Cytokinin Perception in Ancient Plants beyond Angiospermae
Abstract
1. Introduction
2. Results
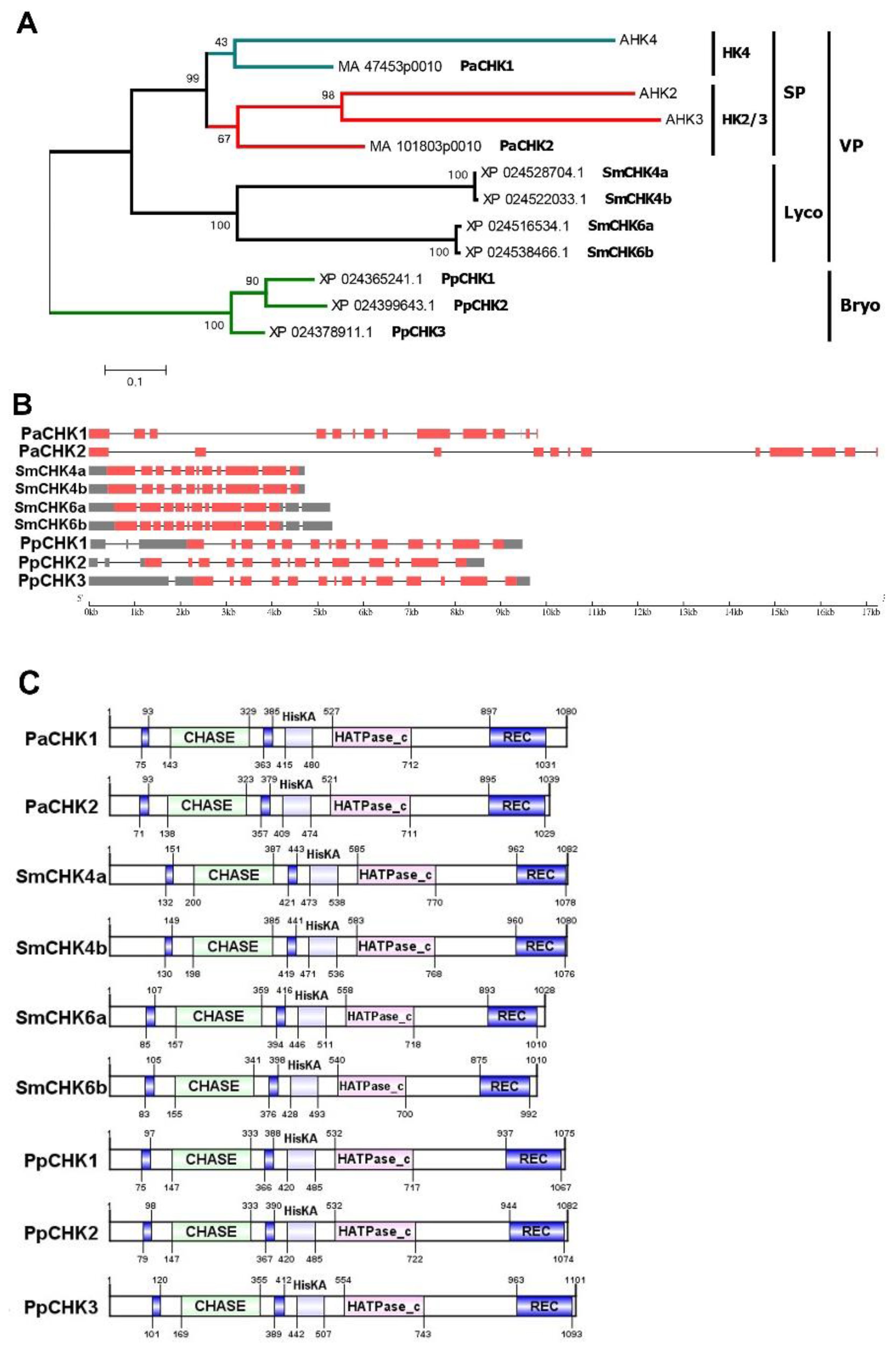
2.1. Ancient CHK Receptors in Comparative and Evolutionary Aspects
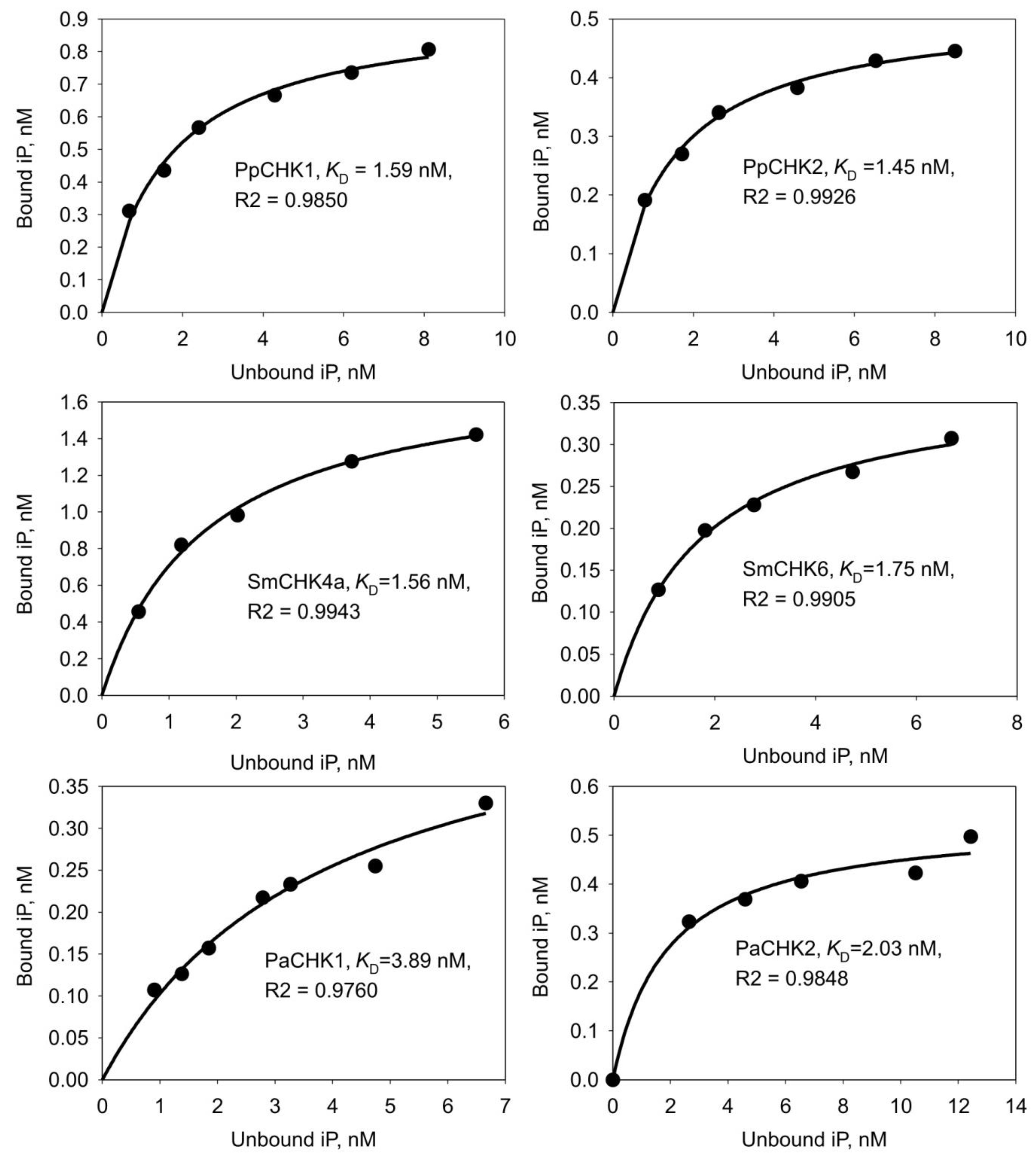
2.2. Ligand Binding Properties of CHK Receptors from Early-Divergent Plants
3. Discussion
3.1. Lessons from Experimental Studies of CHK Receptors from Early-Divergent Lineages
3.1.1. Selection of Representative Receptors
3.1.2. The pH Dependence of Ligand Binding
3.1.3. Ligand Specificity of CHK Receptors
3.2. Non-CK-Binding CHKs Can Participate in CK Signaling
3.3. Probable Scenario for the Evolution of the CK System
4. Materials and Methods
4.1. Bioinformatics Methods
4.2. RNA Isolation from Picea abies Fir-Needles
4.3. DNA Constructs
4.3.1. Cloning Receptor cDNA
4.3.2. Receptors Expression and CK Binding Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. A New Approach for KD Determination in Competition Experiments
References
- Heyl, A.; Riefler, M.; Romanov, G.A.; Schmülling, T. Properties, functions and evolution of cytokinin receptors. Eur. J. Cell. Biol. 2012, 91, 246–256. [Google Scholar] [CrossRef]
- Lomin, S.N.; Krivosheev, D.M.; Steklov, M.Y.; Osolodkin, D.I.; Romanov, G.A. Receptor properties and features of cytokinin signaling. Acta Nat. 2012, 4, 31–45. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Kojima, M.; Yamaya, T.; Sakakibara, H. Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiol. 2004, 134, 1654–1661. [Google Scholar] [CrossRef]
- Lomin, S.N.; Yonekura-Sakakibara, K.; Romanov, G.A.; Sakakibara, H. Ligand-binding properties and subcellular localization of maize cytokinin receptors. J. Exp. Bot. 2011, 62, 5149–5159. [Google Scholar] [CrossRef]
- Lomin, S.N.; Krivosheev, D.M.; Steklov, M.Y.; Arkhipov, D.V.; Osolodkin, D.I.; Schmülling, T.; Romanov, G.A. Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J. Exp. Bot. 2015, 66, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Muszynski, M.G.; Moss-Taylor, L.; Chudalayandi, S.; Cahill, J.; Del Valle-Echevarria, A.R.; Alvarez-Castro, I.; Petefish, A.; Sakakibara, H.; Krivosheev, D.M.; Lomin, S.N.; et al. The maize Hairy Sheath Frayed1 (Hsf1) mutation alters leaf patterning through increased cytokinin signaling. Plant Cell 2020, 32, 1501–1518. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Lee, J.; Kim, K.; Cho, M.; Ryu, H.; An, G.; Hwang, I. Functional identification of OsHk6 as a homotypic cytokinin receptor in rice with preferential affinity for iP. Plant Cell Physiol. 2012, 53, 1334–1343. [Google Scholar] [CrossRef]
