Integrated Physiological, Transcriptomic, and Metabolomic Analyses Revealed Molecular Mechanism for Salt Resistance in Soybean Roots
Abstract
:1. Introduction
2. Results
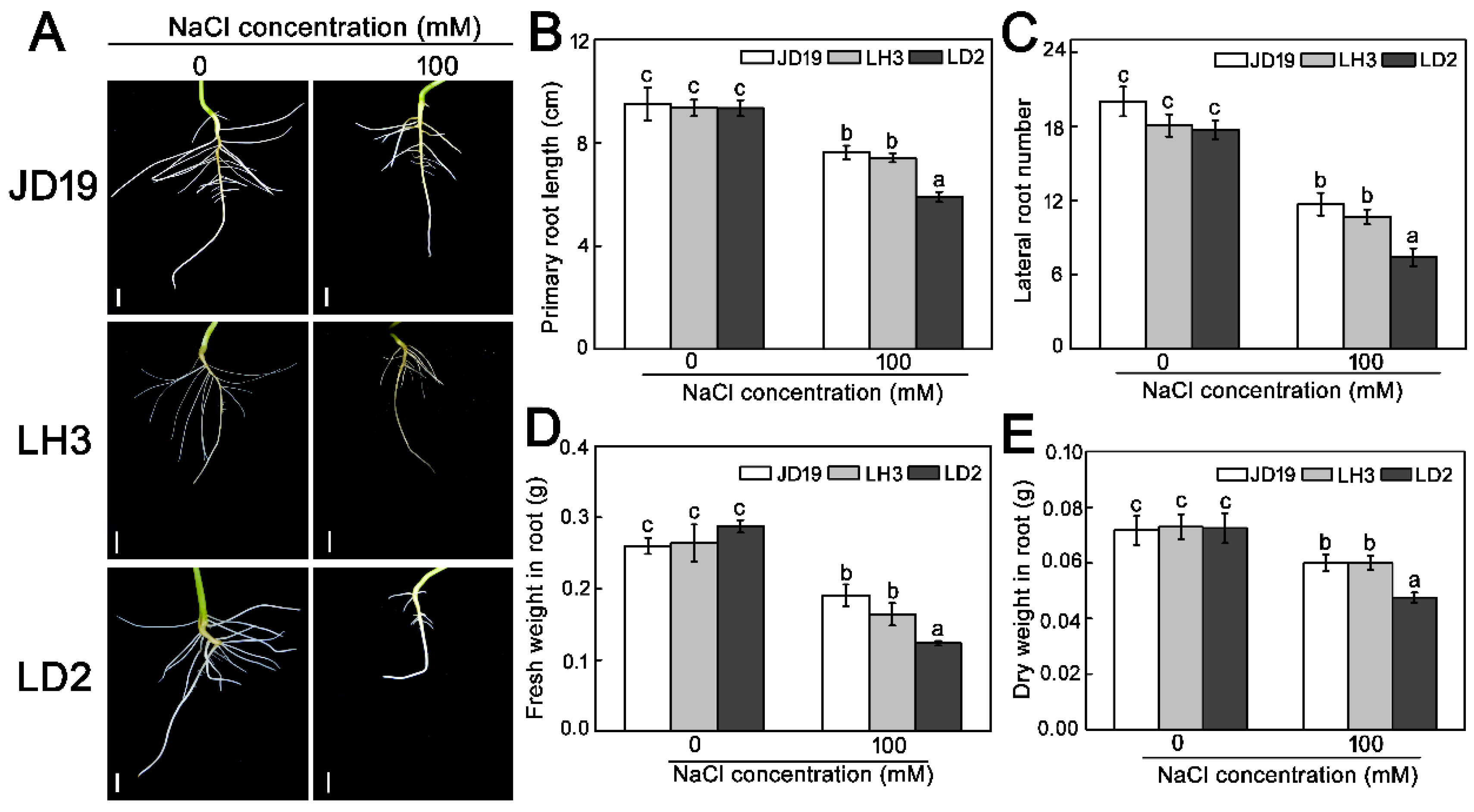
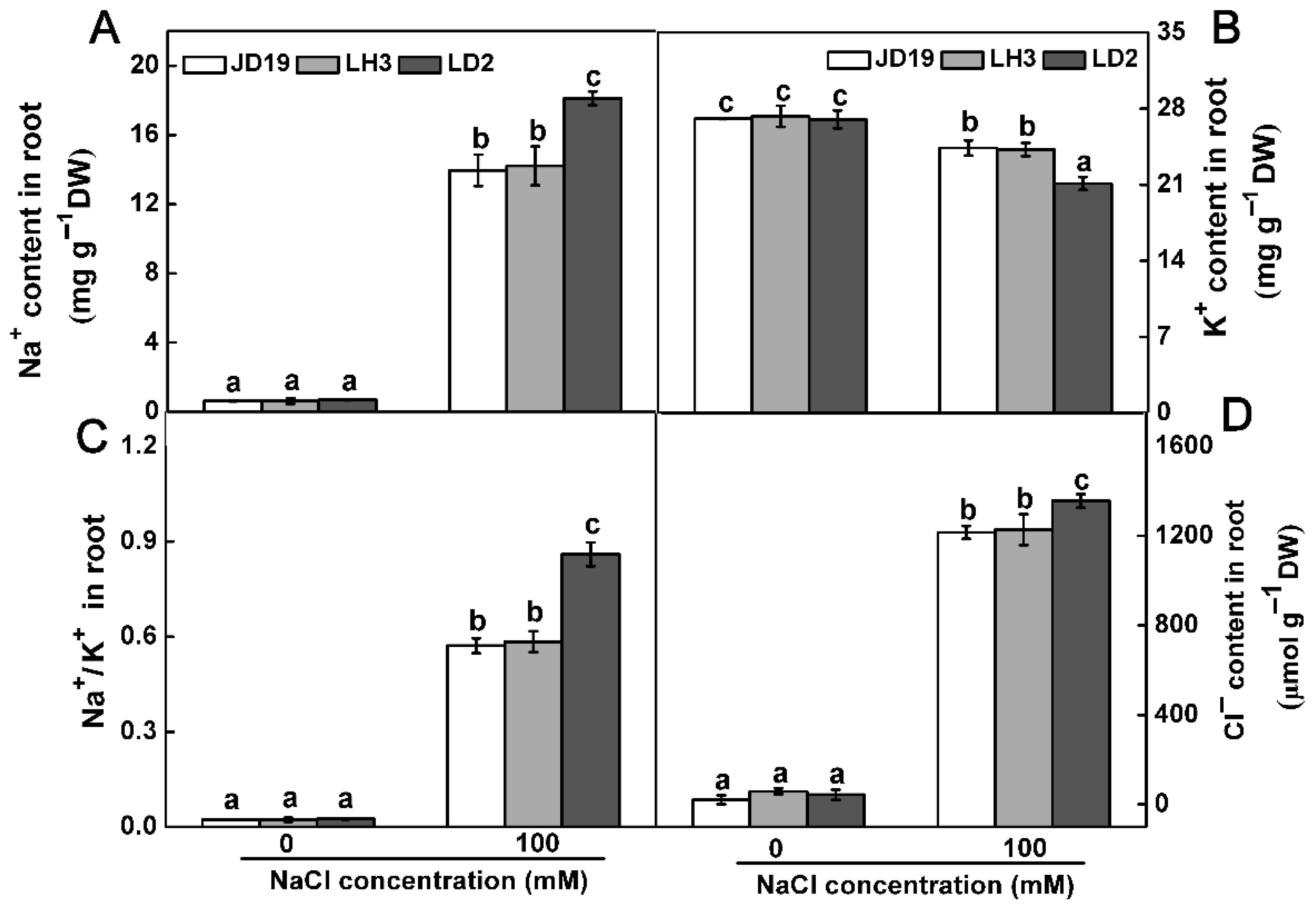
2.1. Differences of Three Soybean Cultivars Response to Salt Stress
2.2. Transcriptional Characteristics of Three Soybean Cultivars Response to Salt Stress
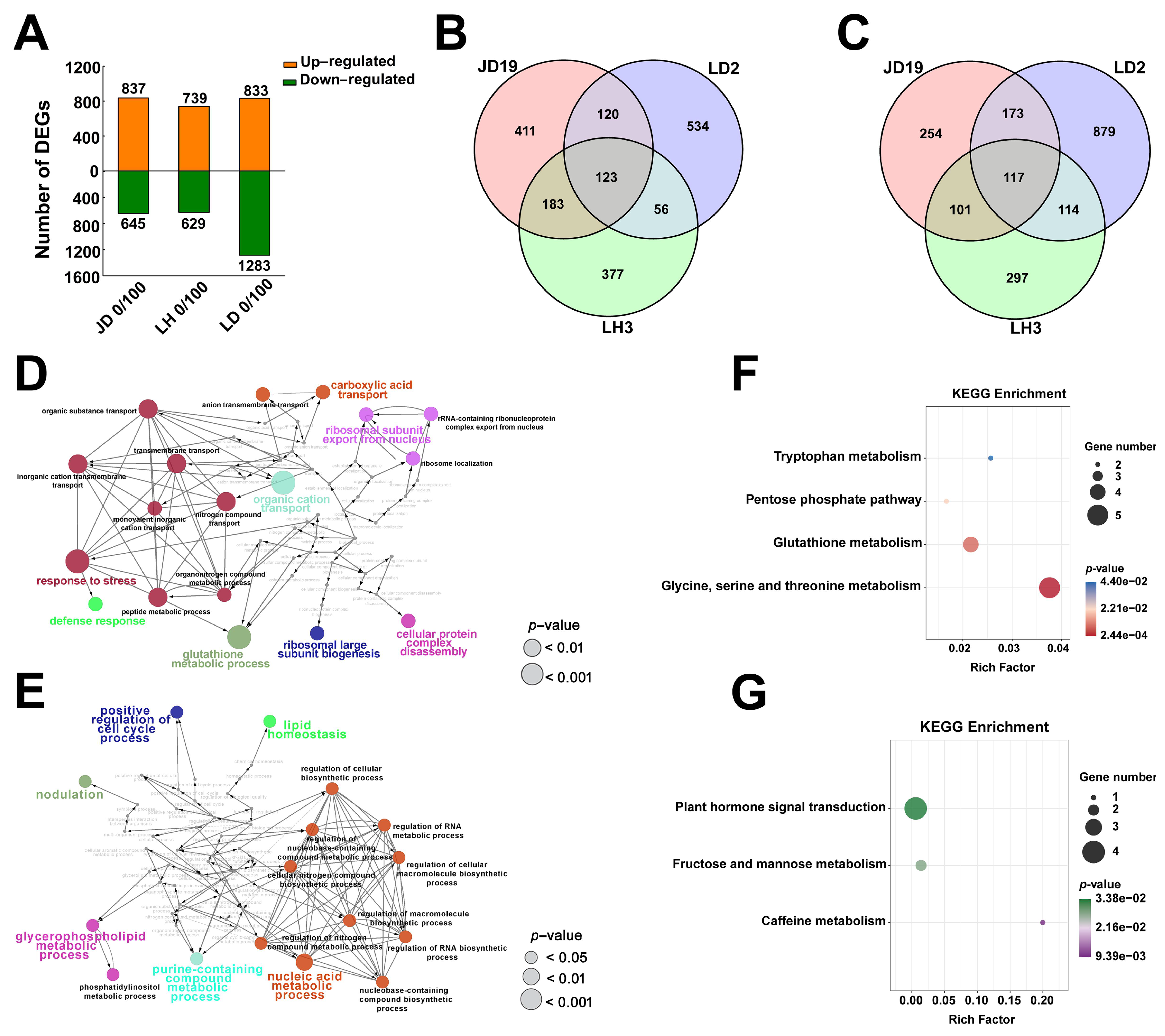
2.3. Differentially Expressed Genes among JD19, LH3, and LD2
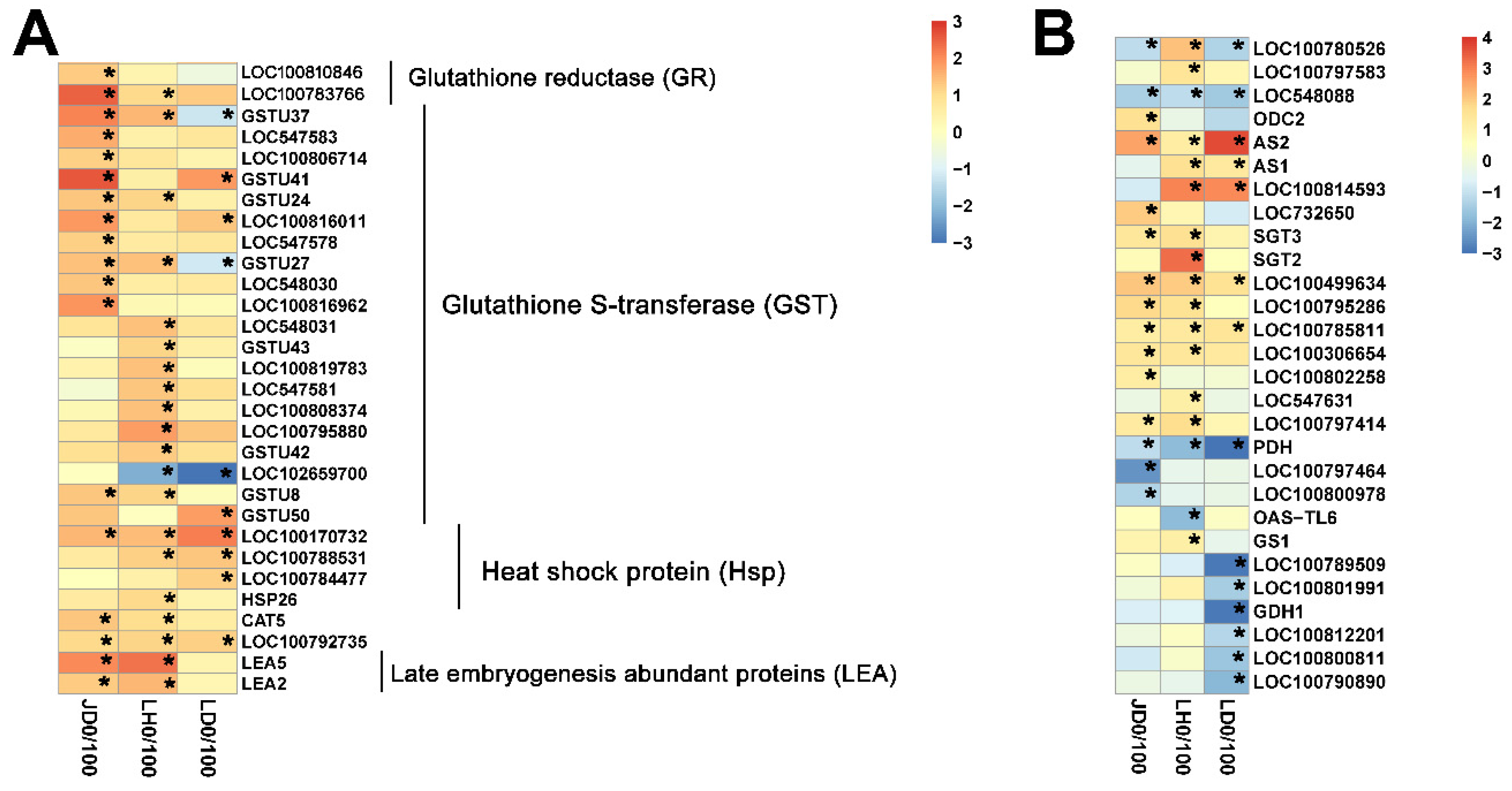
2.4. Different Salt-Responsive Genes among JD19, LH3, and LD2
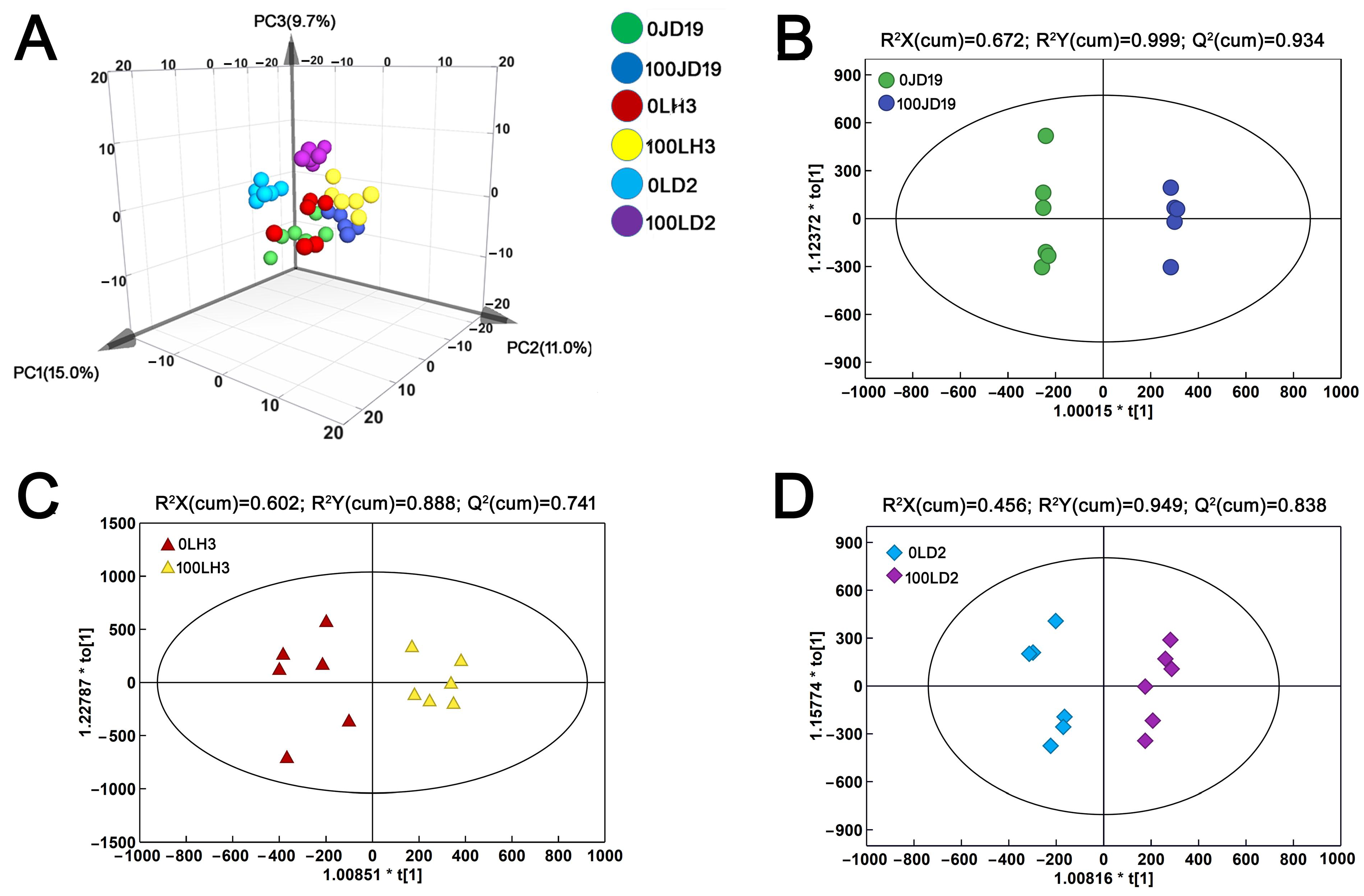
2.5. Metabolic Characteristics of Soybean Response to Salt Stress
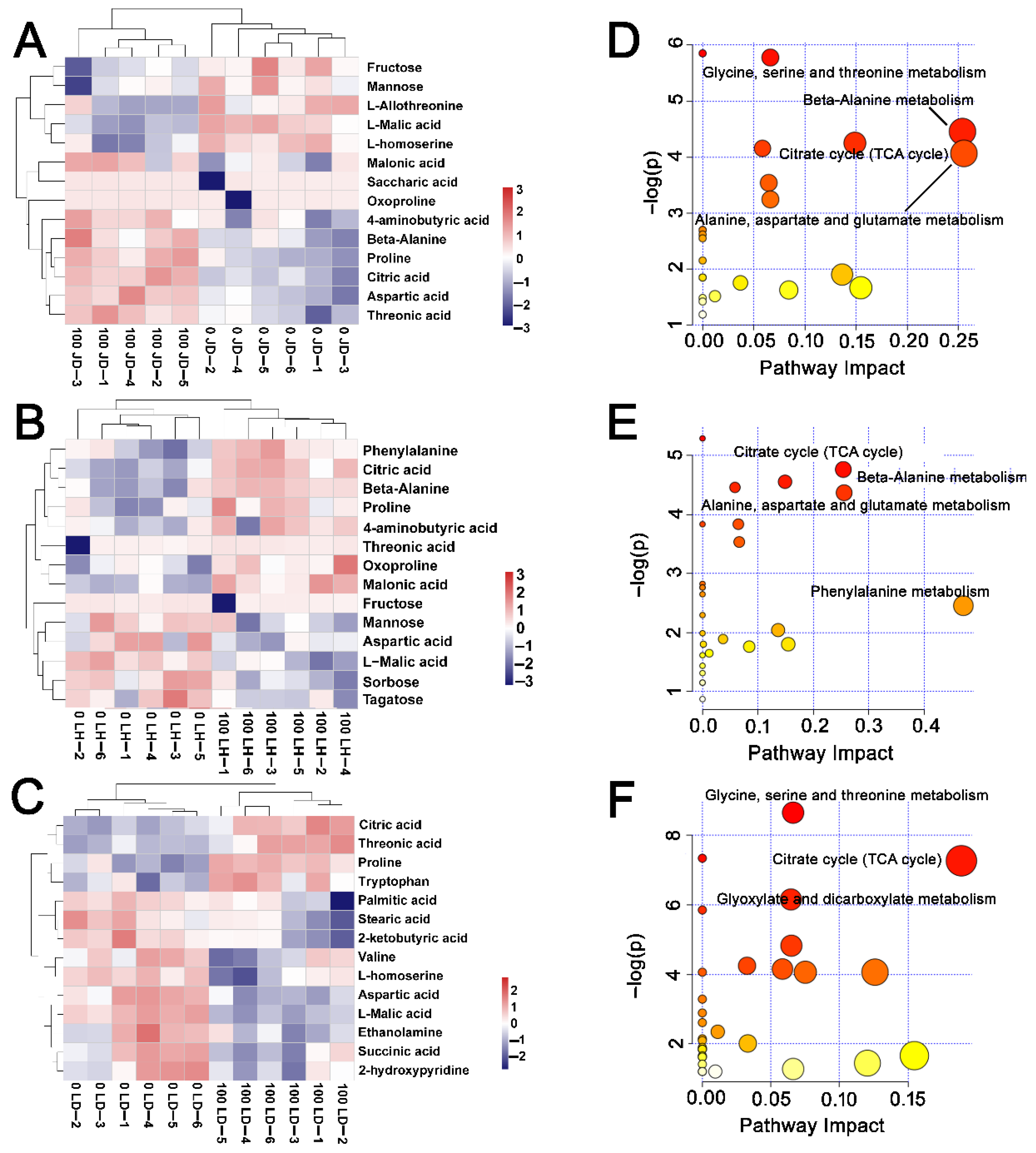
2.6. Differences in Metabolite Content among JD19, LH3, and LD2
2.7. Effects of Salt Stress on Amino Acid Contents and Nitrogen Metabolism for JD19, LH3, and LD2
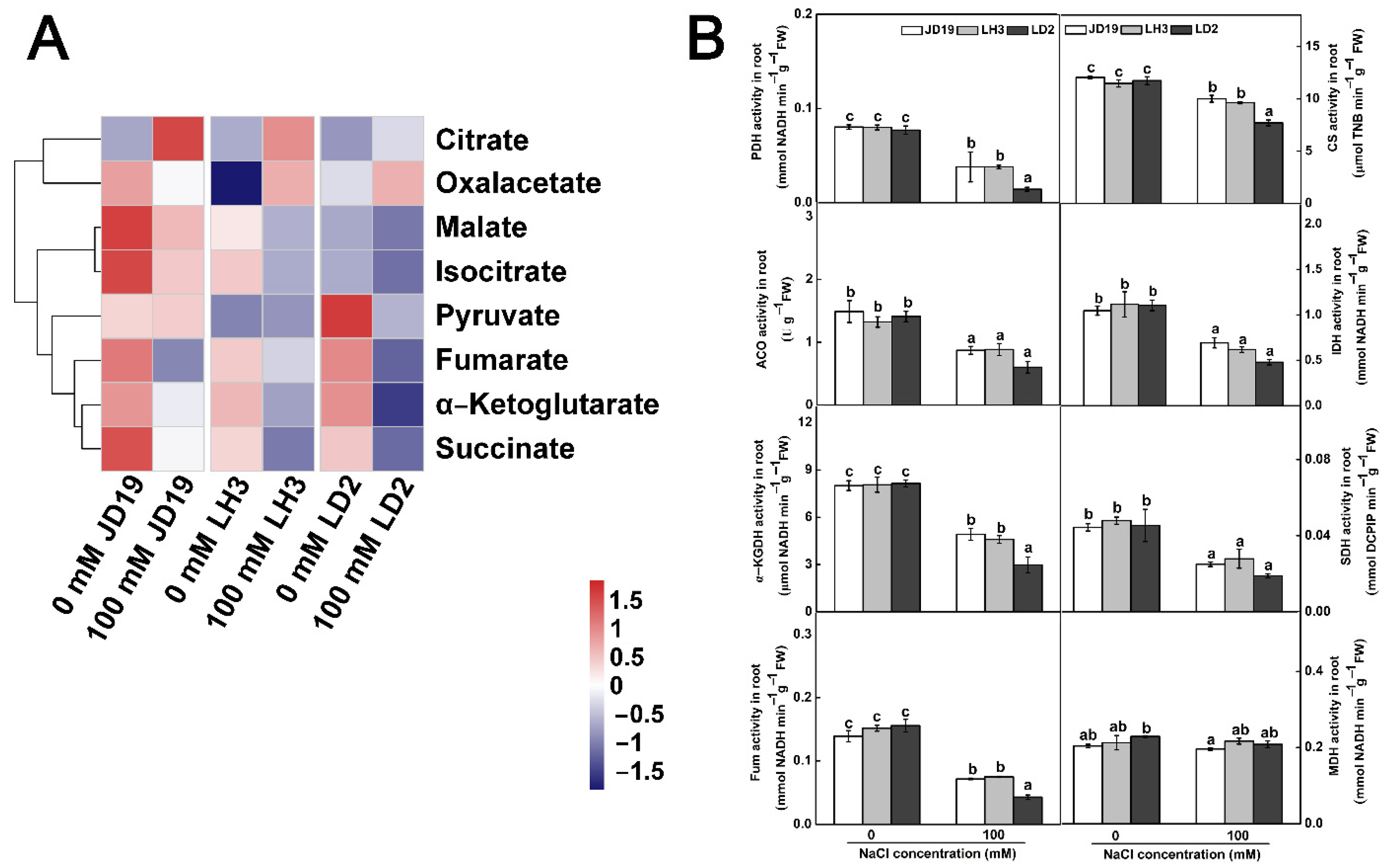
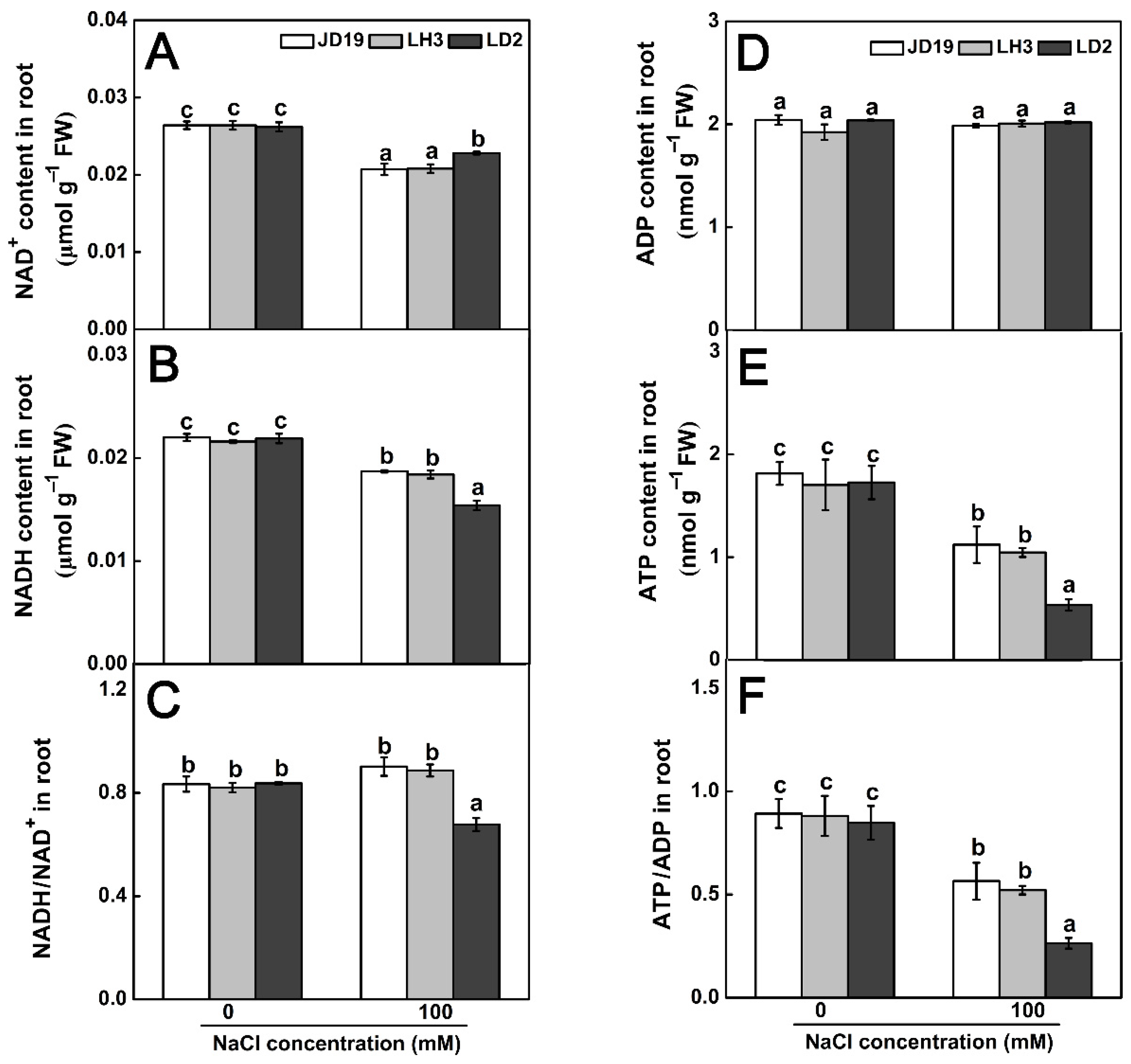
2.8. Effects of Salt Stress on Organic Acid Contents and TCA Cycle for JD19, LH3, and LD2
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and NaCl Treatments
4.2. Determination of MDA and H2O2 Content and the Activities of Antioxidant Enzymes
