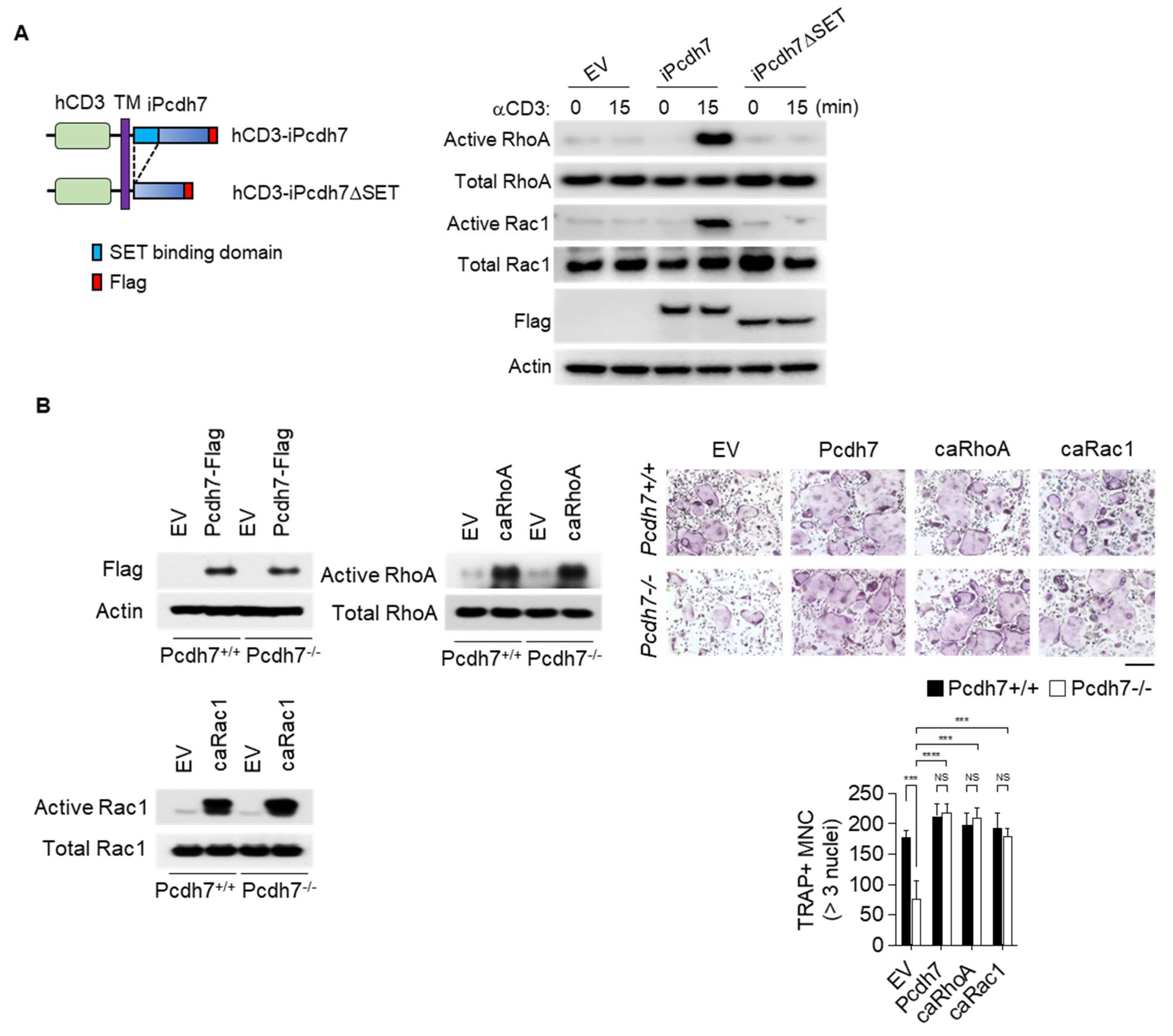
2.4. Crosslink of Pcdh7 Intracellular Region Is Critical for Activation of RhoA and Rac1, and Formation of Osteoclasts
3. Discussion
4. Materials and Methods
4.1. In Vitro Cell Culture, Osteoclast Differentiation, and Tartrate-Resistant Acid Phosphatase (TRAP) Staining
4.2. Coimmunoprecipitation, Pull-Down Assay, and Western Blotting
4.3. Generation of Constructs
4.4. Retrovirus Preparation and Transduction
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Hayashi, S.; Takeichi, M. Emerging roles of protocadherins: From self-avoidance to enhancement of motility. J. Cell Sci. 2015, 128, 1455–1464. [Google Scholar] [CrossRef]
- Bradley, R.S.; Espeseth, A.; Kintner, C. NF-protocadherin, a novel member of the cadherin superfamily, is required for Xenopus ectodermal differentiation. Curr. Biol. 1998, 8, 325–334. [Google Scholar] [CrossRef]
- Yoshida, K.; Yoshitomo-Nakagawa, K.; Seki, N.; Sasaki, M.; Sugano, S. Cloning, expression analysis, and chromosomal localization of BH-protocadherin (PCDH7), a novel member of the cadherin superfamily. Genomics 1998, 49, 458–461. [Google Scholar] [CrossRef]
- Peek, S.L.; Mah, K.M.; Weiner, J.A. Regulation of neural circuit formation by protocadherins. Cell. Mol. Life Sci. 2017, 74, 4133–4157. [Google Scholar] [CrossRef]
- Yoshida, K. Fibroblast cell shape and adhesion in vitro is altered by overexpression of the 7a and 7b isoforms of protocadherin 7, but not the 7c isoform. Cell. Mol. Biol. Lett. 2003, 8, 735–741. [Google Scholar] [PubMed]
- Heggem, M.A.; Bradley, R.S. The cytoplasmic domain of Xenopus NF-protocadherin interacts with TAF1/set. Dev. Cell 2003, 4, 419–429. [Google Scholar] [CrossRef]
- Rashid, D.; Newell, K.; Shama, L.; Bradley, R. A requirement for NF-protocadherin and TAF1/Set in cell adhesion and neural tube formation. Dev. Biol. 2006, 291, 170–181. [Google Scholar] [CrossRef]
- Piper, M.; Dwivedy, A.; Leung, L.; Bradley, R.S.; Holt, C.E. NF-protocadherin and TAF1 regulate retinal axon initiation and elongation in vivo. J. Neurosci. 2008, 28, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Leung, L.C.; Urbancic, V.; Baudet, M.L.; Dwivedy, A.; Bayley, T.G.; Lee, A.C.; Harris, W.A.; Holt, C.E. Coupling of NF-protocadherin signaling to axon guidance by cue-induced translation. Nat. Neurosci. 2013, 16, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Padanad, M.S.; Evers, B.M.; Smith, B.; Novaresi, N.; Suresh, S.; Richardson, J.A.; Stein, E.; Zhu, J.; Hammer, R.E.; et al. Modulation of Mutant Kras(G12D) -Driven Lung Tumorigenesis In Vivo by Gain or Loss of PCDH7 Function. Mol. Cancer Res. 2019, 17, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Yasuda, S.; Tanaka, H.; Yamagata, K.; Kim, H. Non-clustered protocadherin. Cell Adh. Migr. 2011, 5, 97–105. [Google Scholar] [CrossRef]