- Ding, W.; Tong, H.; Zheng, W.; Ye, J.; Pan, Z.; Zhang, B.; Zhu, S. Isolation, characterization and transcriptome analysis of a cytokinin receptor mutant Osckt1 in rice. Front. Plant. Sci. 2017, 8, 88. [Google Scholar] [CrossRef]
- Laffont, C.; Rey, T.; André, O.; Novero, M.; Kazmierczak, T.; Debellé, F.; Bonfante, P.; Jacquet, C.; Frugier, F. The CRE1 cytokinin pathway is differentially recruited depending on Medicago truncatula root environments and negatively regulates resistance to a pathogen. PLoS ONE 2015, 10, e0116819. [Google Scholar] [CrossRef]
- Boivin, S.; Kazmierczak, T.; Brault, M.; Wen, J.; Gamas, P.; Mysore, K.S.; Frugier, F. Different cytokinin histidine kinase receptors regulate nodule initiation as well as later nodule developmental stages in Medicago truncatula. Plant Cell Environ. 2016, 39, 2198–2209. [Google Scholar] [CrossRef]
- Kuderová, A.; Gallová, L.; Kuricová, K.; Nejedlá, E.; Čurdová, A.; Micenková, L.; Plíhal, O.; Šmajs, D.; Spíchal, L.; Hejátko, J. Identification of AHK2- and AHK3-like cytokinin receptors in Brassica napus reveals two subfamilies of AHK2 orthologues. J. Exp. Bot. 2015, 66, 339–353. [Google Scholar] [CrossRef]
- Daudu, D.; Allion, E.; Liesecke, F.; Papon, N.; Courdavault, V.; Dugé de Bernonville, T.; Mélin, C.; Oudin, A.; Clastre, M.; Lanoue, A.; et al. CHASE-Containing histidine kinase receptors in apple tree: From a common receptor structure to divergent cytokinin binding properties and specific functions. Front. Plant. Sci. 2017, 8, 1614. [Google Scholar] [CrossRef]
- Lomin, S.N.; Myakushina, Y.A.; Kolachevskaya, O.O.; Getman, I.A.; Arkhipov, D.V.; Savelieva, E.M.; Osolodkin, D.I.; Romanov, G.A. Cytokinin perception in potato: New features of canonical players. J. Exp. Bot. 2018, 69, 3839–3853. [Google Scholar] [CrossRef]
- Lomin, S.N.; Savelieva, E.M.; Arkhipov, D.V.; Romanov, G.A. Evidences for preferential localization of cytokinin receptors of potato in the endoplasmic reticulum. Biochem. Moscow Suppl. Ser. A Membrane Cell Biol. 2020, 14, 146–153. [Google Scholar] [CrossRef]
- Jaworek, P.; Tarkowski, P.; Hluska, T.; Kouřil, Š.; Vrobel, O.; Nisler, J.; Kopečny, D. Characterization of five CHASE-containing histidine kinase receptors from Populus x canadensis cv. Robusta sensing isoprenoid and aromatic cytokinins. Planta 2020, 251, 1. [Google Scholar] [CrossRef]
- Héricourt, F.; Larcher, M.; Chefdor, F.; Koudounas, K.; Carqueijeiro, I.; Cruz, P.L.; Courdavault, V.; Tanigawa, M.; Maeda, T.; Depierreux, C.; et al. New insight into HPts as hubs in poplar cytokinin and osmosensing multistep phosphorelays: Cytokinin pathway uses specific HPts. Plants 2019, 8, 591. [Google Scholar] [CrossRef] [PubMed]
