hnRNPA2B1-Mediated Extracellular Vesicles Sorting of miR-122-5p Potentially Promotes Lung Cancer Progression
Abstract
:1. Introduction
2. Results
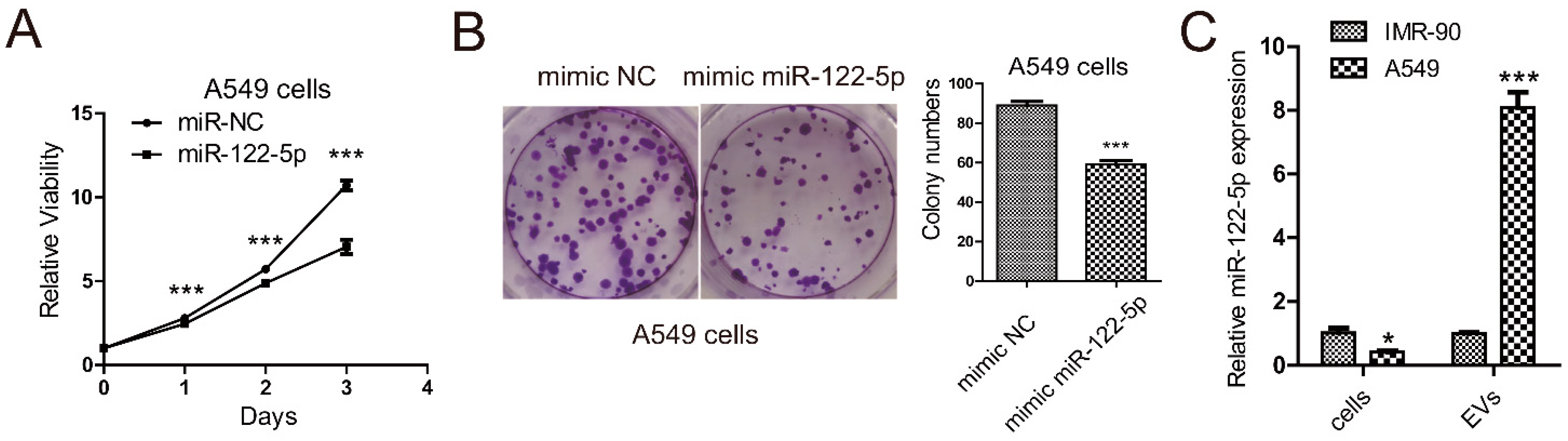
2.1. miR-122-5p Is Highly Enriched in EVs from Lung Cancer Cells
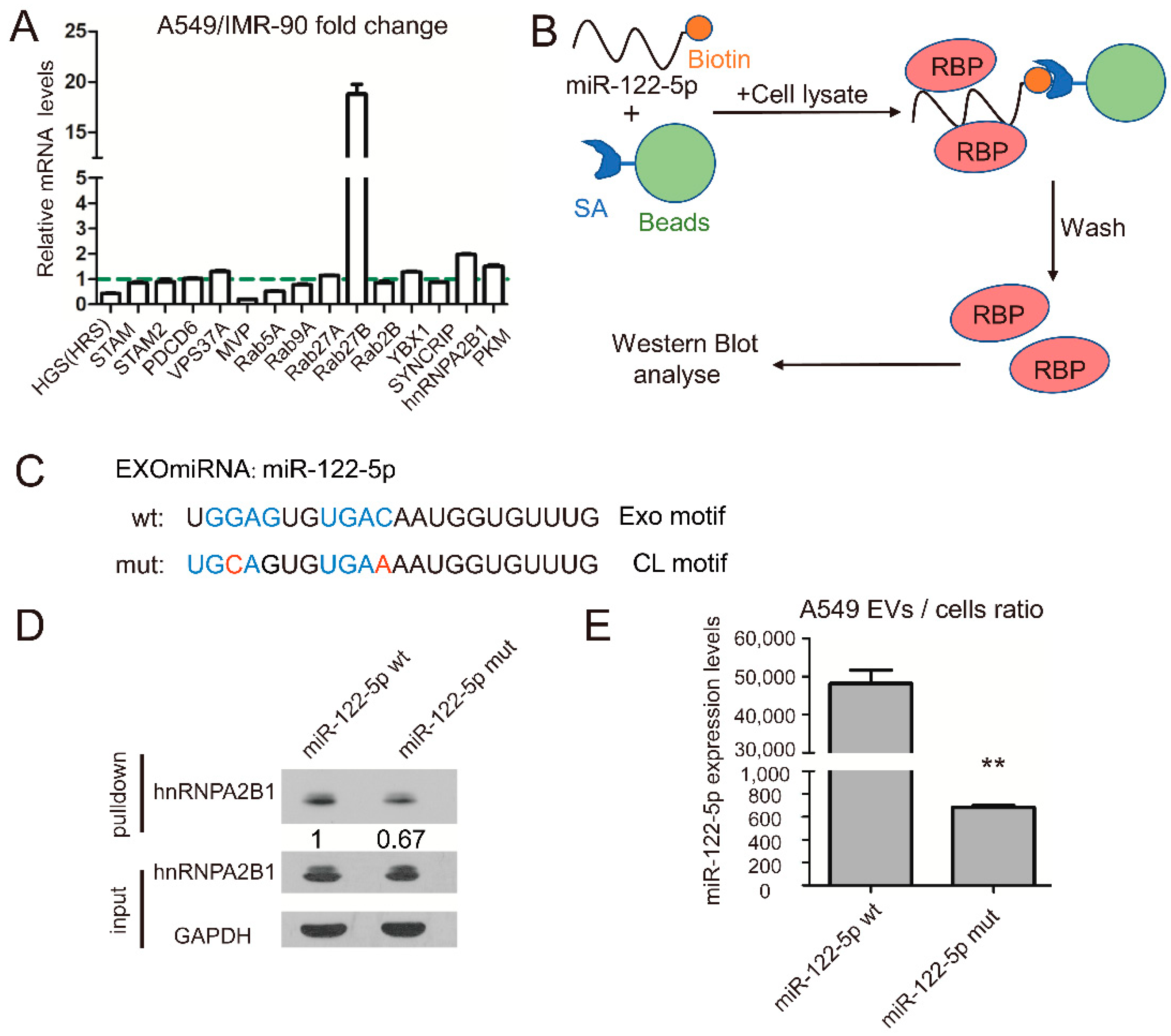
2.2. RNA-Binding Protein hnRNPA2B1 Interact with miR-122 through EXO-Motif
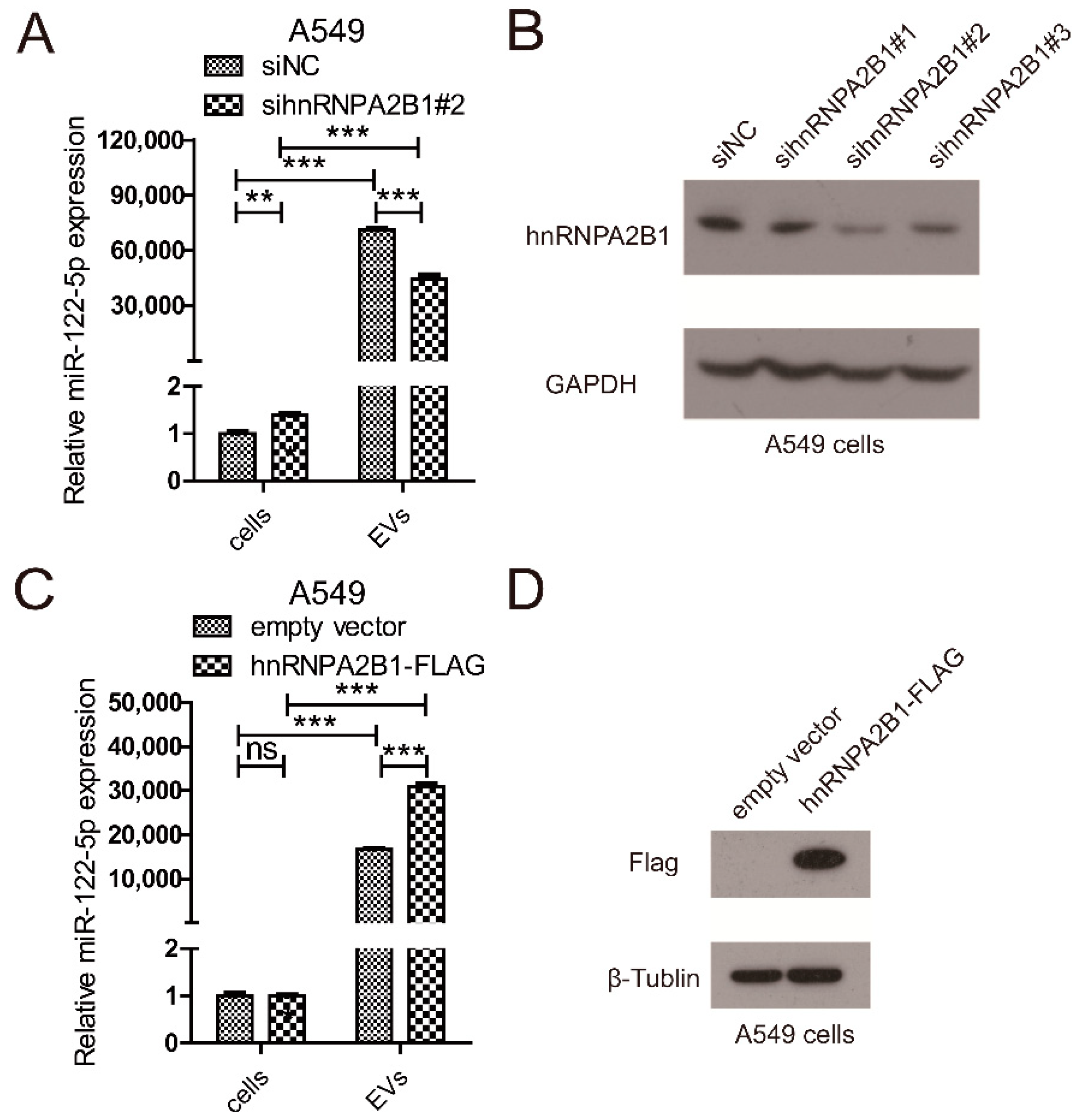
2.3. hnRNPA2B1 Regulates the Selective Secretion of miR-122-5p into Lung Cancer EVs
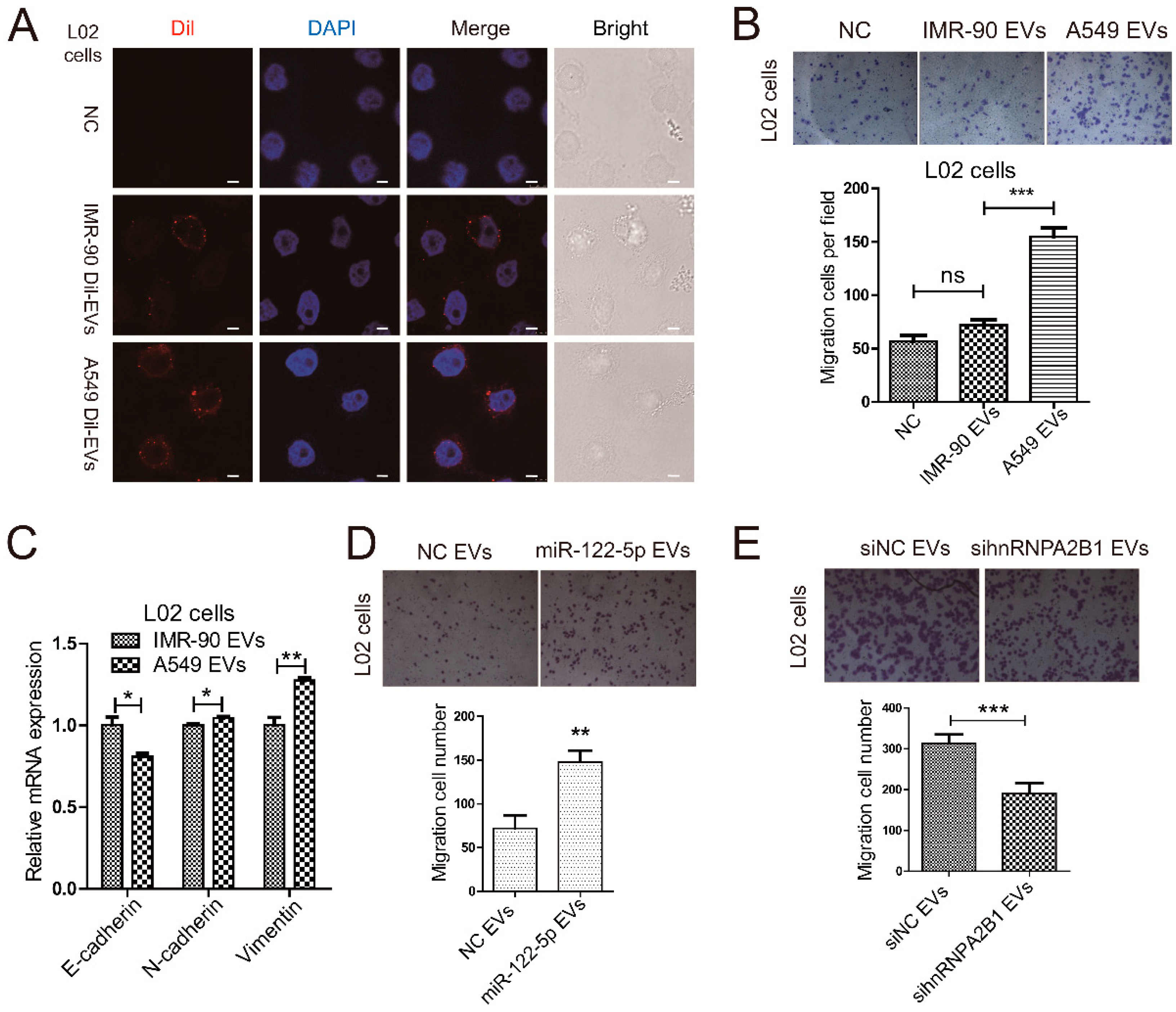
2.4. Lung Cancer Cells Secret miR-122-5p-Enriched EVs to Promote Migration of Normal Liver Cells
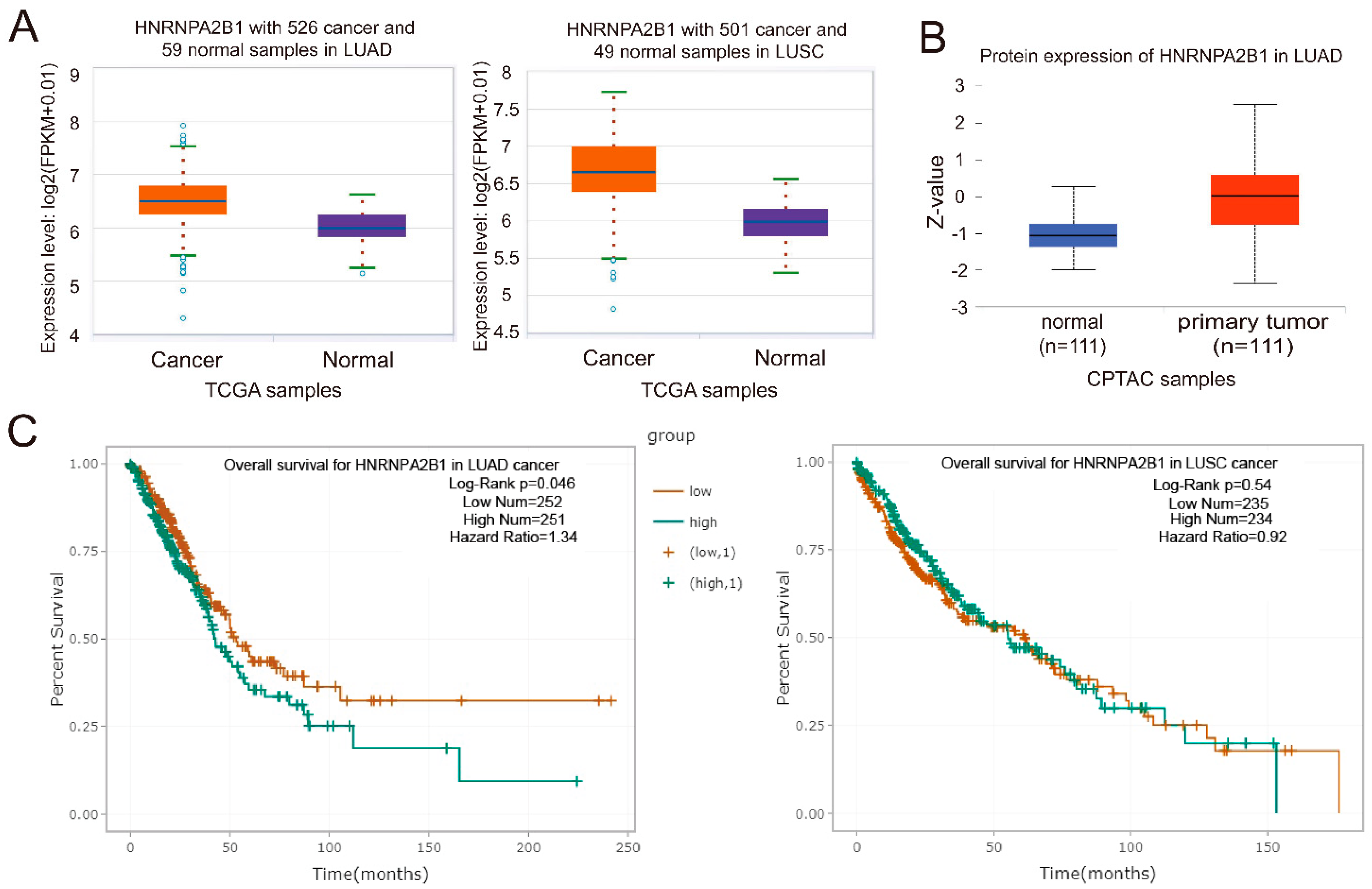
2.5. hnRNPA2B1 Is Closely Associated with the Prognosis of Lung Cancer Patients
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. EVs Isolation
4.3. Transmission Electron Microscopy
4.4. EVs Size Analysis
4.5. Western Blot Analysis
4.6. EVs Uptake Analysis
4.7. Quantitative Real-Time PCR
4.8. Oligonucleotide Transfection
4.9. Cell Migration Assay
4.10. Cell Proliferation Assay
4.11. miRNA Mimic and siRNA
4.12. miRNA-Protein Pulldown
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maas, S.L.; Breakefield, X.O.; Weaver, A.M. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [Green Version]