4.3. Total RNA Extraction and qRT-PCR Analysis
4.4. Transcript Profiling
4.5. Metabolite Extraction and GC-TOF-MS Analysis
4.6. Determination of N and Ion Contents
4.7. Determination of Amino Acid and Organic Acid Contents
4.8. Determination of NO3− and NH4+ Contents and Activities of N Metabolism-Related Enzymes
4.9. Determination of NAD+, NADH, ADP, ATP Contents, and Enzyme Activities Related to TCA Cycle
4.10. Statistics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rengasamy, P. World salinization with emphasis on Australia. J. Exp. Bot. 2006, 57, 1017–1023. [Google Scholar] [CrossRef] [Green Version]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, T.; Blumwald, E. Developing salt-tolerant crop plants: Challenges and opportunities. Trends. Plant Sci. 2005, 10, 615–620. [Google Scholar] [CrossRef]
- Wilson, R.F. Soybean: Market Driven Research Needs; Springer: New York, NY, USA, 2008. [Google Scholar]
- Bellaloui, N.; Bruns, H.A.; Abbas, H.K.; Mengistu, A.; Fisher, D.K.; Reddy, K.N. Agricultural practices altered soybean seed protein, oil, fatty acids, sugars, and minerals in the Midsouth USA. Front. Plant Sci. 2015, 31, 31. [Google Scholar] [CrossRef] [PubMed]
- Garg, B.; Gupta, I. Nodulation and symbiotic nitrogen fixation under salt stress. Curr. Agric. 2000, 24, 23–35. [Google Scholar]
