MicroRNAs Are Involved in Regulating Plant Development and Stress Response through Fine-Tuning of TIR1/AFB-Dependent Auxin Signaling
Abstract
:1. Introduction
2. miRNA-Mediated Regulation of Auxin Signaling and Homeostasis
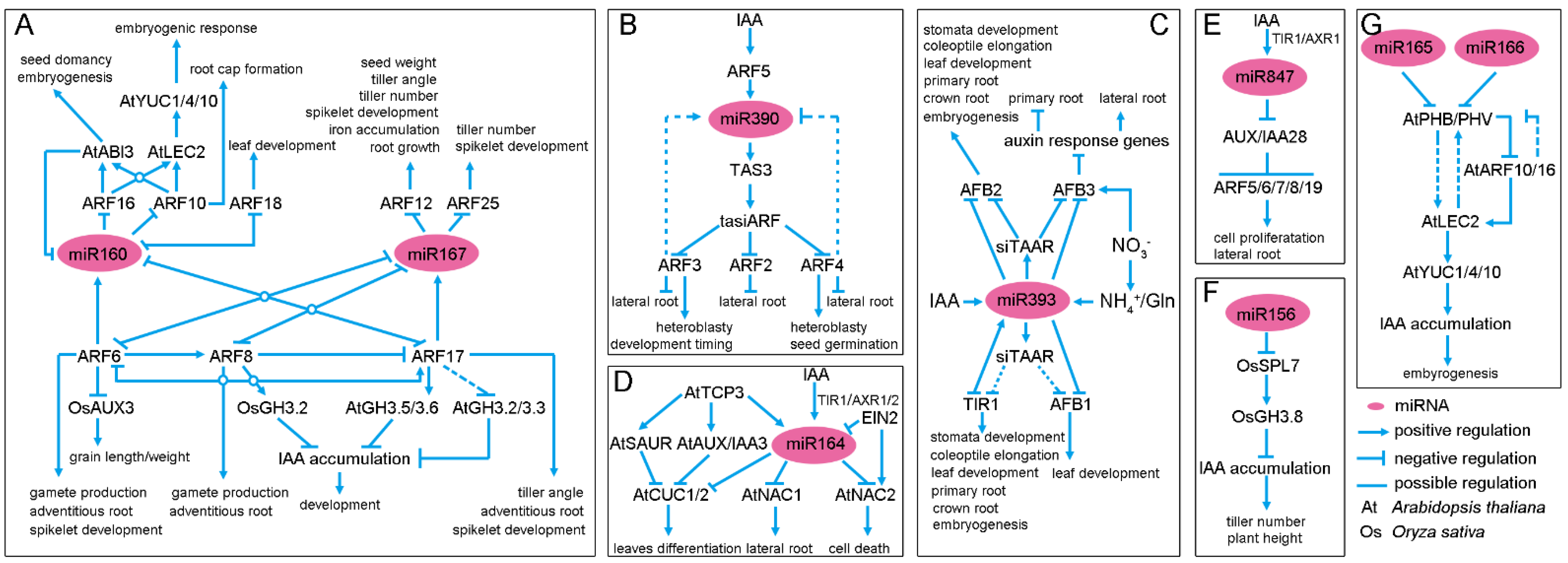
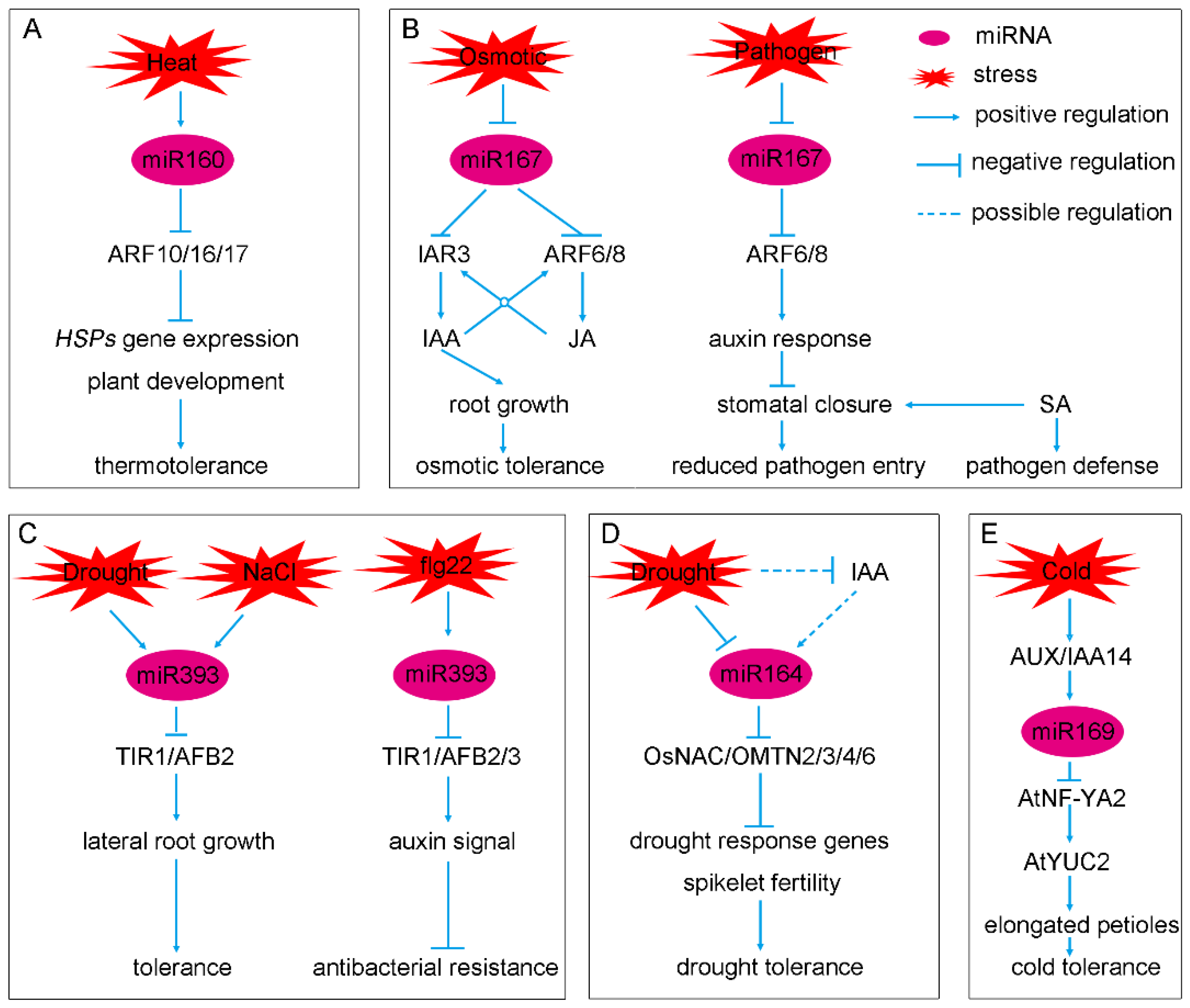
2.1. Two Conserved miRNAs, miR160 and miR167, Constitute a Complex Feedback Loop to Regulate Auxin Signaling and Homeostasis
2.2. The miR390-TAS3-tasiARF Module Is a Regulatory Hub of Auxin Signaling
2.3. miR393, a Repressor of Auxin Signaling
2.4. miR164 and miR169, Two Modulators of Auxin Signaling