- Hertel, N.; Krishna, K.; Nuernberger, M.; Redies, C. A cadherin-based code for the divisions of the mouse basal ganglia. J. Comp. Neurol. 2008, 508, 511–528. [Google Scholar] [CrossRef]
- Yoshida, K.; Watanabe, M.; Kato, H.; Dutta, A.; Sugano, S. BH-protocadherin-c, a member of the cadherin superfamily, interacts with protein phosphatase 1 alpha through its intracellular domain. FEBS Lett. 1999, 460, 93–98. [Google Scholar] [CrossRef]
- Zhou, X.; Updegraff, B.L.; Guo, Y.; Peyton, M.; Girard, L.; Larsen, J.E.; Xie, X.J.; Zhou, Y.; Hwang, T.H.; Xie, Y.; et al. PROTOCADHERIN 7 Acts through SET and PP2A to Potentiate MAPK Signaling by EGFR and KRAS during Lung Tumorigenesis. Cancer Res. 2017, 77, 187–197. [Google Scholar] [CrossRef]
- Walsh, M.C.; Kim, N.; Kadono, Y.; Rho, J.; Lee, S.Y.; Lorenzo, J.; Choi, Y. Osteoimmunology: Interplay between the immune system and bone metabolism. Annu. Rev. Immunol. 2006, 24, 33–63. [Google Scholar] [CrossRef] [PubMed]
- Teitelbaum, S.L. Bone resorption by osteoclasts. Science 2000, 289, 1504–1508. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Hayashi, M.; Fukunaga, T.; Kurata, K.; Oh-Hora, M.; Feng, J.Q.; Bonewald, L.F.; Kodama, T.; Wutz, A.; Wagner, E.F.; et al. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat. Med. 2011, 17, 1231–1234. [Google Scholar] [CrossRef]
- Xiong, J.; Onal, M.; Jilka, R.L.; Weinstein, R.S.; Manolagas, S.C.; O'Brien, C.A. Matrix-embedded cells control osteoclast formation. Nat. Med. 2011, 17, 1235–1241. [Google Scholar] [CrossRef]
- Xiong, J.; Piemontese, M.; Thostenson, J.D.; Weinstein, R.S.; Manolagas, S.C.; O’Brien, C.A. Osteocyte-derived RANKL is a critical mediator of the increased bone resorption caused by dietary calcium deficiency. Bone 2014, 66, 146–154. [Google Scholar] [CrossRef]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef]
- Kim, H.; Walsh, M.C.; Takegahara, N.; Middleton, S.A.; Shin, H.I.; Kim, J.; Choi, Y. The purinergic receptor P2X5 regulates inflammasome activity and hyper-multinucleation of murine osteoclasts. Sci. Rep. 2017, 7, 196. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, M.B.; Nakamura, M.C. A Comprehensive Review of Immunoreceptor Regulation of Osteoclasts. Clin. Rev. Allergy Immunol. 2016, 51, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Cavallaro, U.; Christofori, G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat. Rev. Cancer 2004, 4, 118–132. [Google Scholar] [CrossRef] [PubMed]
- Volkmer, H.; Schreiber, J.; Rathjen, F.G. Regulation of adhesion by flexible ectodomains of IgCAMs. Neurochem. Res. 2013, 38, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Croke, M.; Ross, F.P.; Korhonen, M.; Williams, D.A.; Zou, W.; Teitelbaum, S.L. Rac deletion in osteoclasts causes severe osteopetrosis. J. Cell Sci. 2011, 124, 3811–3821. [Google Scholar] [CrossRef]