- Papiernik, S.K.; Grieve, C.M.; Lesch, S.M.; Yates, S.R. Effects of salinity, imazethapyr, and chlorimuron application on soybean growth and yield. Commun. Soil Sci. Plant Anal. 2005, 36, 951–967. [Google Scholar] [CrossRef]
- Abd-Alla, M.; Issa, A.; Ohyama, T. Impact of harsh environmental conditions on nodule formation and dinitrogen fixation of legumes. In Advances in Biology and Ecology of Nitrogen Fixatio; InTech: London, UK, 2014; pp. 131–193. [Google Scholar]
- Zhu, J.; Fu, X.; Koo, Y.D.; Zhu, J.K.; Jenney, F.E.; Adams, M.W.; Zhu, Y.; Shi, H.; Yun, D.J.; Hasegawa, P.M. An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response. Mol. Cell Biol. 2007, 27, 5214–5224. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Ismail, A.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Brini, F.; Masmoudi, K. Ion transporters and abiotic stress tolerance in plants. ISRN Mol. Biol. 2012, 3, 1–13. [Google Scholar] [CrossRef]
- Wei, W.; Li, Q.T.; Chu, Y.N.; Reiter, R.J.; Yu, X.M.; Zhu, D.H.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, J.S. Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 2015, 66, 695–707. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.Y.; Tong, J.F.; He, X.L.; Xu, Z.L.; Xu, L.; Wei, P.P.; Huang, Y.H.; Brestic, M.; Ma, H.X.; Shao, H.B. A novel soybean intrinsic protein gene, GmTIP2;3, involved in responding to osmotic stress. Front. Plant Sci. 2016, 6, 1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [Green Version]
- Behr, J.H.; Bouchereau, A.; Berardocco, S.; Seal, C.E.; Flowers, T.J.; Zörb, C. Metabolic and physiological adjustment of Suaeda maritima to combined salinity and hypoxia. Ann. Bot. 2017, 119, 965–976. [Google Scholar] [PubMed] [Green Version]
- Kumari, A.; Parida, A.K. Metabolomics and network analysis reveal the potential metabolites and biological pathways involved in salinity tolerance of the halophyte Salvadora persica. Environ. Exp. Bot. 2018, 148, 85–99. [Google Scholar] [CrossRef]
- Gurmani, A.R.; Bano, A.; Najeeb, U.; Zhang, J.; Khan, S.U.; Flowers, T.J. Exogenously applied silicate and abscisic acid ameliorates the growth of salinity stressed wheat Triticum aestivum L. seedlings through Na+ exclusion. Aust. J. Crop Sci. 2013, 7, 1123–1130. [Google Scholar]
- Batista-Silva, W.; Heinemann, B.; Rugen, N.; Nunes-Nesi, A.; Araújo, W.L.; Braun, H.P.; Hildebrandt, T.M. The role of amino acid metabolism during abiotic stress release. Plant Cell Environ. 2019, 42, 1630–1644. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Mao, C.; Shi, Z.; Kou, X. The amino acid metabolic and carbohydrate metabolic pathway play important roles during salt-stress response in tomato. Front. Plant Sci. 2017, 8, 1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Kuang, L.; Li, X.; Wu, L.; Wu, D.; Zhang, G. Metabolomic and transcriptomic analyses reveal the reasons why Hordeum marinum has higher salt tolerance than Hordeum vulgare. Environ. Exp. Bot. 2018, 156, 48–61. [Google Scholar] [CrossRef]
- Xiong, X.; Wei, Y.Q.; Chen, J.H.; Liu, N.; Zhang, Y.J. Transcriptome analysis of genes and pathways associated with salt tolerance in alfalfa under non-uniform salt stress. Plant Physiol. Biochem. 2020, 151, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, D.; Zhou, R.; Wang, X.; Dossa, K.; Wang, L.; Zhang, Y.; Yu, J.; Gong, H.; Zhang, X.; et al. Transcriptome and metabolome analyses of two contrasting sesame genotypes reveal the crucial biological pathways involved in rapid adaptive response to salt stress. BMC Plant Biol. 2019, 191, 66. [Google Scholar] [CrossRef] [Green Version]
- Jia, G.; Sha, K.; Feng, X.; Liu, H. Post-thawing metabolite profile and amino acid oxidation of thawed pork tenderloin by HVEF-A short communication. Food Chem. 2019, 291, 16–21. [Google Scholar] [CrossRef]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nacry, P.; Bouguyon, E.; Gojon, A. Nitrogen acquisition by roots: Physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 2013, 370, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- Shahram, K.; Joseph, O.C.; Jonathan, R.; Yaneth, R.C.; Miercke, L.J.W.; Stroud, R.M. Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.35 A. Science 2004, 305, 1587–1594. [Google Scholar]
- Carlos, O.R.; Mora, S.I.; Jorge, T.; Omar, P. PvAMT1;1, a highly selective ammonium transporter that functions as H+/NH4+ symporter. J. Biol. Chem. 2011, 286, 31113–31122. [Google Scholar]
- Chen, T.X.; Johnson, R.; Chen, S.H.; Lv, H.; Zhou, J.L.; Li, C.J. Infection by the fungal endophyte Epichloë bromicola enhances the tolerance of wild barley Hordeum brevisubulatum to salt and alkali stresses. Plant Soil 2018, 428, 353–370. [Google Scholar] [CrossRef]
- Wang, J.F.; Tian, P.; Christensen, M.J.; Zhang, X.X.; Li, C.J.; Nan, Z.B. Effect of Epichloë gansuensis endophyte on the activity of enzymes of nitrogen metabolism, nitrogen use efficiency and photosynthetic ability of Achnatherum inebrians under various NaCl concentrations. Plant Soil 2019, 435, 57–68. [Google Scholar] [CrossRef]