2.5. miR847, an Activator of Auxin Signaling
2.6. miR156, miR165 and miR166, a New Mode of Action for Regulators of Auxin Homeostasis and Signaling
2.7. Non-Conserved miRNA-Auxin Module in Plants
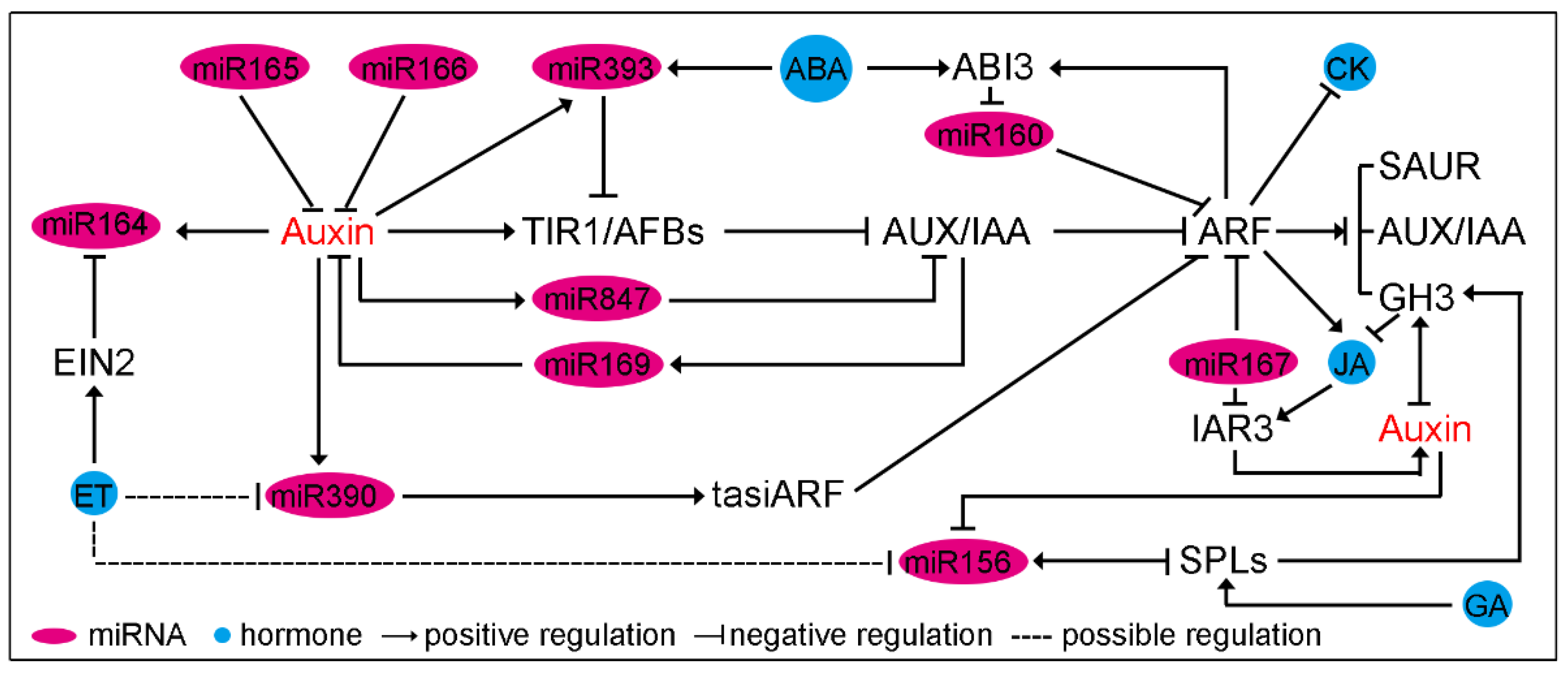
3. The Crosstalk of Auxin, miRNA and Other Hormones during Plant Development and Stress Responses
4. Concluding Remarks
- (i)
- How do plants integrate all these developmental signals or stress responses to control the levels of the individual miRNAs-auxin signaling module?
- (ii)
- Which lineage-, species- and tissue-specific miRNA molecules are also involved in regulating auxin responses?