- Tkach, M.; Thery, C. Communication by extracellular vesicles: Where we are and where we need to go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Et Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Fu, H.; Xu, W.; Zhang, X. Exosomal non-coding RNAs: A promising cancer biomarker. Clin. Chem. Lab. Med. 2016, 54, 1871–1879. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA: A Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [Green Version]
- Heuvers, M.E.; Wisnivesky, J.; Stricker, B.H.; Aerts, J.G. Generalizability of results from the National Lung Screening Trial. Eur. J. Epidemiol. 2012, 27, 669–672. [Google Scholar] [CrossRef]
- Thind, A.; Wilson, C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 2016, 5, 31292. [Google Scholar] [CrossRef]
- Cazzoli, R.; Buttitta, F.; Di Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.N.; Pass, H.I. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2013, 8, 1156–1162. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Sai, B.; Wang, F.; Wang, L.; Wang, Y.; Zheng, L.; Li, G.; Tang, J.; Xiang, J. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer 2019, 18, 40. [Google Scholar] [CrossRef] [Green Version]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Nolte-’t Hoen, E.N.; Buermans, H.P.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.; ’t Hoen, P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Balkom, B.W.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M.C. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J Extracell Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef]
- Qin, X.; Guo, H.; Wang, X.; Zhu, X.; Yan, M.; Wang, X.; Xu, Q.; Shi, J.; Lu, E.; Chen, W.; et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, L.; Giurato, G.; Cicchini, C.; Montaldo, C.; Mancone, C.; Tarallo, R.; Battistelli, C.; Alonzi, T.; Weisz, A.; Tripodi, M. The RNA-binding orotein SYNCRIP is a component of the hepatocyte exosomal machinery controlling MicroRNA sorting. Cell Rep. 2016, 17, 799–808. [Google Scholar] [CrossRef] [Green Version]
- Hobor, F.; Dallmann, A.; Ball, N.J.; Cicchini, C.; Ramos, A. A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets. Nat. Commun. 2018, 9, 831. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff, M.J.; Yao, J.; Qin, Y.; Nottingham, R.M.; Temoche-Diaz, M.M.; Schekman, R. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E8987–E8995. [Google Scholar] [CrossRef] [Green Version]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 2016, 5, e19276. [Google Scholar] [CrossRef]
- Li, C.; Qin, F.; Hu, F.; Xu, H.; Sun, G.; Han, G.; Wang, T.; Guo, M. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. 2018, 8, 2. [Google Scholar] [CrossRef] [Green Version]
- Kunden, R.D.; Khan, J.Q.; Ghezelbash, S.; Wilson, J.A. The role of the liver-specific microRNA, miRNA-122 in the HCV Replication Cycle. Int. J. Mol. Sci. 2020, 21, 5677. [Google Scholar] [CrossRef]
- Bandiera, S.; Pfeffer, S.; Baumert, T.F.; Zeisel, M.B. miR-122--a key factor and therapeutic target in liver disease. J. Hepatol. 2015, 62, 448–457. [Google Scholar] [CrossRef] [Green Version]
- Teng, Y.; Ren, Y.; Hu, X.; Mu, J.; Samykutty, A.; Zhuang, X.; Deng, Z.; Kumar, A.; Zhang, L.; Merchant, M.L.; et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun 2017, 8, 14448. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980–2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Riihimäki, M.; Hemminki, A.; Fallah, M.; Thomsen, H.; Sundquist, K. Metastatic sites and survival in lung cancer. Lung Cancer 2014, 86, 78–84. [Google Scholar] [CrossRef]
