Integration of Light and Brassinosteroid Signaling during Seedling Establishment
Abstract
:1. Introduction
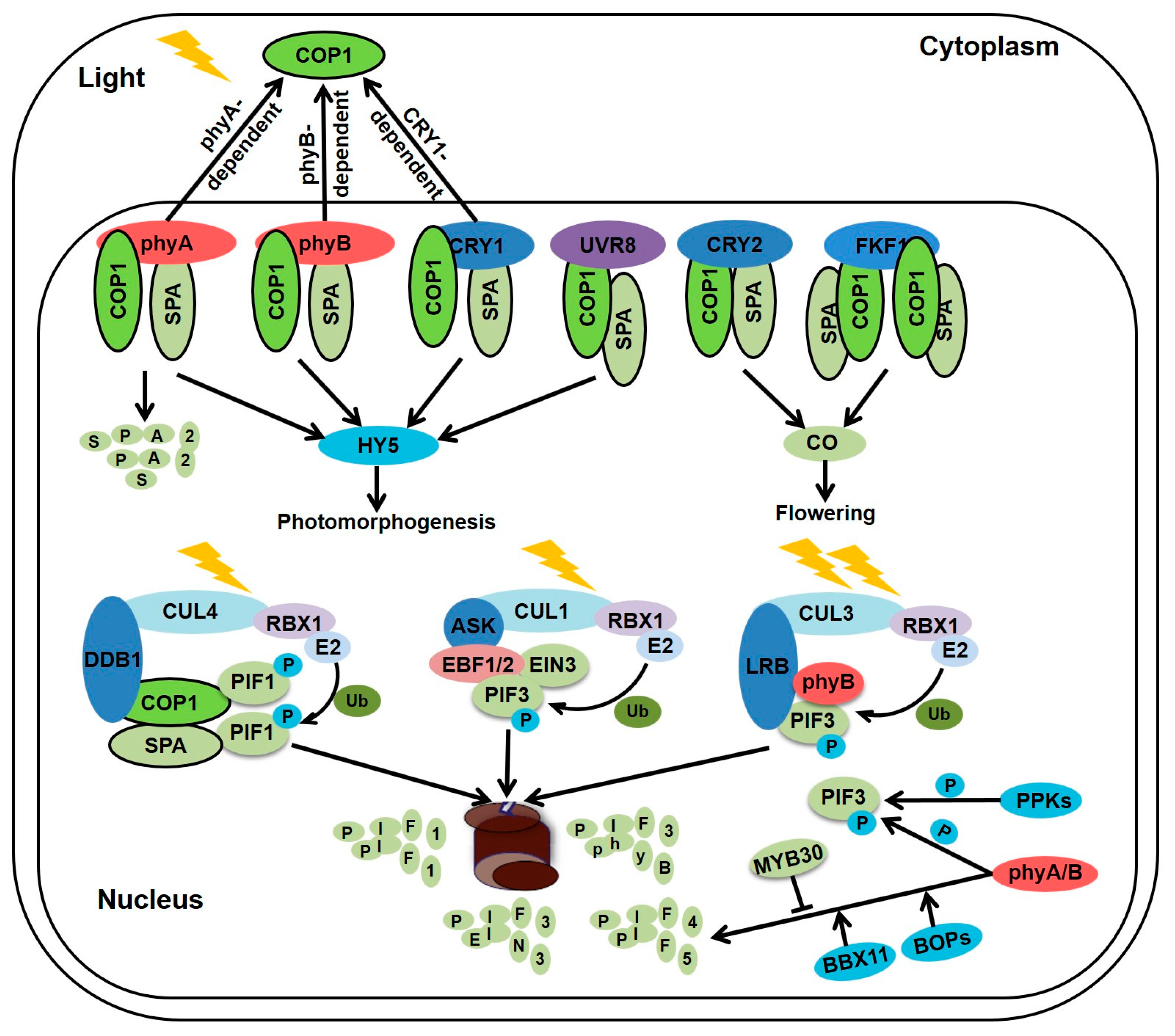
2. Regulation of Seedlings Morphogenesis by Light Information
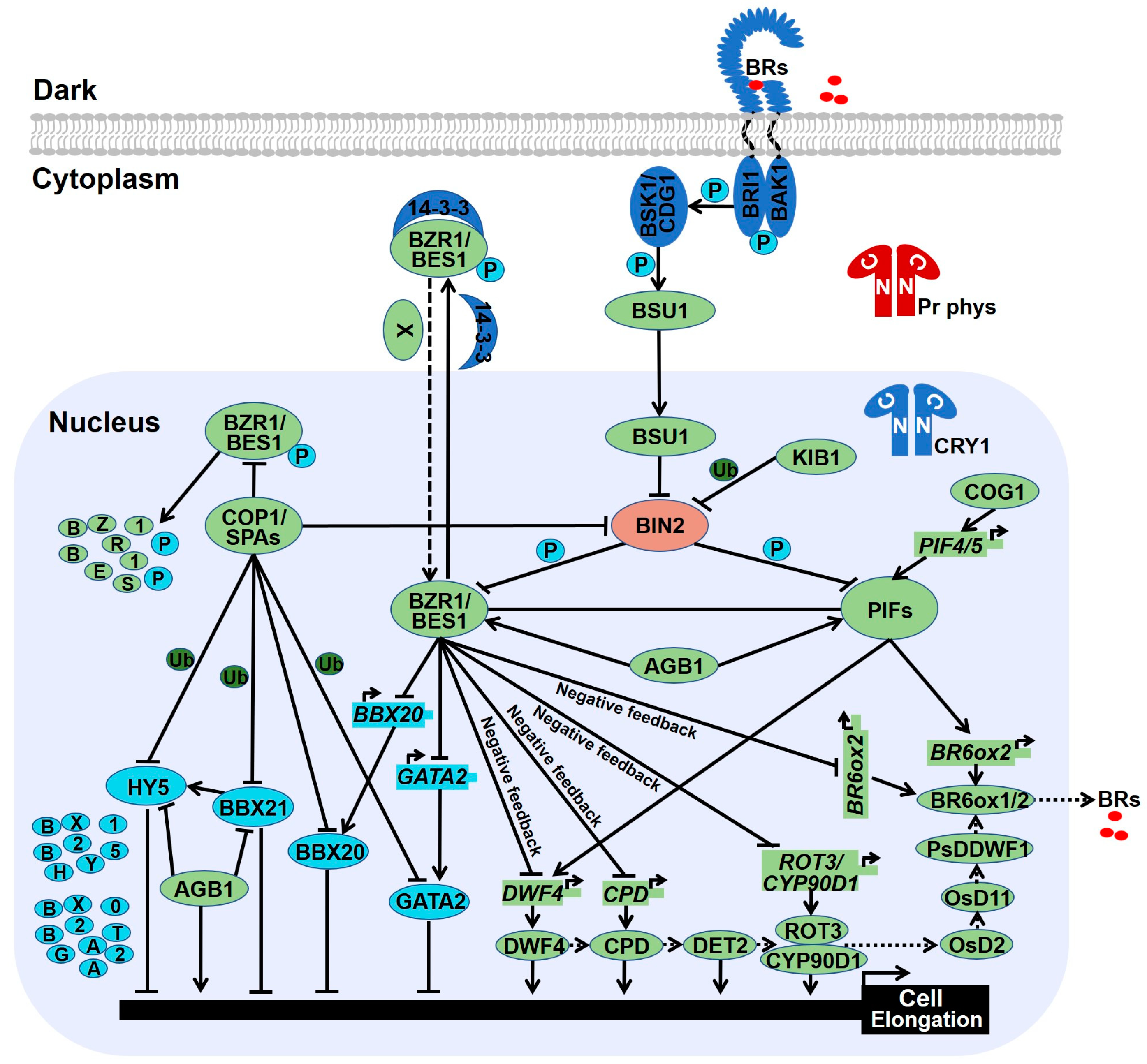
3. Brassinosteroid Biosynthesis and Signal Transduction
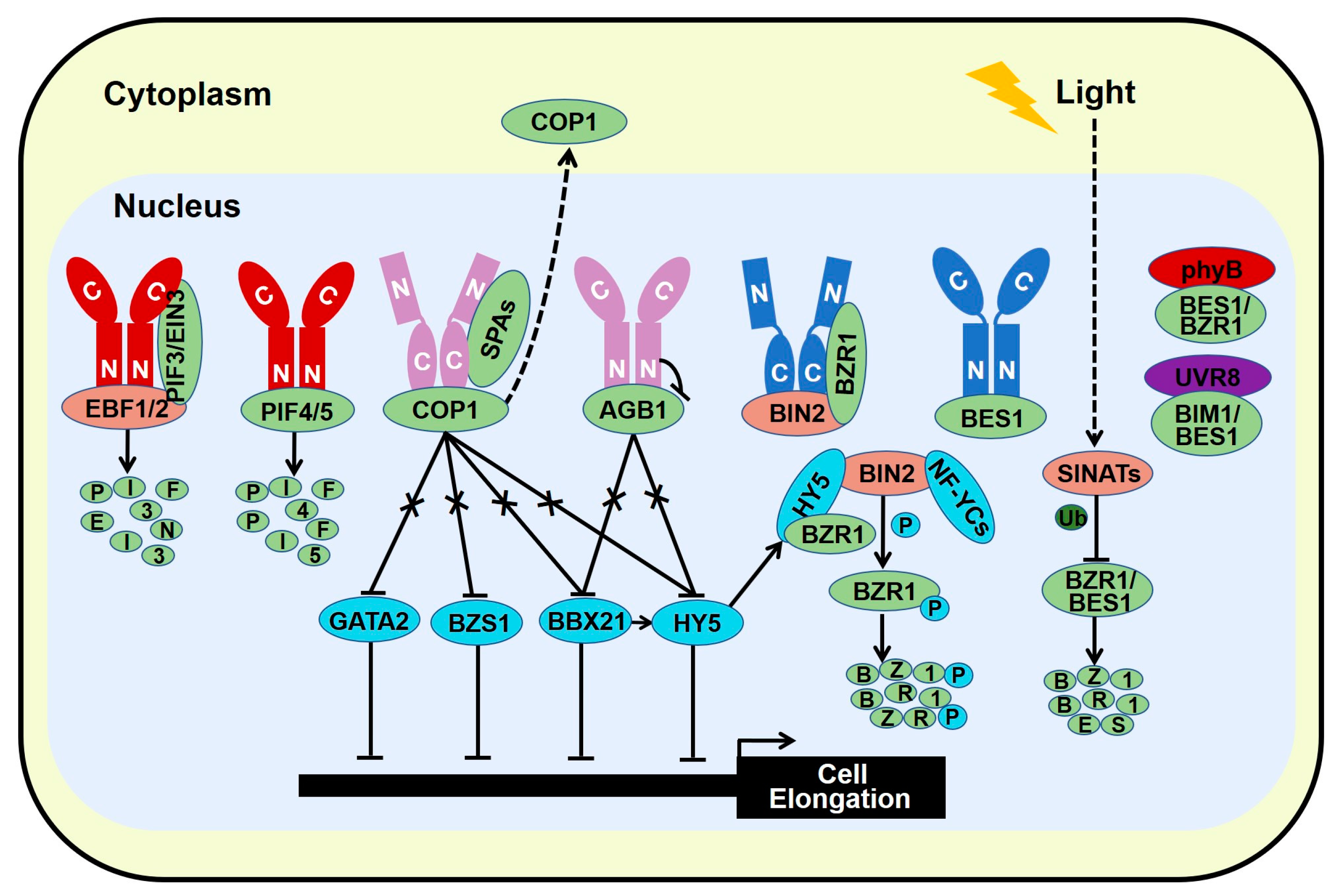
4. Signal Integration of Light and Brassinosteroid
4.1. Photomorphogenic Repressors Elevate Brassinosteroid Response
4.2. Photomorphogenesis-Promoting Factors Repress Brassinosteroid Signal
4.3. MicroRNAs Integrate Light and Brassinosteroid Signals
5. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007, 8, 217–230. [Google Scholar] [CrossRef]
- Von Arnim, A.G.; Deng, X.W. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 1994, 79, 1035–1045. [Google Scholar] [CrossRef]
- Kami, C.; Lorrain, S.; Hornitschek, P.; Fankhauser, C. Light-Regulated Plant Growth and Development. Curr. Top. Dev. Biol. 2010, 91, 29–66. [Google Scholar]
- Clouse, S.D.; Sasse, J.M. BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu. Rev. Plant Biol. 1998, 49, 427–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Nagpal, P.; Vitart, V.; McMorris, T.C.; Chory, J. A Role for Brassinosteroids in Light-Dependent Development of Arabidopsis. Science 1996, 272, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Chaiwanon, J.; Wang, W.; Zhu, J.-Y.; Oh, E.; Wang, Z.-Y. Information Integration and Communication in Plant Growth Regulation. Cell 2016, 164, 1257–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.-Y.; Bai, M.-Y.; Oh, E.; Zhu, J.-Y. Brassinosteroid Signaling Network and Regulation of Photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.-Y.; Wang, Z.-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.-Y.; Sun, Y.; Cao, D.-M.; Bai, M.-Y.; Luo, X.-M.; Yang, H.-J.; Wei, C.-Q.; Zhu, S.-W.; Sun, Y.; Chong, K.; et al. BZS1, a B-box Protein, Promotes Photomorphogenesis Downstream of Both Brassinosteroid and Light Signaling Pathways. Mol. Plant 2012, 5, 591–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, M.-Y.; Fan, M.; Oh, E.; Wang, Z.-Y. A Triple Helix-Loop-Helix/Basic Helix-Loop-Helix Cascade Controls Cell Elongation Downstream of Multiple Hormonal and Environmental Signaling Pathways in Arabidopsis. Plant Cell 2013, 24, 4917–4929. [Google Scholar] [CrossRef] [Green Version]
- Bernardo-García, S.; de Lucas, M.; Martínez, C.; Espinosa-Ruiz, A.; Davière, J.-M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef] [Green Version]
- Liang, T.; Mei, S.; Shi, C.; Yang, Y.; Peng, Y.; Ma, L.; Wang, F.; Li, X.; Huang, X.; Yin, Y.; et al. UVR8 Interacts with BES1 and BIM1 to Regulate Transcription and Photomorphogenesis in Arabidopsis. Dev. Cell 2018, 44, 512–523.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Lu, X.; Li, L.; Lian, H.; Mao, Z.; Xu, P.; Guo, T.; Xu, F.; Du, S.; Cao, X.; et al. Photoexcited CRYPTOCHROME1 Interacts with Dephosphorylated BES1 to Regulate Brassinosteroid Signaling and Photomorphogenesis in Arabidopsis. Plant Cell 2018, 30, 1989–2005. [Google Scholar] [CrossRef] [Green Version]
- He, G.; Liu, J.; Dong, H.; Sun, J. The Blue-Light Receptor CRY1 Interacts with BZR1 and BIN2 to Modulate the Phosphorylation and Nuclear Function of BZR1 in Repressing BR Signaling in Arabidopsis. Mol. Plant 2019, 12, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, W.; Xu, P.; Pan, J.; Zhang, T.; Li, Y.; Li, G.; Yang, H.; Lian, H. phyB Interacts with BES1 to Regulate Brassinosteroid Signaling in Arabidopsis. Plant Cell Physiol. 2018, 60, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Liu, J.; He, G.; Liu, P.; Sun, J. Photoexcited phytochrome B interacts with brassinazole resistant 1 to repress brassinosteroid signaling in Arabidopsis. J. Integr. Plant Biol. 2019, 62, 652–667. [Google Scholar] [CrossRef]
- Li, J.; Terzaghi, W.; Gong, Y.; Li, C.; Ling, J.-J.; Fan, Y.; Qin, N.; Gong, X.; Zhu, D.; Deng, X.W. Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat. Commun. 2020, 11, 1592. [Google Scholar] [CrossRef] [Green Version]
- Liang, T.; Shi, C.; Peng, Y.; Tan, H.; Xin, P.; Yang, Y.; Wang, F.; Li, X.; Chu, J.; Huang, J.; et al. Brassinosteroid-Activated BRI1-EMS-SUPPRESSOR 1 Inhibits Flavonoid Biosynthesis and Coordinates Growth and UV-B Stress Responses in Plants. Plant Cell 2020, 32, 3224–3239. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, Y.; Hu, Y.; Yang, Y.; Cai, J.; Liu, H.; Zhang, C.; Liu, X.; Hou, X. Arabidopsis NF-YCs play dual roles in repressing brassinosteroid biosynthesis and signaling during light-regulated hypocotyl elongation. Plant Cell 2021, 33, 2360–2374. [Google Scholar] [CrossRef]