- (iii)
- How can we use miRNA-auxin modules to create tolerance to stresses and/or high-yield crops?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Báez, R.R.; Nemhauser, J.L. Expansion and innovation in auxin signaling: Where do we grow from here? Development 2021, 148, dev187120. [Google Scholar] [CrossRef]
- Di, D.-W.; Li, G.; Sun, L.; Wu, J.; Wang, M.; Kronzucker, H.J.; Fang, S.; Chu, J.; Shi, W. High ammonium inhibits root growth in Arabidopsis thaliana by promoting auxin conjugation rather than inhibiting auxin biosynthesis. J. Plant Physiol. 2021, 261, 153415. [Google Scholar] [CrossRef]
- Di, D.-W.; Sun, L.; Zhang, X.; Li, G.; Kronzucker, H.J.; Shi, W. Involvement of auxin in the regulation of ammonium tolerance in rice (Oryza sativa L.). Plant Soil 2018, 432, 373–387. [Google Scholar] [CrossRef]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef] [Green Version]
- Sauer, M.; Robert, S.; Kleine-Vehn, J. Auxin: Simply complicated. J. Exp. Bot. 2013, 64, 2565–2577. [Google Scholar] [CrossRef] [Green Version]
- Simon, S.; Petrasek, J. Why plants need more than one type of auxin. Plant Sci. 2011, 180, 454–460. [Google Scholar] [CrossRef] [Green Version]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.H.; Estelle, M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 2014, 21, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Villalobos, L.I.A.C.; Lee, S.; De Oliveira, C.; Ivetac, A.; Brandt, W.; Armitage, L.; Sheard, L.B.; Tan, X.; Parry, G.; Mao, H.; et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 2012, 8, 477–485. [Google Scholar] [CrossRef] [Green Version]
- Goda, H.; Sasaki, E.; Akiyama, K.; Maruyama-Nakashita, A.; Nakabayashi, K.; Li, W.; Ogawa, M.; Yamauchi, Y.; Preston, J.; Aoki, K.; et al. The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 2008, 55, 526–542. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef] [Green Version]
- Jodder, J. miRNA-mediated regulation of auxin signaling pathway during plant development and stress responses. J. Biosci. 2020, 45, 91. [Google Scholar] [CrossRef]
- Matías, B.; Manuel, D.J.; Antonella, F.; Palatnik, J.F. ARF2 represses expression of plant GRF transcription factors in a complementary mechanism to microRNA miR396. Plant Physiol. 2021, 185, 1798–1812. [Google Scholar]
- Li, M.; Yu, B. Recent advances in the regulation of plant miRNA biogenesis. RNA Biol. 2021, 18, 2087–2096. [Google Scholar] [CrossRef]
- Wang, J.; Mei, J.; Ren, G. Plant microRNAs: Biogenesis, homeostasis, and degradation. Front Plant Sci. 2019, 10, 360. [Google Scholar] [CrossRef] [Green Version]
- Mallory, A.C.; Elmayan, T.; Vaucheret, H. MicroRNA maturation and action-the expanding roles of ARGONAUTEs. Curr. Opin. Plant Biol. 2008, 11, 560–566. [Google Scholar] [CrossRef]
- Li, M.; Yu, H.; Liu, K.; Yang, W.; Zhou, B.; Gan, L.; Li, S.; Zhang, C.; Yu, B. Serrate-Associated Protein 1, a splicing-related protein, promotes miRNA biogenesis in Arabidopsis. New Phytol. 2021, 232, 1959–1973. [Google Scholar] [CrossRef]
- Yang, G.; Li, Y.; Wu, B.; Zhang, K.; Gao, L.; Zheng, C. MicroRNAs transcriptionally regulate promoter activity in Arabidopsis thaliana. J. Integr. Plant Biol. 2019, 61, 1128–1133. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Anderson, T.A. Identification of 188 conserved maize microRNAs and their targets. FEBS Lett. 2006, 580, 3753–3762. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Sun, Y.; Shi, R.; Clark, C.; Li, L.; Chiang, V. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 2005, 17, 2186–2203. [Google Scholar] [CrossRef] [Green Version]
- Sanan-Mishra, N.; Kumar, V.; Sopory, S.K.; Mukherjee, S.K. Cloning and validation of novel miRNA from basmati rice indicates cross talk between abiotic and biotic stresses. Mol. Genet. Genom. 2009, 282, 463–474. [Google Scholar] [CrossRef]
- Zhang, B.-H.; Pan, X.-P.; Wang, Q.-L.; Cobb, G.P.; Anderson, T.A. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005, 15, 336–360. [Google Scholar] [CrossRef] [Green Version]
- Di, D.-W.; Zhang, C.; Guo, G.-Q. Involvement of secondary messengers and small organic molecules in auxin perception and signaling. Plant Cell Rep. 2015, 34, 895–904. [Google Scholar] [CrossRef]
- Dubey, S.; Saxena, S.; Chauhan, A.S.; Mathur, P.; Chakrabaroty, D. Identification and expression analysis of conserved microRNAs during short and prolonged chromium stress in rice (Oryza sativa). Environ. Sci. Pollut. Res. Int. 2020, 27, 380–390. [Google Scholar] [CrossRef]
- Singh, S.; Singh, A. A prescient evolutionary model for genesis, duplication and differentiation of MIR160 homologs in Brassicaceae. Mol. Genet. Genom. 2021, 296, 985–1003. [Google Scholar] [CrossRef]
- Gutierrez, L.; Bussell, J.D.; Pacurar, D.I.; Schwambach, J.; Pacurar, M.; Bellini, C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.-P.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef]
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 2005, 17, 1360–1375. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-W.; Wang, L.-J.; Mao, Y.-B.; Cai, W.-J.; Xue, H.-W.; Chen, X.-Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.-F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Martijn, F.; Dekkers, B.; Lena, M.; Wilma, V.; Angenent, G.C.; Zhao, Y.; Kim, B. ABA signalling promotes cell totipotency in the shoot apex of germinating embryos. J. Exp. Bot. 2021, 72, 6418–6436. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.-Q.; Luan, S.; Li, J.; He, Z.-H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490. [Google Scholar] [CrossRef] [Green Version]
- Tian, R.; Wang, F.; Zheng, Q.; Niza, V.M.A.G.E.; Downie, A.B.; Perry, S.E. Direct and indirect targets of the Arabidopsis seed transcription factor ABSCISIC ACID INSENSITIVE3. Plant J. 2020, 103, 1679–1694. [Google Scholar] [CrossRef]