- von Schwartzenberg, K.; Núñez, M.F.; Blaschke, H.; Dobrev, P.I.; Novák, O.; Motyka, V.; Strnad, M. Cytokinins in the bryophyte Physcomitrella patens: Analyses of activity, distribution, and cytokinin oxidase/dehydrogenase overexpression reveal the role of extracellular cytokinins. Plant Physiol. 2007, 145, 786–800. [Google Scholar] [CrossRef] [PubMed]
- von Schwartzenberg, K.; Lindner, A.C.; Gruhn, N.; Šimura, J.; Novák, O.; Strnad, M.; Gonneau, M.; Nogué, F.; Heyl, A. CHASE domain-containing receptors play an essential role in the cytokinin response of the moss Physcomitrella patens. J. Exp. Bot. 2016, 67, 667–679. [Google Scholar] [CrossRef]
- Kaltenegger, E.; Leng, S.; Heyl, A. The effects of repeated whole genome duplication events on the evolution of cytokinin signaling pathway. BMC Evol. Biol. 2018, 18, 76. [Google Scholar] [CrossRef]
- Gruhn, N.; Halawa, M.; Snel, B.; Seidl, M.F.; Heyl, A. A subfamily of putative cytokinin receptors is revealed by an analysis of the evolution of the two-component signaling system of plants. Plant Physiol. 2014, 165, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Aki, S.S.; Mikami, T.; Naramoto, S.; Nishihama, R.; Ishizaki, K.; Kojima, M.; Takebayashi, Y.; Sakakibara, H.; Kyozuka, J.; Kohchi, T.; et al. Cytokinin signaling is essential for organ formation in Marchantia polymorpha. Plant Cell Physiol. 2019, 60, 1842–1854. [Google Scholar] [CrossRef] [PubMed]
- Kakimoto, T. Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 2003, 54, 605–627. [Google Scholar] [CrossRef] [PubMed]
- Heyl, A.; Schmülling, T. Cytokinin signal perception and transduction. Curr. Opin. Plant. Biol. 2003, 6, 480–488. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Schaller, G.E. Cytokinins. Arab. Book 2014, 12, e0168. [Google Scholar] [CrossRef]
- Spíchal, L.; Rakova, N.Y.; Riefler, M.; Mizuno, T.; Romanov, G.A.; Strnad, M.; Schmülling, T. Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol. 2004, 45, 1299–1305. [Google Scholar] [CrossRef]
- Romanov, G.A.; Lomin, S.N.; Schmülling, T. Biochemical characteristics and ligand-properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J. Exp. Bot. 2006, 57, 4051–4058. [Google Scholar] [CrossRef]
- Riefler, M.; Novak, O.; Strnad, M.; Schmülling, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Mähönen, A.P.; Higuchi, M.; Törmäkangas, K.; Miyawaki, K.; Pischke, M.S.; Sussman, M.R.; Helariutta, Y.; Kakimoto, T. Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr. Biol. 2006, 16, 1116–1122. [Google Scholar] [CrossRef]