- Gupta, N.; Nath, U. Integration of light and hormone response during seedling establishment. J. Plant Biochem. Biotechnol. 2020, 29, 652–664. [Google Scholar] [CrossRef]
- De Wit, M.; Galvão, V.C.; Fankhauser, C. Light-Mediated Hormonal Regulation of Plant Growth and Development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef]
- Wu, Y.; Shi, L.; Li, L.; Fu, L.; Liu, Y.; Xiong, Y.; Sheen, J. Integration of nutrient, energy, light, and hormone signalling via TOR in plants. J. Exp. Bot. 2019, 70, 2227–2238. [Google Scholar] [CrossRef] [PubMed]
- Paik, I.; Kathare, P.K.; Kim, J.-I.; Huq, E. Expanding Roles of PIFs in Signal Integration from Multiple Processes. Mol. Plant 2017, 10, 1035–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Li, G.; Wang, H.; Deng, X.W. Phytochrome Signaling Mechanisms. Arab. Book 2011, 9, e0148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvão, V.C.; Fankhauser, C. Sensing the light environment in plants: Photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 2015, 34, 46–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osterlund, M.T.; Hardtke, C.; Wei, N.; Deng, X.W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nat. Cell Biol. 2000, 405, 462–466. [Google Scholar] [CrossRef]
- Xu, X.; Paik, I.; Zhu, L.; Huq, E. Illuminating Progress in Phytochrome-Mediated Light Signaling Pathways. Trends Plant Sci. 2015, 20, 641–650. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Jiang, Y.; Li, J.; Lin, F.; Holm, M.; Deng, X.W. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 7655–7660. [Google Scholar] [CrossRef] [Green Version]
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Lyu, M.; Luo, Y.; Liu, S.; Li, Y.; He, H.; Wei, N.; Deng, X.W.; Zhong, S. Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc. Natl. Acad. Sci. USA 2018, 115, 6482–6487. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.W.; Matsui, M.; Wei, N.; Wagner, D.; Chu, A.M.; Feldmann, K.A.; Quail, P.H. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 1992, 71, 791–801. [Google Scholar] [CrossRef]
- Laubinger, S.; Fittinghoff, K.; Hoecker, U. The SPA Quartet: A Family of WD-Repeat Proteins with a Central Role in Suppression of Photomorphogenesis in Arabidopsis. Plant Cell 2004, 16, 2293–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoecker, U.; Tepperman, J.M.; Quail, P.H. SPA1, a WD-Repeat Protein Specific to Phytochrome A Signal Transduction. Science 1999, 284, 496–499. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Maier, A.; Lee, J.-H.; Laubinger, S.; Saijo, Y.; Wang, H.; Qu, L.-J.; Hoecker, U.; Deng, X.W. Biochemical Characterization of Arabidopsis Complexes Containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA Proteins in Light Control of Plant Development. Plant Cell 2008, 20, 2307–2323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Huang, X.; Gusmaroli, G.; Terzaghi, W.; Lau, O.S.; Yanagawa, Y.; Zhang, Y.; Li, J.; Lee, J.-H.; Zhu, D.; et al. Arabidopsis CULLIN4-Damaged DNA Binding Protein 1 Interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA Complexes to Regulate Photomorphogenesis and Flowering Time. Plant Cell 2010, 22, 108–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Huang, X.; Deng, X.W. The Photomorphogenic Central Repressor COP1: Conservation and Functional Diversification during Evolution. Plant Commun. 2020, 1, 100044. [Google Scholar] [CrossRef]
- Qin, N.; Xu, D.; Li, J.; Deng, X.W. COP9 signalosome: Discovery, conservation, activity, and function. J. Integr. Plant Biol. 2020, 62, 90–103. [Google Scholar] [CrossRef]
- Chen, H.; Shen, Y.; Tang, X.; Yu, L.; Wang, J.; Guo, L.; Zhang, Y.; Zhang, H.; Feng, S.; Strickland, E.; et al. Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development. Plant Cell 2006, 18, 1991–2004. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.-D.; Zhou, C.-M.; Xu, P.-B.; Luo, Q.; Lian, H.-L.; Yang, H.-Q. Red-Light-Dependent Interaction of phyB with SPA1 Promotes COP1–SPA1 Dissociation and Photomorphogenic Development in Arabidopsis. Mol. Plant 2015, 8, 467–478. [Google Scholar] [CrossRef] [Green Version]