- Wojcik, A.M.; Nodine, M.D.; Gaj, M.D. miR160 and miR166/165 contribute to the LEC2-mediated auxin response involved in the somatic embryogenesis induction in Arabidopsis. Front. Plant Sci. 2017, 8, 2024. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.-S.; Kuo, C.-C.; Yang, I.-C.; Tsai, W.-A.; Shen, Y.-H.; Lin, C.-C.; Liang, Y.-C.; Li, Y.-C.; Kuo, Y.-W.; King, Y.-C.; et al. MicroRNA160 modulates plant development and heat shock protein gene expression to mediate heat tolerance in Arabidopsis. Front. Plant Sci. 2018, 9, 68. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; MacPherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.-H. IAA-Ala resistant 3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef] [Green Version]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef] [Green Version]
- Caruana, J.C.; Dhar, N.; Raina, R. Overexpression of Arabidopsis microRNA167 induces salicylic acid-dependent defense against Pseudomonas syringae through the regulation of its targets ARF6 and ARF8. Plant Direct 2020, 4, e00270. [Google Scholar] [CrossRef]
- Huang, J.; Li, Z.Y.; Zhao, D.Z. Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci. Rep. 2016, 6, 29938. [Google Scholar] [CrossRef]
- Yang, J.; Han, S.; Yoon, E.K.; Lee, W.S. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 2006, 34, 1892–1899. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, S.; Shen, C.; Zhang, S.; Chen, Y.; Xu, Y.; Liu, Y.; Wu, Y.; Jiang, D. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 2012, 193, 109–120. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Chen, Z.; Wei, Y.; Qi, Y.; Wu, C. OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice. Plant Biotechnol. J. 2020, 18, 2015–2026. [Google Scholar] [CrossRef] [Green Version]
- Qiao, J.; Jiang, H.; Lin, Y.; Shang, L.; Wang, M.; Li, D.; Fu, X.; Geisler, M.; Qi, Y.; Gao, Z.; et al. A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 2021, 14, 1683–1698. [Google Scholar] [CrossRef]
- Suzaki, T.; Yano, K.; Ito, M.; Umehara, Y.; Suganuma, N.; Kawaguchi, M. Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 2012, 139, 3997–4006. [Google Scholar] [CrossRef] [Green Version]
- Turner, M.; Nizampatnam, N.R.; Baron, M.; Coppin, S.; Damodaran, S.; Adhikari, S.; Arunachalam, S.P.; Yu, O.; Subramanian, S. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 2013, 162, 2042–2055. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, K.; Chen, L.; Zou, Y.; Liu, H.; Tian, Y.; Li, D.; Wang, R.; Zhao, F.; Ferguson, B.J.; et al. MicroRNA167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiol. 2015, 168, 984–999. [Google Scholar] [CrossRef] [Green Version]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [Green Version]
- Xia, R.; Xu, J.; Meyers, B.C. The emergence, evolution, and diversification of the miR390-TAS3-ARF pathway in land plants. Plant Cell 2017, 29, 1232–1247. [Google Scholar] [CrossRef] [Green Version]
- Yoon, E.K.; Yang, J.H.; Lee, W.S. Auxin and abscisic acid responses of auxin response factor 3 in Arabidopsis lateral root development. J. Plant Biol. 2010, 53, 150–154. [Google Scholar] [CrossRef]
- de Felippes, F.F.; Marchais, A.; Sarazin, A.; Oberlin, S.; Voinnet, O. A single miR390 targeting event is sufficient for triggering TAS3-tasiRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2017, 45, 5539–5554. [Google Scholar] [CrossRef]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR Targets Define an Autoregulatory Network Quantitatively Regulating Lateral Root Growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef] [Green Version]
- Williams, L.; Carles, C.C.; Osmont, K.S.; Fletcher, J.C. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA 2005, 102, 9703–9708. [Google Scholar] [CrossRef] [Green Version]
- Dastidar, M.G.; Scarpa, A.; Magele, I.; Ruiz-Duarte, P.; von Born, P.; Bald, L.; Jouannet, V.; Maizel, A. ARF5/MONOPTEROS directly regulates miR390 expression in the Arabidopsis thaliana primary root meristem. Plant Direct 2019, 3, e00116. [Google Scholar] [CrossRef] [Green Version]
- Yoon, E.K.; Yang, J.H.; Lim, J.; Kim, S.H.; Kim, S.K.; Lee, W.S. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 2010, 38, 1382–1391. [Google Scholar] [CrossRef] [Green Version]
- Curaba, J.; Singh, M.B.; Bhalla, P.L. miRNAs in the crosstalk between phytohormone signalling pathways. J. Exp. Bot. 2014, 65, 1425–1438. [Google Scholar] [CrossRef]
- Hunter, C.; Willmann, M.R.; Wu, G.; Yoshikawa, M.; de la Luz Gutierrez-Nava, M.; Poethig, S.R. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 2006, 133, 2973–2981. [Google Scholar] [CrossRef] [Green Version]
- Hobecker, K.V.; Reynoso, M.A.; Bustos-Sanmamed, P.; Wen, J.Q.; Mysore, K.S.; Crespi, M.; Blanco, F.A.; Zanetti, M.E. The microRNA390/TAS3 pathway mediates symbiotic nodulation and lateral toot growth. Plant Physiol. 2017, 174, 2469–2486. [Google Scholar] [CrossRef] [Green Version]
- Wen, F.-L.; Yue, Y.; He, T.-F.; Gao, X.-M.; Zhou, Z.S.; Long, X.-H. Identification of miR390-TAS3-ARF pathway in response to salt stress in Helianthus tuberosus L. Gene 2020, 738, 144460. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef]
- Iglesias, M.J.; Terrile, M.C.; Windels, D.; Lombardo, M.C.; Bartoli, C.G.; Vazquez, F.; Estelle, M.; Casalongue, C.A. MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS ONE 2014, 9, e107678. [Google Scholar]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D.G. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Si-Ammour, A.; Windels, D.; Arn-Bouldoires, E.; Kutter, C.; Ailhas, J.; Meins, F.; Vazquez, F. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 2011, 157, 683–691. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.-H.; Bao, M.-L.; Sun, Y.-Z.; Yang, Y.-J.; Xu, X.-H.; Wang, J.-H.; Han, N.; Bian, H.-W.; Zhu, M.-Y. Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol. Biol. 2011, 77, 619–629. [Google Scholar] [CrossRef]