- Romanov, G.A. How do cytokinins affect the cell? Russ. J. Plant. Physiol. 2009, 56, 268–290. [Google Scholar] [CrossRef]
- Stolz, A.; Riefler, M.; Lomin, S.N.; Achazi, K.; Romanov, G.A.; Schmülling, T. The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J. 2011, 67, 157–168. [Google Scholar] [CrossRef]
- Hothorn, M.; Dabi, T.; Chory, J. Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase AHK4. Nat. Chem. Biol. 2011, 7, 766–768. [Google Scholar] [CrossRef] [PubMed]
- Ueguchi, C.; Sato, S.; Kato, T.; Tabata, S. The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, M.; Pischke, M.S.; Mähönen, A.P.; Miyawaki, K.; Hashimoto, Y.; Seki, M.; Kobayashi, M.; Shinozaki, K.; Kato, T.; Tabata, S.; et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 2004, 101, 8821–8826. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto-Kitano, M.; Kusumoto, T.; Tarkowski, P.; Kinoshita-Tsujimura, K.; Václavíková, K.; Miyawaki, K.; Kakimoto, T. Cytokinins are central regulators of cambial activity. Proc. Natl. Acad. Sci. USA 2008, 105, 20027–20031. [Google Scholar] [CrossRef] [PubMed]
- Hirose, N.; Takei, K.; Kuroha, T.; Kamada-Nobusada, T.; Hayashi, H.; Sakakibara, H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 2008, 59, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Yamaya, T.; Sakakibara, H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J. Biol. Chem. 2004, 279, 41866–41872. [Google Scholar] [CrossRef]
- Kiba, T.; Takei, K.; Kojima, M.; Sakakibara, H. Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell. 2013, 27, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Bartrina, I.; Schmülling, T. Cytokinin regulates vegetative phase change in Arabidopsis thaliana through the miR172/TOE1-TOE2 module. Nat. Commun. 2021, 12, 5816. [Google Scholar] [CrossRef]
- Bishopp, A.; Help, H.; El-Showk, S.; Weijers, D.; Scheres, B.; Friml, J.; Benková, E.; Mähönen, A.P.; Helariutta, Y. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 2011, 21, 917–926. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Y.; Li, S.S.; Han, G.Z. Insights into the origin and evolution of the plant hormone signaling machinery. Plant. Physiol. 2015, 167, 872–886. [Google Scholar] [CrossRef] [PubMed]
- Lindner, A.C.; Lang, D.; Seifert, M.; Podlešáková, K.; Novák, O.; Strnad, M.; Reski, R.; von Schwartzenberg, K. Isopentenyltransferase-1 (IPT1) knockout in Physcomitrella together with phylogenetic analyses of IPTs provide insights into evolution of plant cytokinin biosynthesis. J. Exp. Bot. 2014, 65, 2533–2543. [Google Scholar] [CrossRef]