- Sheerin, D.J.; Menon, C.; zur Oven-Krockhaus, S.; Enderle, B.; Zhu, L.; Johnen, P.; Schleifenbaum, F.; Stierhof, Y.D.; Huq, E.; Hiltbrunner, A. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 2015, 27, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Z.; Liu, H.; Liu, B.; Liu, X.; Lin, C. Blue Light-Dependent Interaction of CRY2 with SPA1 Regulates COP1 activity and Floral Initiation in Arabidopsis. Curr. Biol. 2011, 21, 841–847. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Zuo, Z.; Liu, H.; Liu, X.; Lin, C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011, 25, 1029–1034. [Google Scholar] [CrossRef] [Green Version]
- Lian, H.-L.; He, S.-B.; Zhang, Y.-C.; Zhu, D.-M.; Zhang, J.-Y.; Jia, K.-P.; Sun, S.-X.; Li, L.; Yang, H.-Q. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011, 25, 1023–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponnu, J.; Riedel, T.; Penner, E.; Schrader, A.; Hoecker, U. Cryptochrome 2 competes with COP1 substrates to repress COP1 ubiquitin ligase activity during Arabidopsis photomorphogenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 201909181. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.; Podolec, R.; Chappuis, R.; Ulm, R.; Hothorn, M. Plant photoreceptors and their signaling components compete for COP 1 binding via VP peptide motifs. EMBO J. 2019, 38, e102140. [Google Scholar] [CrossRef]
- Ren, H.; Han, J.; Yang, P.; Mao, W.; Liu, X.; Qiu, L.; Qian, C.; Liu, Y.; Chen, Z.; Ouyang, X.; et al. Two E3 ligases antagonistically regulate the UV-B response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 4722–4731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Ouyang, X.; Yang, P.; Lau, O.S.; Chen, L.; Wei, N.; Deng, X.W. Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc. Natl. Acad. Sci. USA 2013, 110, 16669–16674. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.D.; Kim, M.R.; Kang, M.Y.; Cha, J.Y.; Han, S.H.; Nawkar, G.M.; Sakuraba, Y.; Lee, S.Y.; Imaizumi, T.; McClung, C.R.; et al. The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering. Nat. Commun. 2017, 8, 2259. [Google Scholar] [CrossRef] [Green Version]
- Pacín, M.; Legris, M.; Casal, J.J. Rapid Decline in Nuclear COSTITUTIVE PHOTOMORPHOGENESIS1 Abundance Anticipates the Stabilization of Its Target ELONGATED HYPOCOTYL5 in the Light. Plant Physiol. 2014, 164, 1134–1138. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Lory, N.; Stauber, J.; Hoecker, U. Photoreceptor Specificity in the Light-Induced and COP1-Mediated Rapid Degradation of the Repressor of Photomorphogenesis SPA2 in Arabidopsis. PLoS Genet. 2015, 11, e1005516. [Google Scholar] [CrossRef] [Green Version]
- Podolec, R.; Ulm, R. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 2018, 45, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Al-Sady, B.; Ni, W.; Kircher, S.; Schäfer, E.; Quail, P.H. Photoactivated Phytochrome Induces Rapid PIF3 Phosphorylation Prior to Proteasome-Mediated Degradation. Mol. Cell 2006, 23, 439–446. [Google Scholar] [CrossRef]
- Ma, D.; Li, X.; Guo, Y.; Chu, J.; Fang, S.; Yan, C.; Noel, J.P.; Liu, H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 2016, 113, 224–229. [Google Scholar] [CrossRef] [Green Version]
- Ni, W.; Xu, S.-L.; Tepperman, J.M.; Stanley, D.J.; Maltby, D.A.; Gross, J.D.; Burlingame, A.L.; Wang, Z.-Y.; Quail, P.H. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 2014, 344, 1160–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedmale, U.V.; Huang, S.-S.C.; Zander, M.; Cole, B.J.; Hetzel, J.; Ljung, K.; Reis, P.A.; Sridevi, P.; Nito, K.; Nery, J.R.; et al. Cryptochromes Interact Directly with PIFs to Control Plant Growth in Limiting Blue Light. Cell 2016, 164, 233–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, A.-Y.; Han, Y.-J.; Baek, A.; Ahn, T.; Kim, S.Y.; Nguyen, T.S.; Son, M.; Lee, K.W.; Shen, Y.; Song, P.-S.; et al. Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun. 2016, 7, 11545. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Xu, S.-L.; Grandío, E.G.; Chalkley, R.; Huhmer, A.F.R.; Burlingame, A.L.; Wang, Z.-Y.; Quail, P.H. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 2017, 8, 15236. [Google Scholar] [CrossRef]
- Dong, J.; Ni, W.; Yu, R.; Deng, X.W.; Chen, H.; Wei, N. Light-Dependent Degradation of PIF3 by SCF(EBF1/2) Promotes a Photomorphogenic Response in Arabidopsis. Curr. Biol. 2017, 27, 2420–2430.e6. [Google Scholar] [CrossRef] [Green Version]