- Vidal, E.A.; Araus, V.; Lu, C.; Parry, G.; Green, P.J.; Coruzzi, G.M.; Gutierrez, R.A. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 4477–4482. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Li, Z.F.; Xiong, L.M. A plant microRNA regulates the adaptation of roots to drought stress. Febs Lett. 2012, 586, 1742–1747. [Google Scholar] [CrossRef]
- Wojcik, A.M.; Gaj, M.D. miR393 contributes to the embryogenic transition induced in vitro in Arabidopsis via the modification of the tissue sensitivity to auxin treatment. Planta 2016, 244, 231–243. [Google Scholar] [CrossRef] [Green Version]
- Archak, S.; Nagaraju, J. Computational prediction of rice (Oryza sativa) miRNA targets. Genom. Proteom. Bioinf. 2007, 5, 196–206. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.-F.; Zheng, Y.; Addo-Quaye, C.; Zhang, L.; Saini, A.; Jagadeeswaran, G.; Axtell, M.J.; Zhang, W.; Sunkar, R. Transcriptome-wide identification of microRNA targets in rice. Plant J. 2010, 62, 742–759. [Google Scholar] [CrossRef]
- Xia, K.-F.; Wang, R.; Ou, X.-J.; Fang, Z.-M.; Tian, C.-G.; Duan, J.; Wang, Y.-Q.; Zhang, M.-Y. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, 364–373. [Google Scholar] [CrossRef]
- Bian, H.W.; Xie, Y.K.; Guo, F.; Han, N.; Ma, S.; Zeng, Z.; Wang, J.; Yang, Y.; Zhu, M. Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 2012, 196, 149–161. [Google Scholar] [CrossRef]
- Guo, F.; Han, N.; Xie, Y.; Fang, K.; Yang, Y.; Zhu, M.; Wang, J.; Bian, H. The miR393a/target module regulates seed germination and seedling establishment under submergence in rice (Oryza sativa L.). Plant Cell Environ. 2016, 39, 2288–2302. [Google Scholar] [CrossRef]
- Damodharan, S.; Zhao, D.Z.; Arazi, T. A common miRNA160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 2016, 86, 458–471. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Lai, L.; Lin, R.; Jin, C.; Chen, J. Differential effects of Cucumber mosaic virus satellite RNAs in the perturbation of microRNA-regulated gene expression in tomato. Mol. Biol. Rep. 2012, 39, 775–784. [Google Scholar] [CrossRef]
- Feng, J.; Lin, R.; Chen, J. Alteration of tomato microRNAs expression during fruit development upon Cucumber mosaic virus and Tomato aspermy virus infection. Mol. Biol. Rep. 2013, 40, 3713–3722. [Google Scholar] [CrossRef]
- Jin, W.B.; Wu, F.L.; Xiao, L.; Liang, G.W.; Zhen, Y.X.; Guo, Z.K.; Guo, A.G. Microarray-based analysis of tomato miRNA regulated by Botrytis cinerea. J. Plant Growth Regul. 2012, 31, 38–46. [Google Scholar] [CrossRef]
- Liu, X.; Dong, X.; Liu, Z.; Shi, Z.; Jiang, Y.; Qi, M.F.; Xu, T.; Li, T. Repression of ARF10 by microRNA160 plays an important role in the mediation of leaf water loss. Plant Mol. Biol. 2016, 92, 313–336. [Google Scholar] [CrossRef]
- Naqvi, A.R.; Haq, Q.M.R.; Mukherjee, S.K. MicroRNA profiling of tomato leaf curl new delhi virus (tolcndv) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol. J. 2010, 7, 281. [Google Scholar] [CrossRef] [Green Version]
- Tsushima, D.; Adkar-Purushothama, C.R.; Taneda, A.; Sano, T. Changes in relative expression levels of viroid-specific small RNAs and microRNAs in tomato plants infected with severe and mild symptom-inducing isolates of Potato spindle tuber viroid. J. Gen. Plant Pathol. 2015, 81, 49–62. [Google Scholar] [CrossRef]
- Kruszka, K.; Pacak, A.; Swida-Barteczka, A.; Nuc, P.; Alaba, S.; Wroblewska, Z.; Karlowski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J. Exp. Bot. 2014, 65, 6123–6135. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.R.; Pathak, H.; Sharma, S.K.; Kala, Y.K.; Nirjal, M.K.; Singh, G.P.; Goswami, S.; Rai, R.D. Novel and conserved heat-responsive microRNAs in wheat (Triticum aestivum L.). Funct. Integr. Genomic 2015, 15, 323–348. [Google Scholar] [CrossRef]
- Aravind, J.; Rinku, S.; Pooja, B.; Shikha, M.; Kaliyugam, S.; Mallikarjuna, M.G.; Kumar, A.; Rao, A.R.; Nepolean, T. Identification, characterization, and functional validation of drought-responsive microRNAs in subtropical maize inbreds. Front. Plant Sci. 2017, 8, 941. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, B.; Kalsi, H.S.; Godbole, P.; Malankar, N.; Thiagarayaselvam, A.; Siddappa, S.; Thulasiram, H.V.; Chakrabarti, S.K.; Banerjee, A.K. MiRNA160 is associated with local defense and systemic acquired resistance against Phytophthora infestans infection in potato. J. Exp. Bot. 2018, 69, 2023–2036. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, N.; Zhang, J.; Jin, X.; Si, H. Knockdown of MicroRNA160a/b by STTM leads to root architecture changes via auxin signaling in Solanum tuberosum. Plant Physiol. Bioch. 2021, 166, 939–949. [Google Scholar] [CrossRef]
- Ding, Y.; Ma, Y.; Liu, N.; Xu, J.; Hu, Q.; Li, Y.; Wu, Y.; Xie, S.; Zhu, L.; Min, L.; et al. microRNAs involved in auxin signalling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J. 2017, 91, 977–994. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Shen, J.; Xu, Q.; Dong, J.; Song, L.; Wang, W.; Shen, F. Long noncoding RNA lncRNA354 functions as a competing endogenous RNA of miR160b to regulate ARF genes in response to salt stress in upland cotton. Plant Cell Environ. 2021, 44, 3302–3321. [Google Scholar] [CrossRef]
- Lin, Y.; Lin, L.; Lai, R.; Liu, W.; Chen, Y.; Zhang, Z.; XuHan, X.; Lai, Z. MicroRNA390-directed TAS3 cleavage leads to the production of tasiRNA-ARF3/4 during somatic embryogenesis in Dimocarpus longan Lour. Front. Plant Sci. 2015, 6, 1119. [Google Scholar] [CrossRef]