- Bian, X.H.; Li, W.; Niu, C.F.; Wei, W.; Hu, Y.; Han, J.Q.; Lu, X.; Tao, J.J.; Jin, M.; Qin, H.; et al. A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol. 2020, 1, 268–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S. Redox modulation matters: Emerging functions for glutaredoxins in plant development and stress responses. Plants 2014, 3, 559–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, J.; Zhang, Y.; Liu, A.; Li, D.; Zhang, X. Transcriptomic and metabolobmic profiling of drought-tolerant and susceptible sesame genotypes in response to drought stress. BMC Plant Biol. 2019, 19, 267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Huang, C.; Xie, Y.; Song, F.; Zhou, X. A tomato glutaredoxin gene SlGRX1 regulates plant responses to oxidative, drought and salt stresses. Planta 2010, 2326, 1499–1509. [Google Scholar] [CrossRef]
- Wu, Q.; Hu, Y.; Sprague, S.A.; Kakeshpour, T.; Park, J.; Nakata, P.A.; Cheng, N.; Hirschi, K.D.; White, F.F.; Park, S. Expression of a monothiol glutaredoxin, AtGRXS17, in tomato Solanum lycopersicum enhances drought tolerance. Biochem. Bioph. Res. Commun. 2017, 491, 1034–1039. [Google Scholar] [CrossRef] [PubMed]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell Rep. 2017, 366, 791–805. [Google Scholar] [CrossRef]