- Lorrain, S.; Allen, T.; Duek, P.D.; Whitelam, G.C.; Fankhauser, C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2007, 53, 312–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Bu, Q.; Xu, X.; Paik, I.; Huang, X.; Hoecker, U.; Deng, X.W.; Huq, E. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 2015, 6, 7245. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.; Park, E.; Song, K.; Bae, G.; Choi, G. PHYTOCHROME INTERACTING FACTOR8 Inhibits Phytochrome A-Mediated Far-Red Light Responses in Arabidopsis. Plant Cell 2020, 32, 186–205. [Google Scholar] [CrossRef]
- Xu, X.; Paik, I.; Zhu, L.; Bu, Q.; Huang, X.; Deng, X.W.; Huq, E. PHYTOCHROME INTERACTING FACTOR1 Enhances the E3 Ligase Activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to Synergistically Repress Photomorphogenesis in Arabidopsis. Plant Cell 2014, 26, 1992–2006. [Google Scholar] [CrossRef] [Green Version]
- Paik, I.; Chen, F.; Ngoc Pham, V.; Zhu, L.; Kim, J.I.; Huq, E. A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat. Commun. 2019, 10, 4216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorrain, S.; Trevisan, M.; Pradervand, S.; Fankhauser, C. Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 2009, 60, 449–461. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Holmlund, M.; Lorrain, S.; Norberg, M.; Bakó, L.; Fankhauser, C.; Nilsson, O. BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. eLife 2017, 6, e26759. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Khanna, R.; Carle, C.M.; Quail, P.H. Phytochrome Induces Rapid PIF5 Phosphorylation and Degradation in Response to Red-Light Activation. Plant Physiol. 2007, 145, 1043–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Li, C.; Dong, X.; Li, H.; Zhang, D.; Zhou, Y.; Jiang, B.; Peng, J.; Qin, X.; Cheng, J.; et al. MYB30 Is a Key Negative Regulator of Arabidopsis Photomorphogenic Development That Promotes PIF4 and PIF5 Protein Accumulation in the Light. Plant Cell 2020, 32, 2196–2215. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.Q.; Heng, Y.Q.; Bian, Y.T.; Xiao, Y.T.; Liu, J.J.; Zhao, X.H.; Jiang, Y.; Deng, X.W.; Xu, D.Q. BBX11 promotes red light-mediated photomorphogenic development by modulating phyB-PIF4 signaling. aBIOTECH 2021, 2, 117–130. [Google Scholar] [CrossRef]
- Yue, J.; Qin, Q.; Meng, S.; Jing, H.; Gou, X.; Li, J.; Hou, S. TOPP4 Regulates the Stability of PHYTOCHROME INTERACTING FACTOR5 during Photomorphogenesis in Arabidopsis. Plant Physiol. 2016, 170, 1381–1397. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Dong, J.; Deng, Z.; Jiang, Y.; Wu, C.; Qin, X.; Terzaghi, W.; Chen, H.; Dai, M.; Deng, X.W. Arabidopsis PP6 phosphatases dephosphorylate PIF proteins to repress photomorphogenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 20218–20225. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Xun, Q.; Zhang, D.; Lv, M.; Ou, Y.; Li, J. TCP Transcription Factors Associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to Regulate Thermomorphogenesis in Arabidopsis thaliana. iScience 2019, 15, 600–610. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Yu, H.; Yuan, R.; Yang, Y.; An, F.; Qin, G. Arabidopsis Transcription Factor TCP5 Controls Plant Thermomorphogenesis by Positively Regulating PIF4 Activity. iScience 2019, 15, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Zhang, D.; An, J.; Yin, H.; Fang, S.; Chu, J.; Zhao, Y.; Li, J. TCP Transcription Factors Regulate Shade Avoidance via Directly Mediating the Expression of Both PHYTOCHROME INTERACTING FACTORs and Auxin Biosynthetic Genes. Plant Physiol. 2018, 176, 1850–1861. [Google Scholar] [CrossRef] [Green Version]
- Zhai, H.; Xiong, L.; Li, H.; Lyu, X.; Yang, G.; Zhao, T.; Liu, J.; Liu, B. Cryptochrome 1 Inhibits Shoot Branching by Repressing the Self-Activated Transciption Loop of PIF4 in Arabidopsis. Plant Commun. 2020, 1, 100042. [Google Scholar] [CrossRef]
- Li, L.; Ljung, K.; Breton, G.; Schmitz, R.J.; Pruneda-Paz, J.; Cowing-Zitron, C.; Cole, B.J.; Ivans, L.J.; Pedmale, U.V.; Jung, H.-S.; et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012, 26, 785–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leivar, P.; Monte, E.; Al-Sady, B.; Carle, C.; Storer, A.; Alonso, J.M.; Ecker, J.R.; Quail, P.H. The Arabidopsis Phytochrome-Interacting Factor PIF7, Together with PIF3 and PIF4, Regulates Responses to Prolonged Red Light by Modulating phyB Levels. Plant Cell 2008, 20, 337–352. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Zhang, Q.; Jiang, Y.; Yang, C.; Wang, Z.-Y.; Li, L. Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. eLife 2018, 7, e31636. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677. [Google Scholar] [CrossRef] [Green Version]