- Sattar, S.; Addo-Quaye, C.; Thompson, G.A. miRNA-mediated auxin signalling repression during Vat-mediated aphid resistance in Cucumis melo. Plant Cell Environ. 2016, 39, 1216–1227. [Google Scholar] [CrossRef] [Green Version]
- Bustos-Sanmamed, P.; Mao, G.H.; Deng, Y.; Elouet, M.; Khan, G.A.; Bazin, J.; Turner, M.; Subramanian, S.; Yu, O.; Crespi, M.; et al. Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Funct. Plant Biol. 2013, 40, 1208–1220. [Google Scholar] [CrossRef]
- Pinweha, N.; Asvarak, T.; Viboonjun, U.; Narangajavana, J. Involvement of miR160/miR393 and their targets in cassava responses to anthracnose disease. J. Plant Physiol 2015, 174, 26–35. [Google Scholar] [CrossRef]
- Shi, M.; Hu, X.; Wei, Y.; Hou, X.; Yuan, X.; Liu, J.; Liu, Y. Genome-wide profiling of small RNAs and degradome revealed conserved regulations of miRNAs on auxin-responsive genes during fruit enlargement in Peaches. Inter. J. Mol. Sci. 2017, 18, 2599. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Dai, J.; Chen, R.; Song, C.; Wei, P.; Cai, Y.; Wang, Y.; Han, B. miRNA-based drought regulation in the important medicinal plant Dendrobium huoshanense. J. Plant Growth Regul. 2021, 1–10. [Google Scholar] [CrossRef]
- Jin, W.; Wu, F. Characterization of miRNAs associated with Botrytis cinerea infection of tomato leaves. BMC Plant Biol. 2015, 15, 1–14. [Google Scholar] [CrossRef]
- Li, Z.-X.; Zhang, L.-F.; Li, W.-F.; Qi, L.-W.; Han, S.-Y. MIR166a affects the germination of somatic embryos in Larixleptolepis by modulating IAA biosynthesis and signaling genes. J. Plant Growth Regul. 2017, 36, 889–896. [Google Scholar] [CrossRef]
- Diermann, N.; Matousek, J.; Junge, M.; Riesner, D.; Steger, G. Characterization of plant miRNAs and small RNAs derived from potato spindle tuber viroid (PSTVd) in infected tomato. Biol. Chem. 2010, 391, 1379–1390. [Google Scholar] [CrossRef]
- Lang, Q.-L.; Zhou, X.-C.; Zhang, X.-L.; Drabek, R.; Zuo, Z.-X.; Ren, Y.-L.; Li, T.-B.; Chen, J.-S.; Gao, X.-L. Microarray-based identification of tomato microRNAs and time course analysis of their response to Cucumber mosaic virus infection. J. Zhejiang Univ. Sci. 2011, 12, 116–125. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Yang, C.; Liu, S.; Qi, H.; Wu, L.; Xu, L.A.; Xu, M. MiRNA-target pairs regulate adventitious rooting in Populus: A functional role for miR167a and its target auxin response factor 8. Tree Physiol. 2019, 39, 1922–1936. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Z.; Shi, G.; Bai, Q.; Guo, C.; Xiao, K. MIR167a transcriptionally regulates ARF6 and ARF8 and mediates drastically plant Pi-starvation response via modulation of various biological processes. Plant Cell Tiss. Org. 2018, 133, 177–191. [Google Scholar] [CrossRef]
- Ding, B.; Xia, R.; Lin, Q.; Gurung, V.; Sagawa, J.; Stanley, L.E.; Strobel, M.; Diggle, P.K.; Meyers, B.C.; Yuan, Y. Developmental genetics of corolla tube formation: Role of the tasiRNA-ARF pathway and a conceptual model. Plant Cell 2020, 32, 3452–3468. [Google Scholar] [CrossRef]
- Cho, S.H.; Coruh, C.; Axtell, M.J. miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 2012, 24, 4837–4849. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Wang, Y.; Jia, Y.; Yang, Y.; Wang, C. Ectopic overexpression of bol-miR390a from broccoli (B. oleracea L var. italica) increases lateral branches in Arabidopsis. Plant Growth Regul. 2020, 92, 1–12. [Google Scholar] [CrossRef]
- Fu, C.; Shen, H.; Guo, Y.; Leng, L.; Luo, M. The MicroRNA390/TRANS-ACTING SHORT INTERFERING RNA3 module mediates lateral root growth under salt stress via the auxin pathway. Plant Physiol. 2018, 177, 775–791. [Google Scholar]
- Pradhan, M.; Rocha, C.; Halitschke, R.; Baldwin, I.T.; Pandey, S.P. microRNA390 modulates Nicotiana attenuata’s tolerance response to Manduca sexta herbivory. Plant Direct 2021, 5, e350. [Google Scholar] [CrossRef]
- Cai, Z.; Wang, Y.; Zhu, L.; Tian, Y.; Chen, L.; Sun, Z.; Ullah, I.; Li, X. GmTIR1/GmAFB3-based auxin perception regulated by miR393 modulates soybean nodulation. New Phytol. 2017, 215, 672–686. [Google Scholar] [CrossRef] [Green Version]
- Bai, B.; Bian, H.; Zeng, Z.H.; Hou, N.; Shi, B.; Wang, J.H.; Zhu, M.Y.; Han, N. miR393-mediated auxin signaling regulation is involved in root elongation inhibition in response to toxic aluminum stress in barley. Plant Cell Physiol. 2017, 58, 426–439. [Google Scholar] [CrossRef]
- Xu, J.; Li, J.; Cui, L.; Zhang, T.; Wu, Z.; Zhu, P.; Meng, Y.; Zhang, K.; Yu, X.; Lou, Q.; et al. New insights into the roles of cucumber TIR1 homologs and miR393 in regulating fruit/seed set development and leaf morphogenesis. BMC Plant Biol. 2017, 17, 130. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Wei, Y.; Wang, R.; Mao, J.; Tian, H.; Chen, S.; Li, S.; Tahir, M.M.; Zhang, D. Mdm-MIR393b-mediated adventitious root formation by targeted regulation of MdTIR1A expression and weakened sensitivity to auxin in apple rootstock. Plant Sci. 2021, 308, 110909. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, K.; Li, D.; Yan, J.; Zhang, W. Enhanced cold tolerance and tillering in Switchgrass (Panicum virgatum L.) by heterologous expression of Osa-miR393a. Plant Cell Physiol. 2017, 58, 2226–2240. [Google Scholar] [CrossRef] [Green Version]
- Cui, G.; Zhao, M.; Zhang, S.; Wang, Z.; Xi, Y. MicroRNA and regulation of auxin and cytokinin signalling during post-mowing regeneration of winter wheat (Triticum aestivum L.). Plant Physiol. Bioch. 2020, 155, 769–779. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, Z.; Chen, C.; Li, C.; Xia, R.; Li, J. Genome-wide characterization of the auxin response factor (ARF) gene family of litchi (Litchi chinensis Sonn.): Phylogenetic analysis, miRNA regulation and expression changes during fruit abscission. PeerJ 2019, 7, e6677. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Xie, K.; Xiong, L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2014, 65, 2119–2135. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef] [Green Version]