- Chen, R.; Xu, X.; Qian, Z.; Zhang, C.; Niu, Y.; Wang, Z.; Sun, J.; Zhang, X.; Yu, Y. The biological functions and clinical applications of exosomes in lung cancer. Cell. Mol. Life Sci. 2019, 76, 4613–4633. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Lv, T.; Zhang, Q.; Zhu, Q.; Zhan, P.; Zhu, S.; Zhang, J.; Song, Y. The biology, function and clinical implications of exosomes in lung cancer. Cancer Lett. 2017, 407, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannafon, B.N.; Ding, W.Q. Intercellular communication by exosome-derived microRNAs in cancer. Int. J. Mol. Sci. 2013, 14, 14240–14269. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Xu, Y.; Hao, J.; Wang, S.; Li, C.; Meng, S. MiR-122 in hepatic function and liver diseases. Protein Cell 2012, 3, 364–371. [Google Scholar] [CrossRef] [Green Version]
- Luna, J.M.; Scheel, T.K.; Danino, T.; Shaw, K.S.; Mele, A.; Fak, J.J.; Nishiuchi, E.; Takacs, C.N.; Catanese, M.T.; de Jong, Y.P.; et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell 2015, 160, 1099–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakral, S.; Ghoshal, K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr. Gene Ther. 2015, 15, 