- Zhong, S.; Shi, H.; Xue, C.; Wang, L.; Xi, Y.; Li, J.; Quail, P.H.; Deng, X.W.; Guo, H. A Molecular Framework of Light-Controlled Phytohormone Action in Arabidopsis. Curr. Biol. 2012, 22, 1530–1535. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Liu, R.; Xue, C.; Shen, X.; Wei, N.; Deng, X.W.; Zhong, S. Seedlings Transduce the Depth and Mechanical Pressure of Covering Soil Using COP1 and Ethylene to Regulate EBF1/EBF2 for Soil Emergence. Curr. Biol. 2016, 26, 139–149. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Ma, M.; Feng, Y.; Li, H.; Wang, Y.; Ma, Y.; Li, M.; An, F.; Guo, H. EIN2-Directed Translational Regulation of Ethylene Signaling in Arabidopsis. Cell 2015, 163, 670–683. [Google Scholar] [CrossRef] [Green Version]
- Potuschak, T.; Lechner, E.; Parmentier, Y.; Yanagisawa, S.; Grava, S.; Koncz, C.; Genschik, P. EIN3-Dependent Regulation of Plant Ethylene Hormone Signaling by Two Arabidopsis F Box Proteins: EBF1 and EBF2. Cell 2003, 115, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Ji, Y.; Xue, C.; Ma, H.; Xi, Y.; Huang, P.; Wang, H.; An, F.; Li, B.; Wang, Y.; et al. Integrated Regulation of Apical Hook Development by Transcriptional Coupling of EIN3/EIL1 and PIFs in Arabidopsis. Plant Cell 2018, 30, 1971–1988. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Shen, X.; Liu, R.; Xue, C.; Wei, N.; Deng, X.W.; Zhong, S. The Red Light Receptor Phytochrome B Directly Enhances Substrate-E3 Ligase Interactions to Attenuate Ethylene Responses. Dev. Cell 2016, 39, 597–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Z.; Bian, Y.; Liu, J.; Sun, Y.; Xu, D. B-box proteins: Pivotal players in light-mediated development in plants. J. Integr. Plant Biol. 2020, 62, 1293–1309. [Google Scholar] [CrossRef] [PubMed]
- Xu, D. COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol. 2020, 228, 1748–1753. [Google Scholar] [CrossRef]
- Bursch, K.; Toledo-Ortiz, G.; Pireyre, M.; Lohr, M.; Braatz, C.; Johansson, H. Identification of BBX proteins as rate-limiting cofactors of HY5. Nat Plants 2020, 6, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huai, J.; Shang, F.; Xu, G.; Tang, W.; Jing, Y.; Lin, R. A PIF1/PIF3-HY5-BBX23 Transcription Factor Cascade Affects Photomorphogenesis. Plant Physiol. 2017, 174, 2487–2500. [Google Scholar] [CrossRef] [Green Version]
- Lin, F.; Jiang, Y.; Li, J.; Yan, T.; Fan, L.; Liang, J.; Chen, Z.J.; Xu, D.; Deng, X.W. B-BOX DOMAIN PROTEIN28 Negatively Regulates Photomorphogenesis by Repressing the Activity of Transcription Factor HY5 and Undergoes COP1-Mediated Degradation. Plant Cell 2018, 30, 2006–2019. [Google Scholar] [CrossRef] [Green Version]
- Heng, Y.; Lin, F.; Jiang, Y.; Ding, M.; Yan, T.; Lan, H.; Zhou, H.; Zhao, X.; Xu, D.; Deng, X.W. B-Box Containing Proteins BBX30 and BBX31, Acting Downstream of HY5, Negatively Regulate Photomorphogenesis in Arabidopsis. Plant Physiol. 2019, 180, 497–508. [Google Scholar] [CrossRef]
- Song, Z.; Yan, T.; Liu, J.; Bian, Y.; Heng, Y.; Lin, F.; Jiang, Y.; Wang Deng, X.; Xu, D. BBX28/BBX29, HY5 and BBX30/31 form a feedback loop to fine-tune photomorphogenic development. Plant J. 2020, 104, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Q.; Sarmast, M.K.; Jiang, J.; Dehesh, K. The Transcriptional Regulator BBX19 Promotes Hypocotyl Growth by Facilitating COP1-Mediated EARLY FLOWERING3 Degradation in Arabidopsis. Plant Cell 2015, 27, 1128–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.M.; Zeng, J.X.; Deng, K.Q.; Tu, X.J.; Zhao, X.Y.; Tang, D.Y.; Liu, X.M. DBB1a, involved in gibberellin homeostasis, functions as a negative regulator of blue light-mediated hypocotyl elongation in Arabidopsis. Planta 2011, 233, 13–23. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Li, J. Regulation of Brassinosteroid Biosynthesis and InactivationF. J. Integr. Plant Biol. 2012, 54, 746–759. [Google Scholar] [CrossRef]