- Sanders, H.L.; Langdale, J.A. Conserved transport mechanisms but distinct auxin responses govern shoot patterning in Selaginella kraussiana. New Phytol. 2013, 198, 419–428. [Google Scholar] [CrossRef] [PubMed]
- de Vries, J.; Fischer, A.M.; Roettger, M.; Rommel, S.; Schluepmann, H.; Bräutigam, A.; Carlsbecker, A.; Gould, S.B. Cytokinin-induced promotion of root meristem size in the fern Azolla supports a shoot-like origin of euphyllophyte roots. New Phytol. 2016, 209, 705–720. [Google Scholar] [CrossRef]
- Valdés, A.E.; Fernández, B.; Centeno, M.L. Alterations in endogenous levels of cytokinins following grafting of Pinus radiata support ratio of cytokinins as an index of ageing and vigour. J. Plant. Physiol. 2003, 160, 1407–1410. [Google Scholar] [CrossRef]
- Zhang, H.; Horgan, K.J.; Stewart Reynolds, P.H.; Norris, G.E.; Jameson, P.E. Novel cytokinins: The predominant forms in mature buds of Pinus radiata. Physiol. Plant. 2001, 112, 127–134. [Google Scholar] [CrossRef]
- Montalbán, I.A.; De Diego, N.; Moncaleán, P. Testing novel cytokinins for improved in vitro adventitious shoots formation and subsequent ex vitro performance in Pinus radiata. Forestry 2011, 84, 363–373. [Google Scholar] [CrossRef]
- Strnad, M. The aromatic cytokinins. Physiol. Plant 1997, 101, 674–688. [Google Scholar] [CrossRef]
- Nieminen, K.; Immanen, J.; Laxell, M.; Kauppinen, L.; Tarkowski, P.; Dolezal, K.; Tähtiharju, S.; Elo, A.; Decourteix, M.; Ljung, K.; et al. Cytokinin signaling regulates cambial development in poplar. Proc. Natl. Acad. Sci. USA 2008, 105, 20032–20037. [Google Scholar] [CrossRef] [PubMed]
- De Diego, N.; Montalbán, I.A.; Moncaleán, P. In Vitro regeneration of adult Pinus sylvestris L. trees. S. Afr. J. Bot. 2010, 76, 158–162. [Google Scholar] [CrossRef]
- Pullman, G.S.; Bucalo, K. Pine somatic embryogenesis using zygotic embryos as explants. Methods Mol. Biol. 2011, 710, 267–291. [Google Scholar] [CrossRef]
- Uggla, C.; Mellerowicz, E.J.; Sundberg, B. Indole-3-acetic acid controls cambial growth in scots pine by positional signaling. Plant Physiol. 1998, 117, 113–121. [Google Scholar] [CrossRef]
- Uggla, C.; Magel, E.; Moritz, T.; Sundberg, B. Function and dynamics of auxin and carbohydrates during earlywood/latewood transition in scots pine. Plant Physiol 2001, 125, 2029–2039. [Google Scholar] [CrossRef]
- Hejnowicz, A.; Tomaszewski, M. Growth regulators and wood formation in Pinus silvestris. Physiol. Plant. 1969, 22, 984–992. [Google Scholar] [CrossRef]