- Razzouk, S.; Lieberherr, M.; Cournot, G. Rac-GTPase, osteoclast cytoskeleton and bone resorption. Eur. J. Cell Biol. 1999, 78, 249–255. [Google Scholar] [CrossRef]
- Kim, H.; Takegahara, N.; Walsh, M.C.; Ueda, J.; Fujihara, Y.; Ikawa, M.; Choi, Y. Protocadherin-7 contributes to maintenance of bone homeostasis through regulation of osteoclast multinucleation. BMB Rep. 2020, 53, 472–477. [Google Scholar] [CrossRef]
- Nakamura, H.; Nakashima, T.; Hayashi, M.; Izawa, N.; Yasui, T.; Aburatani, H.; Tanaka, S.; Takayanagi, H. Global epigenomic analysis indicates protocadherin-7 activates osteoclastogenesis by promoting cell-cell fusion. Biochem. Biophys. Res. Commun. 2014, 455, 305–311. [Google Scholar] [CrossRef]
- Yin, L.; Zeng, Y.; Xiao, Y.; Chen, Y.; Shen, H.; Dong, J. Cyclin-dependent kinase 1-mediated phosphorylation of SET at serine 7 is essential for its oncogenic activity. Cell Death Dis. 2019, 10, 385. [Google Scholar] [CrossRef]
- Adachi, Y.; Pavlakis, G.N.; Copeland, T.D. Identification of in vivo phosphorylation sites of SET, a nuclear phosphoprotein encoded by the translocation breakpoint in acute undifferentiated leukemia. FEBS Lett. 1994, 340, 231–235. [Google Scholar] [CrossRef]
- ten Klooster, J.P.; Leeuwen, I.; Scheres, N.; Anthony, E.C.; Hordijk, P.L. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J. 2007, 26, 336–345. [Google Scholar] [CrossRef]
- Zhu, M.; Sun, B.H.; Saar, K.; Simpson, C.; Troiano, N.; Dallas, S.L.; Tiede-Lewis, L.M.; Nevius, E.; Pereira, J.P.; Weinstein, R.S.; et al. Deletion of Rac in Mature Osteoclasts Causes Osteopetrosis, an Age-Dependent Change in Osteoclast Number, and a Reduced Number of Osteoblasts In Vivo. J. Bone Miner. Res. 2016, 31, 864–873. [Google Scholar] [CrossRef]
- Strzelecka-Kiliszek, A.; Mebarek, S.; Roszkowska, M.; Buchet, R.; Magne, D.; Pikula, S. Functions of Rho family of small GTPases and Rho-associated coiled-coil kinases in bone cells during differentiation and mineralization. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Chellaiah, M.A.; Soga, N.; Swanson, S.; McAllister, S.; Alvarez, U.; Wang, D.; Dowdy, S.F.; Hruska, K.A. Rho-A is critical for osteoclast podosome organization, motility, and bone resorption. J. Biol. Chem. 2000, 275, 11993–12002. [Google Scholar] [CrossRef] [PubMed]
- Ory, S.; Munari-Silem, Y.; Fort, P.; Jurdic, P. Rho and Rac exert antagonistic functions on spreading of macrophage-derived multinucleated cells and are not required for actin fiber formation. J. Cell Sci. 2000, 113 Pt 7, 1177–1188. [Google Scholar] [CrossRef] [PubMed]
- Bokoch, G.M. Regulation of innate immunity by Rho GTPases. Trends. Cell Biol. 2005, 15, 163–171. [Google Scholar] [CrossRef]