- Ohnishi, T.; Godza, B.; Watanabe, B.; Fujioka, S.; Hategan, L.; Ide, K.; Shibata, K.; Yokota, T.; Szekeres, M.; Mizutani, M. CYP90A1/CPD, a Brassinosteroid Biosynthetic Cytochrome P450 of Arabidopsis, Catalyzes C-3 Oxidation. J. Biol. Chem. 2012, 287, 31551–31560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Chory, J. A Putative Leucine-Rich Repeat Receptor Kinase Involved in Brassinosteroid Signal Transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.; Walker, J.C. BAK1, an Arabidopsis LRR Receptor-like Protein Kinase, Interacts with BRI1 and Modulates Brassinosteroid Signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Nam, K.H.; Li, J. BRI1/BAK1, a Receptor Kinase Pair Mediating Brassinosteroid Signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, T.; Canodelgado, A.I.; Seto, H.; Hiranuma, S.; Fujioka, S.; Yoshida, S.; Chory, J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nat. Cell Biol. 2005, 433, 167–171. [Google Scholar] [CrossRef]
- Hothorn, M.; Belkhadir, Y.; Dreux, M.; Dabi, T.; Noel, J.P.; Wilson, I.A.; Chory, J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nat. Cell Biol. 2011, 474, 467–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- She, J.; Han, Z.; Kim, T.-W.; Wang, J.; Cheng, W.; Chang, J.; Shi, S.; Wang, J.; Yang, M.; Wang, Z.-Y.; et al. Structural insight into brassinosteroid perception by BRI1. Nat. Cell Biol. 2011, 474, 472–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-W.; Guan, S.; Burlingame, A.L.; Wang, Z.-Y. The CDG1 Kinase Mediates Brassinosteroid Signal Transduction from BRI1 Receptor Kinase to BSU1 Phosphatase and GSK3-like Kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.-X.; Sun, Y.; Burlingame, A.L.; Wang, Z.-Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009, 11, 1254–1260. [Google Scholar] [CrossRef]
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar] [CrossRef]
- Zhu, J.-Y.; Li, Y.; Cao, D.-M.; Yang, H.; Oh, E.; Bi, Y.; Zhu, S.; Wang, Z.-Y. The F-box Protein KIB1 Mediates Brassinosteroid-Induced Inactivation and Degradation of GSK3-like Kinases in Arabidopsis. Mol. Cell 2017, 66, 648–657.e4. [Google Scholar] [CrossRef] [Green Version]
- He, J.-X.; Gendron, J.M.; Yang, Y.; Li, J.; Wang, Z.-Y. The GSK3-Like Kinase BIN2 Phosphorylates and Destabilizes BZR1, a Positive Regulator of the Brassinosteroid Signaling Pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-Localized BZR1 Mediates Brassinosteroid-Induced Growth and Feedback Suppression of Brassinosteroid Biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Wang, Z.-Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 Accumulates in the Nucleus in Response to Brassinosteroids to Regulate Gene Expression and Promote Stem Elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef] [Green Version]
- He, J.-X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.L.; Gendron, N.; Sun, C.Q.; Wang, Z.-Y. BZR1 Is a Transcriptional Repressor with Dual Roles in Brassinosteroid Homeostasis and Growth Responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Fan, X.-Y.; Cao, D.-M.; Tang, W.; He, K.; Zhu, J.-Y.; He, J.-X.; Bai, M.-Y.; Zhu, S.; Oh, E.; et al. Integration of Brassinosteroid Signal Transduction with the Transcription Network for Plant Growth Regulation in Arabidopsis. Dev. Cell 2010, 19, 765–777. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Li, L.; Zola, J.; Aluru, M.; Ye, H.; Foudree, A.; Guo, H.; Anderson, S.; Aluru, S.; Liu, P.; et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2010, 65, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.-Y.; Zhang, L.-Y.; Gampala, S.S.; Zhu, S.-W.; Song, W.-Y.; Chong, K.; Wang, Z.-Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844. [Google Scholar] [CrossRef] [Green Version]
- Gampala, S.S.; Kim, T.-W.; He, J.-X.; Tang, W.; Deng, Z.; Bai, M.-Y.; Guan, S.; Lalonde, S.; Sun, Y.; Gendron, J.M.; et al. An Essential Role for 14-3-3 Proteins in Brassinosteroid Signal Transduction in Arabidopsis. Dev. Cell 2007, 13, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Liu, M.; Yuan, M.; Oses-Prieto, J.A.; Cai, X.; Sun, Y.; Burlingame, A.L.; Wang, Z.Y.; Tang, W. The Brassinosteroid-Activated BRI1 Receptor Kinase Is Switched off by Dephosphorylation Mediated by Cytoplasm-Localized PP2A B’ Subunits. Mol. Plant 2016, 9, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, R.; Liu, M.; Yuan, W.; Zhao, Z.; Liu, X.; Peng, Y.; Yang, X.; Sun, Y.; Tang, W. Nucleocytoplasmic trafficking and turnover mechanisms of BRASSINAZOLE RESISTANT1 in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chory, J.; Yi, P.; Han, Z.; Li, X.; Olson, E.N. Brassinosteroids Regulate Dissociation of BKI1, a Negative Regulator of BRI1 Signaling, from the Plasma Membrane. Science 2006, 313, 1118–1122. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.; Meisenhelder, J.; Hunter, T.; Yoshida, S.; Asami, T.; Chory, J. Autoregulation and Homodimerization Are Involved in the Activation of the Plant Steroid Receptor BRI1. Dev. Cell 2005, 8, 855–865. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Zhou, X.-Y.; Li, L.; Xue, L.-J.; Yang, X.; Xue, H.-W. Genome-Wide Analysis Revealed the Complex Regulatory Network of Brassinosteroid Effects in Photomorphogenesis. Mol. Plant 2009, 2, 755–772. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-G.; Yun, J.; Kim, D.-H.; Chung, K.-S.; Fujioka, S.; Kim, J.-I.; Dae, H.-W.; Yoshida, S.; Takatsuto, S.; Song, P.-S.; et al. Light and Brassinosteroid Signals Are Integrated via a Dark-Induced Small G Protein in Etiolated Seedling Growth. Cell 2001, 105, 625–636. [Google Scholar] [CrossRef] [Green Version]
- Park, D.H.; Lim, P.O.; Kim, J.S.; Cho, D.S.; Hong, S.H.; Gil Nam, H. The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 2003, 34, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Yuan, T.; Tarkowská, D.; Kim, J.; Gil Nam, H.; Novak, O.; He, K.; Gou, X.; Li, J. Brassinosteroid Biosynthesis Is Modulated via a Transcription Factor Cascade of COG1, PIF4, and PIF5. Plant Physiol. 2017, 174, 1260–1273. [Google Scholar] [CrossRef] [PubMed]
- Lease, K.A.; Wen, J.; Li, J.; Doke, J.T.; Liscum, E.; Walker, J.C. A mutant Arabidopsis heterotrimeric G-protein beta subunit affects leaf, flower, and fruit development. Plant Cell 2001, 13, 2631–2641. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.M.; Ecker, J.; Chen, J.-G. A Reevaluation of the Role of the Heterotrimeric G Protein in Coupling Light Responses in Arabidopsis. Plant Physiol. 2003, 131, 1623–1627. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.B.; Gao, S.Q.; Ma, Y.N.; Wang, X.T.; Feng, L.; Li, L.C.; Xu, Z.S.; Chen, Y.F.; Chen, M.; Ma, Y.Z. The G-Protein beta Subunit AGB1 Promotes Hypocotyl Elongation through Inhibiting Transcription Activation Function of BBX21 in Arabidopsis. Mol. Plant 2017, 10, 1206–1223. [Google Scholar] [CrossRef] [Green Version]
- Lian, H.L.; Xu, P.B.; He, S.B.; Wu, J.; Pan, J.; Wang, W.X.; Xu, F.; Wang, S.; Pan, J.S.; Huang, J.R.; et al. Photoexcited CRYPTOCHROME 1 Interacts Directly with G-Protein beta Subunit AGB1 to Regulate the DNA-Binding Activity of HY5 and Photomorphogenesis in Arabidopsis. Mol. Plant 2018, 11, 1248–1263. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Lian, H.; Xu, F.; Zhang, T.; Wang, S.; Wang, W.; Du, S.; Huang, J.; Yang, H.-Q. Phytochrome B and AGB1 Coordinately Regulate Photomorphogenesis by Antagonistically Modulating PIF3 Stability in Arabidopsis. Mol. Plant 2019, 12, 229–247. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Xu, P.; Wang, W.; Wang, S.; Caruana, J.C.; Yang, H.Q.; Lian, H. Arabidopsis G-Protein beta Subunit AGB1 Interacts with BES1 to Regulate Brassinosteroid Signaling and Cell Elongation. Front. Plant Sci. 2017, 8, 2225. [Google Scholar] [CrossRef] [Green Version]
- Holtan, H.E.; Bandong, S.; Marion, C.M.; Adam, L.; Tiwari, S.; Shen, Y.; Maloof, J.N.; Maszle, D.R.; Ohto, M.A.; Preuss, S.; et al. BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011, 156, 2109–2123. [Google Scholar] [CrossRef] [Green Version]
- Ravindran, N.; Ramachandran, H.; Job, N.; Yadav, A.; Vaishak, K.; Datta, S. B-box protein BBX32 integrates light and brassinosteroid signals to inhibit cotyledon opening. Plant Physiol. 2021, 187, 446–461. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, S.; Zhu, W.; Jia, K.; Yang, H.-Q.; Wang, X. Strigolactone/MAX2-Induced Degradation of Brassinosteroid Transcriptional Effector BES1 Regulates Shoot Branching. Dev. Cell 2013, 27, 681–688. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.J.; Lee, S.H.; Park, C.H.; Kim, S.H.; Hsu, C.C.; Xu, S.L.; Wang, Z.Y.; Kim, S.K.; Kim, T.W. Plant U-Box40 Mediates Degradation of the Brassinosteroid-Responsive Transcription Factor BZR1 in Arabidopsis Roots. Plant Cell 2019, 31, 791–808. [Google Scholar] [CrossRef]