- Carvalho, A.; Paiva, J.; Louzada, J.; Lima-Brito, J. The transcriptomics of secondary growth and wood formation in conifers. Mol. Biol. Int. 2013, 2013, 974324. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, F.P.; Rathgeber, C.B.K.; Badel, É.; Fournier, M.; Moulia, B. Modelling the spatial crosstalk between two biochemical signals explains wood formation dynamics and tree-ring structure. J. Exp. Bot. 2021, 72, 1727–1737. [Google Scholar] [CrossRef]
- Steklov, M.Y.; Lomin, S.N.; Osolodkin, D.I.; Romanov, G.A. Structural basis for cytokinin receptor signaling: An evolutionary approach. Plant. Cell. Rep. 2013, 32, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Lomin, S.N.; Myakushina, Y.A.; Arkhipov, D.V.; Leonova, O.G.; Popenko, V.I.; Schmülling, T.; Romanov, G.A. Studies of cytokinin receptor-phosphotransmitter interaction provide evidences for the initiation of cytokinin signalling in the endoplasmic reticulum. Funct. Plant. Biol. 2018, 45, 192–202. [Google Scholar] [CrossRef]
- Anantharaman, V.; Aravind, L. The CHASE domain: A predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends. Biochem. Sci. 2001, 26, 579–582. [Google Scholar] [CrossRef]
- Mougel, C.; Zhulin, I.B. CHASE: An extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends. Biochem. Sci. 2001, 26, 582–584. [Google Scholar] [CrossRef]
- Heyl, A.; Brault, M.; Frugier, F.; Kuderova, A.; Lindner, A.C.; Motyka, V.; Rashotte, A.M.; von Schwartzenberg, K.; Vankova, R.; Schaller, G.E. Nomenclature for members of the two-component signaling pathway of plants. Plant. Physiol. 2013, 161, 1063–1065. [Google Scholar] [CrossRef] [PubMed]
- Pils, B.; Heyl, A. Unraveling the evolution of cytokinin signaling. Plant. Physiol. 2009, 151, 782–791. [Google Scholar] [CrossRef]
- Gruhn, N.; Seidl, M.F.; Halawa, M.; Heyl, A. Members of a recently discovered subfamily of cytokinin receptors display differences and similarities to their classical counterparts. Plant. Signal. Behav. 2015, 10, 2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cheng, Y.C.; Prusoff, W.H. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [CrossRef]
- Spíchal, L. Cytokinins—Recent news and views of evolutionally old molecules. Func. Plant Biol. 2012, 39, 267–284. [Google Scholar] [CrossRef]
- Morris, J.L.; Puttick, M.N.; Clark, J.W.; Edwards, D.; Kenrick, P.; Pressel, S.; Wellman, C.H.; Yang, Z.; Schneider, H.; Donoghue, P.C.J. The timescale of early land plant evolution. Proc. Natl. Acad. Sci. USA 2018, 115, E2274–E2283. [Google Scholar] [CrossRef]