142–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Gao, F.; Wang, J.; Tao, L.; Ye, J.; Ding, L.; Ji, W.; Chen, X. MiR-122-5p inhibits cell migration and invasion in gastric cancer by down-regulating DUSP4. Cancer Biol. Ther. 2018, 19, 427–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, Y.; Dong, Y.; Dang, R.; Hu, Z.; Yang, Y.; Hu, Y.; Cheng, J. MiR-122 inhibits epithelial mesenchymal transition by regulating P4HA1 in ovarian cancer cells. Cell Biol. Int. 2018, 42, 1564–1574. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Sha, J.; Jiang, C.; Gao, X.; Qu, L.; Yan, H.; Xu, T.; Jiang, Q.; Gao, H. miR-122 inhibits metastasis and epithelial-mesenchymal transition of non-small-cell lung cancer cells. OncoTargets Ther. 2015, 8, 3175–3184. [Google Scholar] [CrossRef] [Green Version]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; Simpson, D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012, 13, 357. [Google Scholar] [CrossRef] [Green Version]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [Green Version]
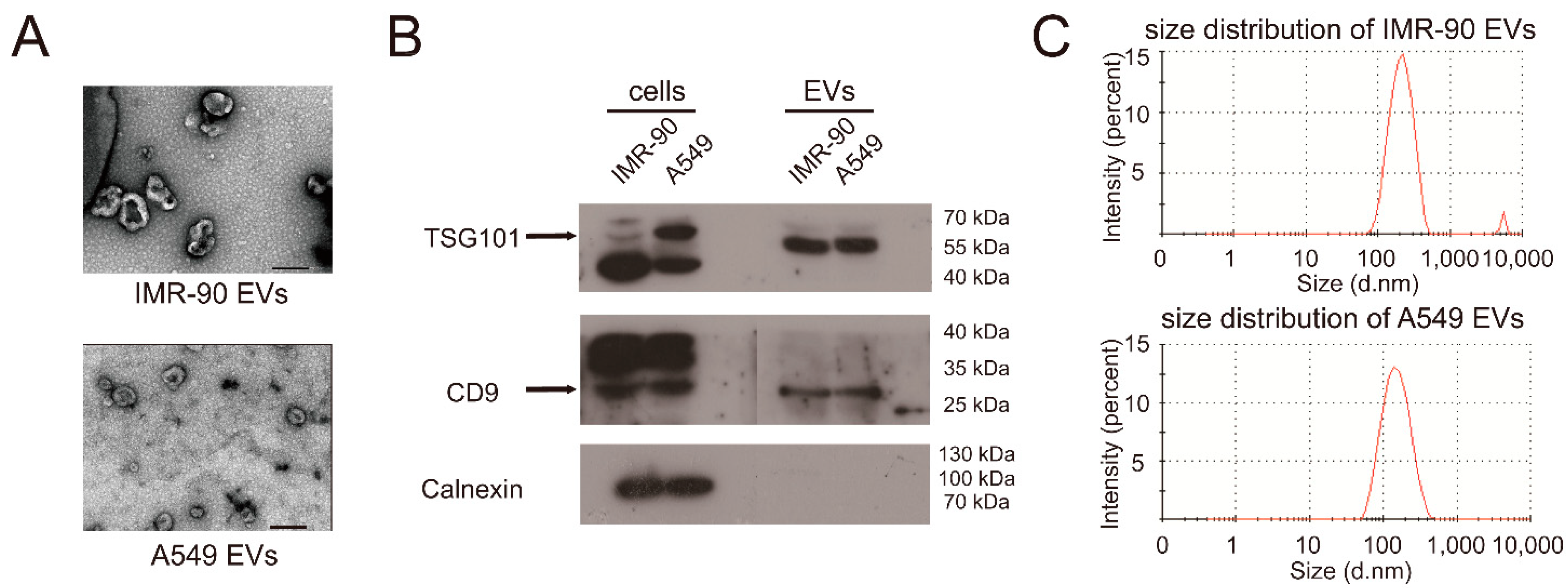
| Samples | PDI | Major Peak (nm) | Percentage of Major Peak (%) |
|---|---|---|---|
| IMR-90 EVs | 0.384 | 221.3 ± 6.31 | 97.9 |