- Sunkar, R.; Zhou, X.; Zheng, Y.; Zhang, W.; Zhu, J.-K. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008, 8, 25. [Google Scholar] [CrossRef] [Green Version]
- Xie, Q.; Frugis, G.; Colgan, D.; Chua, N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Gene Dev. 2000, 14, 3024–3036. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.-S.; Xie, Q.; Fei, J.-F.; Chua, N.-H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef] [Green Version]
- Koyama, T.; Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. TCP Transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 2010, 22, 3574–3588. [Google Scholar] [CrossRef] [Green Version]
- Aslam, M.; Sugita, K.; Qin, Y.; Rahman, A. Aux/IAA14 regulates microRNA-mediated cold stress response in Arabidopsis roots. Int. J. Mol.Sci. 2020, 21, 8441. [Google Scholar] [CrossRef]
- Gyula, P.; Baksa, I.; Toth, T.; Mohorianu, I.; Dalmay, T.; Szittya, G. Ambient temperature regulates the expression of a small set of sRNAs influencing plant development through NF-YA2 and YUC2. Plant Cell Environ. 2018, 41, 2404–2417. [Google Scholar] [CrossRef]
- Fahlgren, N.; Jogdeo, S.; Kasschau, K.D.; Sullivan, C.M.; Chapman, E.J.; Laubinger, S.; Smith, L.M.; Dasenko, M.; Givan, S.A.; Weigel, D.; et al. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 2010, 22, 1074–1089. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Westfall, C.S.; Hicks, L.M.; Wang, S.P.; Jez, J.M. Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J. Biol. Chem. 2010, 285, 29780–29786. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Wang, J.; Yang, X.; Lu, H.; Miao, X.; Shi, Z. Modulation of plant architecture by the miR156f-OsSPL7-OsGH3.8 pathway in rice. J. Exp. Bot. 2018, 69, 5117–5130. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Li, L. Comparative analysis of microRNA promoters in Arabidopsis and rice. Genom. Proteom. Bioinf. 2013, 11, 56–60. [Google Scholar] [CrossRef] [Green Version]
- Betti, F.; Ladera-Carmona, M.J.; Weits, D.A.; Ferri, G.; Iacopino, S.; Novi, G.; Svezia, B.; Kunkowska, A.B.; Santaniello, A.; Piaggesi, A.; et al. Exogenous miRNAs induce post-transcriptional gene silencing in plants. Nat. Plants 2021, 7, 1379–1388. [Google Scholar] [CrossRef]
- Strzyz, P. microRNA communication in plants. Nat. Rev. Mol. Cell Bio. 2021, 22, 775. [Google Scholar] [CrossRef]
- Cheng, Z.; Wang, L.; Sun, W.; Zhang, Y.; Zhou, C.; Su, Y.; Li, W.; Sun, T.; Zhao, X.; Li, X.; et al. Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR 3. Plant Physiol. 2013, 161, 240–251. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Andersen, S.U.; Ljung, K.; Dolezal, K.; Miotk, A.; Schultheiss, S.J.; Lohmann, J.U. Hormonal control of the shoot stem-cell niche. Nature 2010, 465, 1089–1092. [Google Scholar] [CrossRef]
- Liu, Z.; Li, J.; Wang, L.; Li, Q.; Lu, Q.; Yu, Y.; Li, S.; Bai, M.; Hu, Y.; Xiang, F. Repression of callus initiation by the miRNA-directed interaction of auxin-cytokinin in Arabidopsis thaliana. Plant J. 2016, 87, 391–402. [Google Scholar] [CrossRef]
- Yu, S.; Galvao, V.C.; Zhang, Y.-C.; Horrer, D.; Zhang, T.-Q.; Hao, Y.-H.; Feng, Y.-Q.; Wang, S.; Schmid, M.; Wang, J.-W. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING-LIKE transcription factors. Plant Cell 2012, 24, 3320–3332. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.-W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [Green Version]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development 2004, 131, 3357–3365. [Google Scholar] [CrossRef] [Green Version]
- Zuo, J.; Zhu, B.; Fu, D.; Zhu, Y.; Ma, Y.; Chi, L.; Ju, Z.; Wang, Y.; Zhai, B.; Luo, Y. Sculpting the maturation, softening and ethylene pathway: The influences of microRNAs on tomato fruits. BMC Genom. 2012, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chetelat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [Green Version]
- Adams, E.; Turner, J. COI1, a jasmonate receptor, is involved in ethylene-induced inhibition of Arabidopsis root growth in the light. J. Exp. Bot. 2010, 61, 4373–4386. [Google Scholar] [CrossRef] [Green Version]
- Di, D.-W.; Sun, L.; Wang, M.; Wu, J.; Kronzucker, H.J.; Fang, S.; Chu, J.; Shi, W.; Li, G. WRKY46 promotes ammonium tolerance in Arabidopsis by repressing NUDX9 and indole-3-acetic acid-conjugating genes and by inhibiting ammonium efflux in the root elongation zone. New Phytol. 2021, 232, 190–207. [Google Scholar] [CrossRef]
- Di, D.-W.; Wu, L.; Luo, P.; Zhang, L.; Zhang, T.-Z.; Sun, X.; Wei, S.-D.; An, C.-W.; Guo, G.-Q. Analysis the role of Arabidopsis CKRC6/ASA1 in auxin and cytokinin biosynthesis. J. Plant Biol. 2016, 59, 162–171. [Google Scholar] [CrossRef]
- Di, D.-W.; Wu, L.; Zhang, L.; An, C.-W.; Zhang, T.-Z.; Luo, P.; Gao, H.-H.; Kriechbaumer, V.; Guo, G.-Q. Functional roles of Arabidopsis CKRC2/YUCCA8 gene and the involvement of PIF4 in the regulation of auxin biosynthesis by cytokinin. Sci. Rep. 2016, 6, 36866. [Google Scholar] [CrossRef] [Green Version]
- Datta, R.; Paul, S. Plant microRNAs: Master regulator of gene expression mechanism. Cell Biol.Int. 2015, 39, 1185–1190. [Google Scholar] [CrossRef]
- Chitwood, D.H.; Nogueira, F.T.S.; Howell, M.D.; Montgomery, T.A.; Carrington, J.C.; Timmermans, M.C.P. Pattern formation via small RNA mobility. Gene. Dev. 2009, 23, 549–554. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.-L.; Qu, L.-H. Application of microRNA gene resources in the improvement of agronomic traits in rice. Plant Biotechnol. J. 2015, 13, 329–336. [Google Scholar] [CrossRef]