- Felle, H.H. pH regulation in anoxic plants. Ann. Bot. 2005, 96, 519–532. [Google Scholar] [CrossRef]
- Sweeney, E.G.; Henderson, J.N.; Goers, J.; Wreden, C.; Hicks, K.G.; Foster, J.K.; Parthasarathy, R.; Remington, S.J.; Guillemin, K. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure 2012, 20, 1177–1188. [Google Scholar] [CrossRef]
- Dortay, H.; Mehnert, N.; Bürkle, L.; Schmülling, T.; Heyl, A. Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J. 2006, 273, 4631–4644. [Google Scholar] [CrossRef]
- Caesar, K.; Thamm, A.M.; Witthöft, J.; Elgass, K.; Huppenberger, P.; Grefen, C.; Horak, J.; Harter, K. Evidence for the localization of the Arabidopsis cytokinin receptors AHK3 and AHK4 in the endoplasmic reticulum. J. Exp. Bot. 2011, 62, 5571–5580. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.P.; Chickarmane, V.S.; Ohno, C.; Meyerowitz, E.M. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristems. Proc. Nat. Acad. Sci. USA 2009, 160, 16529–16534. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E.; Street, I.H.; Kieber, J.J. Cytokinin and the cell cycle. Curr. Opin. Plant. Biol. 2014, 21, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Kolachevskaya, O.O.; Myakushina, Y.A.; Getman, I.A.; Lomin, S.N.; Deyneko, I.V.; Deigraf, S.V.; Romanov, G.A. Hormonal regulation and crosstalk of auxin/cytokinin signaling pathways in potatoes in vitro and in relation to vegetation or tuberization stages. Int. J. Mol. Sci. 2021, 22, 8207. [Google Scholar] [CrossRef] [PubMed]
- Zdarska, M.; Cuyacot, A.R.; Tarr, P.T.; Yamoune, A.; Szmitkowska, A.; Hrdinová, V.; Gelová, Z.; Meyerowitz, E.M.; Hejátko, J. ETR1 integrates response to ethylene and cytokinins into a single multistep phosphorelay pathway to control root growth. Mol. Plant. 2019, 12, 1338–1352. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F.; Nei, M. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1984, 1, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2014, 47, 5.6.1–5.6.32. [Google Scholar] [CrossRef]
- Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Adiyaman, R.; Maghrabi, A.H.A.; Shuid, A.N.; Brackenridge, D.A.; Nealon, J.O.; Philomina, L.S. IntFOLD: An integrated web resource for high performance protein structure and function prediction. Nucleic Acids Res. 2019, 47, W408–W413. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Kolosova, N.; Miller, B.; Ralph, S.; Ellis, B.E.; Douglas, C.; Ritland, K.; Bohlmann, J. Isolation of high-quality RNA from gymnosperm and angiosperm trees. Nat. Rev. Genet. 