| A549 EVs | 0.236 | 161.5 ± 3.65 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Qin, F.; Wang, W.; Ni, Y.; Gao, M.; Guo, M.; Sun, G. hnRNPA2B1-Mediated Extracellular Vesicles Sorting of miR-122-5p Potentially Promotes Lung Cancer Progression. Int. J. Mol. Sci. 2021, 22, 12866. https://doi.org/10.3390/ijms222312866
Li C, Qin F, Wang W, Ni Y, Gao M, Guo M, Sun G. hnRNPA2B1-Mediated Extracellular Vesicles Sorting of miR-122-5p Potentially Promotes Lung Cancer Progression. International Journal of Molecular Sciences. 2021; 22(23):12866. https://doi.org/10.3390/ijms222312866
Chicago/Turabian StyleLi, Chuang, Fang Qin, Wei Wang, Yifan Ni, Mingyu Gao, Mingxiong Guo, and Guihong Sun. 2021. "hnRNPA2B1-Mediated Extracellular Vesicles Sorting of miR-122-5p Potentially Promotes Lung Cancer Progression" International Journal of Molecular Sciences 22, no. 23: 12866. https://doi.org/10.3390/ijms222312866
APA StyleLi, C., Qin, F., Wang, W., Ni, Y., Gao, M., Guo, M., & Sun, G. (2021). hnRNPA2B1-Mediated Extracellular Vesicles Sorting of miR-122-5p Potentially Promotes Lung Cancer Progression. International Journal of Molecular Sciences, 22(23), 12866. https://doi.org/10.3390/ijms222312866