- Yang, M.; Li, C.; Cai, Z.; Hu, Y.; Nolan, T.; Yu, F.; Yin, Y.; Xie, Q.; Tang, G.; Wang, X. SINAT E3 Ligases Control the Light-Mediated Stability of the Brassinosteroid-Activated Transcription Factor BES1 in Arabidopsis. Dev. Cell 2017, 41, 47–58.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.; Jeong, Y.J.; Corvalan, C.; Fujioka, S.; Cho, S.; Park, T.; Choe, S. Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis. Plant J. 2014, 77, 737–747. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Tian, Y.; Shi, W.; Yu, P.; Hu, Y.; Lv, J.; Fu, C.; Fan, M.; Bai, M.-Y. The miR396-GRFs Module Mediates the Prevention of Photo-oxidative Damage by Brassinosteroids during Seedling De-Etiolation in Arabidopsis. Plant Cell 2020, 32, 2525–2542. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yan, B.; Dong, H.; He, G.; Zhou, Y.; Sun, J. BIC 1 acts as a transcriptional coactivator to promote brassinosteroid signaling and plant growth. EMBO J. 2021, 40, e104615. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.J.; Li, J.; Zhu, D.; Deng, X.W. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. USA 2017, 114, 3539–3544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibañez, C.; Delker, C.; Martinez, C.; Bürstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M.; et al. Brassinosteroids Dominate Hormonal Regulation of Plant Thermomorphogenesis via BZR1. Curr. Biol. 2018, 28, 303–310.e3. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-F.; He, J.-X. BZR1 Interacts with HY5 to Mediate Brassinosteroid- and Light-Regulated Cotyledon Opening in Arabidopsis in Darkness. Mol. Plant 2016, 9, 113–125. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.-M.; Lin, W.-H.; Zhu, S.; Zhu, J.-Y.; Sun, Y.; Fan, X.-Y.; Cheng, M.; Hao, Y.; Oh, E.; Tian, M.; et al. Integration of Light- and Brassinosteroid-Signaling Pathways by a GATA Transcription Factor in Arabidopsis. Dev. Cell 2010, 19, 872–883. [Google Scholar] [CrossRef] [Green Version]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.-H. The Arabidopsis B-Box Zinc Finger Family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F., 3rd; Mantovani, R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.-L.; Wu, C.-C.; Huang, H.-C.; Chen, H.-J.; Hsieh, H.-L.; Juan, H.-F. Identification of MicroRNA 395a in 24-Epibrassinolide-Regulated Root Growth of Arabidopsis thaliana Using MicroRNA Arrays. Int. J. Mol. Sci. 2013, 14, 14270–14286. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Zheng, Y.; Tang, Q.; Zhong, S.; Su, W.; Zheng, B. Brassinosteroids inhibit miRNA-mediated translational repression by decreasing AGO1 on the endoplasmic reticulum. J. Integr. Plant. Biol. 2021, 63, 1475–1490. [Google Scholar] [CrossRef]
- Xia, K.; Ou, X.; Tang, H.; Wang, R.; Wu, P.; Jia, Y.; Wei, X.; Xu, X.; Kang, S.H.; Kim, S.K.; et al. Rice microRNA osa-miR1848 targets the obtusifoliol 14alpha-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress. New Phytol. 2015, 208, 790–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, C.; Luo, D.; Lin, A.; Zhang, C.; Shan, L.; He, P.; Li, B.; Zhang, Q.; Hua, B.; Yuan, Z.; et al. A tomato B-box protein SlBBX20 modulates carotenoid biosynthesis by directly activating PHYTOENE SYNTHASE 1, and is targeted for 26S proteasome-mediated degradation. New Phytol. 2019, 221, 279–294. [Google Scholar] [CrossRef] [Green Version]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Hao, Y.J. MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol. J. 2019, 17, 2231–2233. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 2014, 240, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef] [Green Version]
- Bai, S.; Tao, R.; Yin, L.; Ni, J.; Yang, Q.; Yan, X.; Yang, F.; Guo, X.; Li, H.; Teng, Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef]
- Preuss, S.B.; Meister, R.; Xu, Q.; Urwin, C.P.; Tripodi, F.A.; Screen, S.E.; Anil, V.S.; Zhu, S.; Morrell, J.A.; Liu, G.; et al. Expression of the Arabidopsis thaliana BBX32 Gene in Soybean Increases Grain Yield. PLoS ONE 2012, 7, e30717. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, F.; Cao, J.; Yuan, J.; Liang, Y.; Li, J. Integration of Light and Brassinosteroid Signaling during Seedling Establishment. Int. J. Mol. Sci. 2021, 22, 12971. https://doi.org/10.3390/ijms222312971
Lin F, Cao J, Yuan J, Liang Y, Li J. Integration of Light and Brassinosteroid Signaling during Seedling Establishment. International Journal of Molecular Sciences. 2021; 22(23):12971. https://doi.org/10.3390/ijms222312971
Chicago/Turabian StyleLin, Fang, Jing Cao, Jiale Yuan, Yuxia Liang, and Jia Li. 2021. "Integration of Light and Brassinosteroid Signaling during Seedling Establishment" International Journal of Molecular Sciences 22, no. 23: 12971. https://doi.org/10.3390/ijms222312971
APA StyleLin, F., Cao, J., Yuan, J., Liang, Y., & Li, J. (2021). Integration of Light and Brassinosteroid Signaling during Seedling Establishment. International Journal of Molecular Sciences, 22(23), 12971. https://doi.org/10.3390/ijms222312971