2004, 2, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Pashkovskiy, P.P.; Vankova, R.; Zlobin, I.E.; Dobrev, P.; Ivanov, Y.V.; Kartashov, A.V.; Kuznetsov, V.V. Comparative analysis of abscisic acid levels and expression of abscisic acid-related genes in Scots pine and Norway spruce seedlings under water deficit. Plant Physiol. Biochem. 2019, 140, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Sidorov, G.V.; Myasoedov, N.F.; Lomin, S.N.; Romanov, G.A. Synthesis of tritium- and deuterium-labeled isopentenyladenine. Radiochemistry 2015, 57, 108–110. [Google Scholar] [CrossRef]
- Romanov, G.A.; Lomin, S.N. Hormone-binding assay using living bacteria expressing eukaryotic receptors. Methods Mol. Biol. 2009, 495, 111–120. [Google Scholar] [CrossRef] [PubMed]


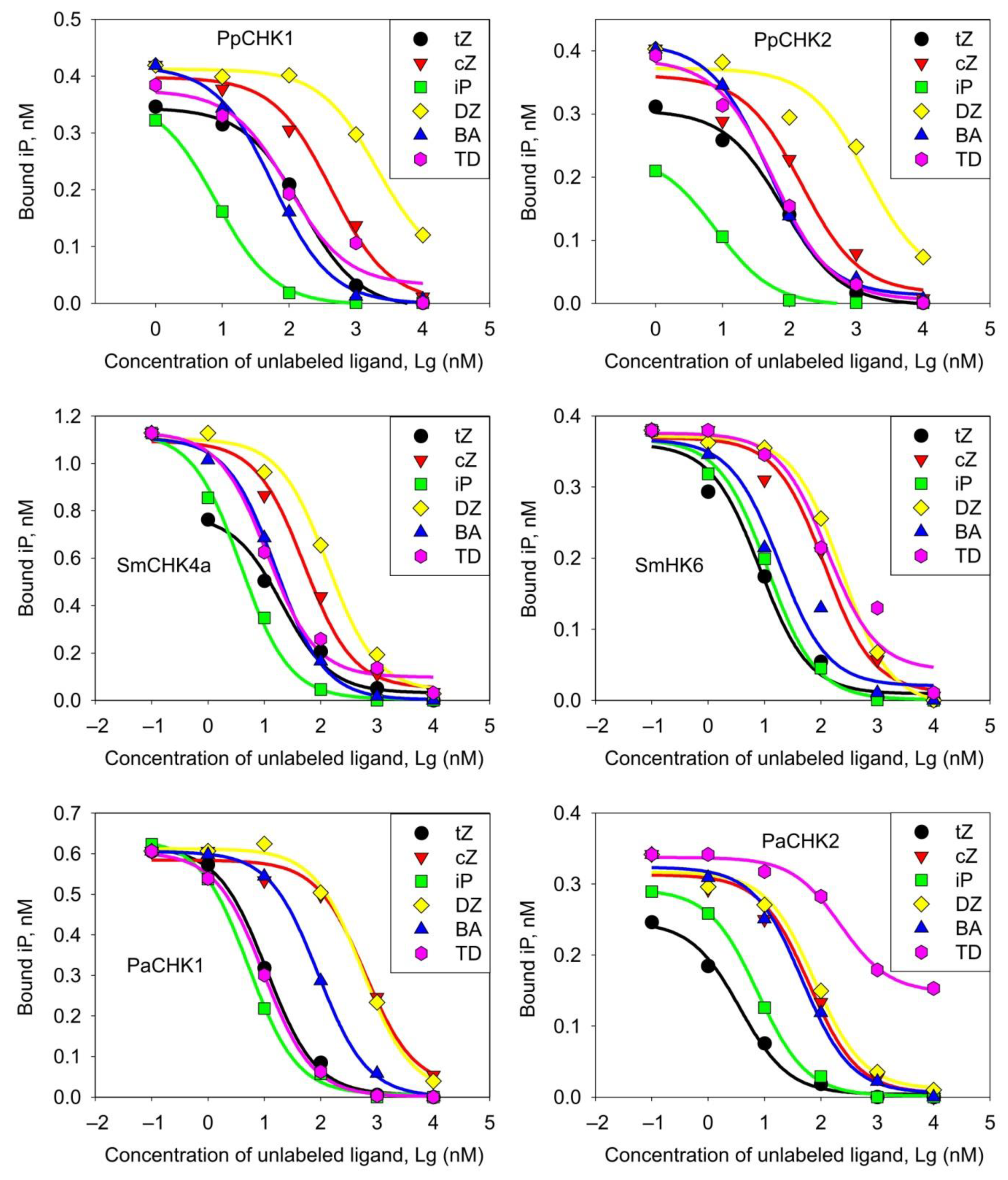
| CK Version | KD of CK-Receptor Complexes (nM ± SE) for: | |||||
|---|---|---|---|---|---|---|
| PpCHK1 | PpCHK2 | SmCHK4a | SmCHK6 | PaCHK1 | PaCHK2 | |
| tZ | 36.9 ± 2 | 18.1 ± 1.5 | 7.99 ± 1.34 | 1.15 ± 0.20 | 7.57 ± 0.37 | 1.05 ± 0.09 |
| cZ | 103 ± 15 | 34.2 ± 9.3 | 20.6 ± 2.6 | 16.6 ± 2.4 | 419 ± 56 | 17.9 ± 2.9 |
| iP | 1.92 ± 0.06 | 1.78 ± 0.14 | 1.60 ± 0.13 | 1.70 ± 0.16 | 3.60 ± 0.17 | 2.20 ± 0.08 |
| DZ | 507 ± 42 | 323 ± 97 | 54.8 ± 6.2 | 36.0 ± 1.6 | 377 ± 27 | 22.4 ± 2.9 |
| BA | 13.9 ± 0.1 | 10.7 ± 0.5 | 6.91 ± 0.34 | 2.28 ± 0.70 | 61.9 ± 1.3 | 13.5 ± 1.6 |
| TD | 23.8 ± 5.4 | 13.02 ± 0.2 | 4.52 ± 0.62 | 16.6 ± 4.7 | 6.70 ± 0.21 | 64.5 ± 6.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lomin, S.N.; Savelieva, E.M.; Arkhipov, D.V.; Pashkovskiy, P.P.; Myakushina, Y.A.; Heyl, A.; Romanov, G.A. Cytokinin Perception in Ancient Plants beyond Angiospermae. Int. J. Mol. Sci. 2021, 22, 13077. https://doi.org/10.3390/ijms222313077
Lomin SN, Savelieva EM, Arkhipov DV, Pashkovskiy PP, Myakushina YA, Heyl A, Romanov GA. Cytokinin Perception in Ancient Plants beyond Angiospermae. International Journal of Molecular Sciences. 2021; 22(23):13077. https://doi.org/10.3390/ijms222313077
Chicago/Turabian StyleLomin, Sergey N., Ekaterina M. Savelieva, Dmitry V. Arkhipov, Pavel P. Pashkovskiy, Yulia A. Myakushina, Alexander Heyl, and Georgy A. Romanov. 2021. "Cytokinin Perception in Ancient Plants beyond Angiospermae" International Journal of Molecular Sciences 22, no. 23: 13077. https://doi.org/10.3390/ijms222313077
APA StyleLomin, S. N., Savelieva, E. M., Arkhipov, D. V., Pashkovskiy, P. P., Myakushina, Y. A., Heyl, A., & Romanov, G. A. (2021). Cytokinin Perception in Ancient Plants beyond Angiospermae. International Journal of Molecular Sciences, 22(23), 13077. https://doi.org/10.3390/ijms222313077