- Lacy, P.; Mahmudi-Azer, S.; Bablitz, B.; Gilchrist, M.; Fitzharris, P.; Cheng, D.; Man, S.F.; Bokoch, G.M.; Moqbel, R. Expression and translocation of Rac2 in eosinophils during superoxide generation. Immunology 1999, 98, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Carnevale, K.A.; Cathcart, M.K. Human monocytes use Rac1, not Rac2, in the NADPH oxidase complex. J. Biol. Chem. 2003, 278, 40788–40792. [Google Scholar] [CrossRef]
- Kwong, C.H.; Adams, A.G.; Leto, T.L. Characterization of the effector-specifying domain of Rac involved in NADPH oxidase activation. J. Biol. Chem. 1995, 270, 19868–19872. [Google Scholar] [CrossRef]
- Lee, N.K.; Choi, Y.G.; Baik, J.Y.; Han, S.Y.; Jeong, D.W.; Bae, Y.S.; Kim, N.; Lee, S.Y. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood 2005, 106, 852–859. [Google Scholar] [CrossRef]
- Pan, W.; Nagpal, K.; Suarez-Fueyo, A.; Ferretti, A.; Yoshida, N.; Tsokos, M.G.; Tsokos, G.C. The Regulatory Subunit PPP2R2A of PP2A Enhances Th1 and Th17 Differentiation through Activation of the GEF-H1/RhoA/ROCK Signaling Pathway. J. Immunol. 2021, 206, 1719–1728. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, E.; Calvayrac, O.; Mazieres, J.; Lajoie-Mazenc, I.; Boubekeur, N.; Favre, G.; Pradines, A. RhoB loss induces Rac1-dependent mesenchymal cell invasion in lung cells through PP2A inhibition. Oncogene 2016, 35, 1760–1769. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Takegahara, N.; Walsh, M.C.; Middleton, S.A.; Yu, J.; Shirakawa, J.; Ueda, J.; Fujihara, Y.; Ikawa, M.; Ishii, M.; et al. IgSF11 regulates osteoclast differentiation through association with the scaffold protein PSD-95. Bone Res. 2020, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Walsh, M.C.; Yu, J.; Laskoski, P.; Takigawa, K.; Takegahara, N.; Choi, Y. Methylosome protein 50 associates with the purinergic receptor P2X5 and is involved in osteoclast maturation. FEBS Lett. 2020, 594, 144–152. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Takegahara, N.; Choi, Y. Protocadherin-7 Regulates Osteoclast Differentiation through Intracellular SET-Binding Domain-Mediated RhoA and Rac1 Activation. Int. J. Mol. Sci. 2021, 22, 13117. https://doi.org/10.3390/ijms222313117
Kim H, Takegahara N, Choi Y. Protocadherin-7 Regulates Osteoclast Differentiation through Intracellular SET-Binding Domain-Mediated RhoA and Rac1 Activation. International Journal of Molecular Sciences. 2021; 22(23):13117. https://doi.org/10.3390/ijms222313117
Chicago/Turabian StyleKim, Hyunsoo, Noriko Takegahara, and Yongwon Choi. 2021. "Protocadherin-7 Regulates Osteoclast Differentiation through Intracellular SET-Binding Domain-Mediated RhoA and Rac1 Activation" International Journal of Molecular Sciences 22, no. 23: 13117. https://doi.org/10.3390/ijms222313117
APA StyleKim, H., Takegahara, N., & Choi, Y. (2021). Protocadherin-7 Regulates Osteoclast Differentiation through Intracellular SET-Binding Domain-Mediated RhoA and Rac1 Activation. International Journal of Molecular Sciences, 22(23), 13117. https://doi.org/10.3390/ijms222313117