- Chang, H.; Yi, B.; Ma, R.; Zhang, X.; Zhao, H.; Xi, Y. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci. Rep. 2016, 6, 22312. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Cheng, X.; Cai, J.; Zhan, L.; Wu, X.; Liu, Q.; Wu, X. Multiple virus resistance using artificial trans-acting siRNAs. J. Virol. Methods 2016, 228, 16–20. [Google Scholar] [CrossRef]
- Djami-Tchatchou, A.T.; Sanan-Mishra, N.; Ntushelo, K.; Dubery, I.A. Functional roles of microRNAs in agronomically important plants-potential as targets for crop improvement and protection. Front. Plant Sci. 2017, 8, 378. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Chen, Y.-Q. A new mechanism in plant engineering: The potential roles of microRNAs in molecular breeding for crop improvement. Biotechnol. Adv. 2010, 28, 301–307. [Google Scholar] [CrossRef]
- Tiwari, M.; Sharma, D.; Trivedi, P.K. Artificial microRNA mediated gene silencing in plants: Progress and perspectives. Plant Mol. Biol. 2014, 86, 1–18. [Google Scholar] [CrossRef]
- Zhao, Y.; Dai, Z.; Liang, Y.; Yin, M.; Ma, K.; He, M.; Ouyang, H.; Teng, C. Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci. Rep. 2014, 4, 3943. [Google Scholar] [CrossRef] [Green Version]
| miRNA | Species | Direct Targets | Secondary Targets | Target Functions | References |
|---|---|---|---|---|---|
| miR160 | Solanum lycopersicum | SlARF10/17 | Ovary patterning; floral organ abscission; lamina outgrowth; leaf water loss; pathogen defense | [74,75,76,77,78,79,80] | |
| Glycine max L. | GmARF | Nodule development | [45] | ||
| Hordeum vulgare | HvARF13/17 | Heat stress | [81] | ||
| Triticum aestivum L. | TaARF | Abiotic stress | [82] | ||
| Zea mays L. | ZmARF | Drought stress | [83] | ||
| Solanum tuberosum | StARF10 | StGH3.6 | Pathogen defense; root architecture | [84,85] | |
| Gossypium hirsutum | GhARF10/17 | Heat stress; salt stress | [86,87] | ||
| Dimocarpus longan | DlARF10/16/17 | Somatic embryogenesis | [88] | ||
| Cucumis melo | CmARF10/16/17 | Aphid resistance | [89] | ||
| Medicago truncatula | MeARF10/16/17 | Nodule number and development | [90] | ||
| Manihot esculenta Crantz | MaARF10 | Pathogen defense | [91] | ||
| Prunus persica L. | PpARF17 | Fruit enlargement | [92] | ||
| Dendrobium huoshanense | Dhu-20362/-11346 | Drought stress | [93] | ||
| miR164 | Solanum lycopersicum | SlNAC1 | NA | Pathogen defense | [79,80,94] |
| miR166 | Larix leptolepis | LaHDZIPIII | LaNIT | Somatic embryogenesis | [95] |
| miR167 | Solanum lycopersicum | SlARF6/8 | Pathogen defence | [75,79,96,97] | |
| Glycine max | GmARF8a/8b | Nodulation; lateral root development | [46] | ||
| Cucumis melo | CmARF6/8 | Aphid resistance | [89] | ||
| Hordeum vulgare | HvARF8 | Heat stress | [81] | ||
| Populus spp. | PeARF8 | Adventitious rooting | [98] | ||
| Nicotiana tabacum L. | NtARF6/8 | Phosphorus starvation | [99] | ||
| miR390 | Zea mays | NA | NA | Drought stress | [83] |
| Dimocarpus longan | DlTAS3 | tasiDlARF3/ARF4 | Somatic embryogenesis | [88] | |
| Cucumis melo | NA | tasiCmARF | Aphids resistance | [89] | |
| Medicago truncatula | MeTAS3 | tasiMeARF2/3/4 | Nodulation; lateral root growth | [57] | |
| Mimulus lewisii | MlTAS3 | tasiMlARF3/4 | Corolla tube formation | [100] | |
| Physcomitrella patens | PpTAS3 | tasiPpARF | Developmental timing | [101] | |
| Helianthus tuberosus L. | HtTAS3a/b/c | tasiHtARF1/2/3 | Salt stress | [58] | |
| B. oleracea L. var. italica | BoTAS3 | tasiBoARF2/3/4 | Lateral root | [102] | |
| Populus spp. | NA | tasiPsARF3.1/3.2/4 | Salt stress | [103] | |
| Nicotiana tabacum L. | NA | tasiNtARF | Manduca sexta resistance | [104] | |
| miR393 | Glycine max | GmTIR1/AFB3 | Nodulation | [105] | |
| Hordeum vulgare | HvTIR1/AFB | Aluminum stress | [106] | ||
| Zea mays | ZmTIR1/AFB | Drought stress | [83] | ||
| Cucumis sativus L. | CsTIR1/AFB2 | Fruit/seed set development and leaf morphogenesis | [107] | ||
| Cucumis melo | TIR1/AFB2 | Aphids resistance | [89] | ||
| Manihot esculenta Crantz | MaTIR1 | Pathogen defence | [91] | ||
| Prunus persica L. | PpAFB2 | Fruit enlargement | [92] | ||
| Malus × domestica Borkh. | MdTIR1A | Adventitious root formation | [108] | ||
| Panicum virgatum L. | PvTIR1/AFB1/2/3 | Cold stress; tillering | [109] | ||
| miR1153-y | Triticum aestivum. | TaGH3.7 | Post-mowing regeneration | [110] | |
| miRN43 | Litchi chinensis Sonn. | LcARF9 | Fruit abscission | [111] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, P.; Di, D.; Wu, L.; Yang, J.; Lu, Y.; Shi, W. MicroRNAs Are Involved in Regulating Plant Development and Stress Response through Fine-Tuning of TIR1/AFB-Dependent Auxin Signaling. Int. J. Mol. Sci. 2022, 23, 510. https://doi.org/10.3390/ijms23010510
Luo P, Di D, Wu L, Yang J, Lu Y, Shi W. MicroRNAs Are Involved in Regulating Plant Development and Stress Response through Fine-Tuning of TIR1/AFB-Dependent Auxin Signaling. International Journal of Molecular Sciences. 2022; 23(1):510. https://doi.org/10.3390/ijms23010510
Chicago/Turabian StyleLuo, Pan, Dongwei Di, Lei Wu, Jiangwei Yang, Yufang Lu, and Weiming Shi. 2022. "MicroRNAs Are Involved in Regulating Plant Development and Stress Response through Fine-Tuning of TIR1/AFB-Dependent Auxin Signaling" International Journal of Molecular Sciences 23, no. 1: 510. https://doi.org/10.3390/ijms23010510
APA StyleLuo, P., Di, D., Wu, L., Yang, J., Lu, Y., & Shi, W. (2022). MicroRNAs Are Involved in Regulating Plant Development and Stress Response through Fine-Tuning of TIR1/AFB-Dependent Auxin Signaling. International Journal of Molecular Sciences, 23(1), 510. https://doi.org/10.3390/ijms23010510