- Shabala, S. Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef]
- Zhao, C.Z.; Wang, X.M.; Wang, X.Y.; Wu, K.L.; Li, P.; Chang, N.; Wang, J.F.; Wang, F.; Li, J.L.; Bi, Y.R. Glucose-6-phosphate dehydrogenase and alternative oxidase are involved in the cross tolerance of highland barley to salt stress and UV-B radiation. J. Plant Physiol. 2015, 181, 83–95. [Google Scholar] [CrossRef]
- Chen, C.; Liu, H.; Wang, C.; Liu, Z.; Liu, X.; Zou, L.; Zhao, H.; Yan, Y.; Shi, J.; Chen, S. Metabolomics characterizes metabolic changes of Apocyni Veneti Folium in response to salt stress. Plant Physiol. Bioch. 2019, 144, 187–196. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, D.; Li, M.; Shi, L. Metabolic profiles reveal changes in wild and cultivated soybean seedling leaves under salt stress. PLoS ONE 2016, 11, e0159622. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhang, Y.; Du, Y.; Chen, S.; Tang, H. Dynamic metabonomic responses of tobacco (Nicotiana tabacum) plants to salt stress. J. Proteome Res. 2011, 10, 1904–1914. [Google Scholar] [CrossRef] [PubMed]
- Khedr, A.H.A.; Abbas, M.A.; Wahid, A.A.A.; Quick, W.P.; Abogadallah, G.M. Proline induces the expression of salt-stress-responsive proteins and may improve the adaptation of Pancratium maritimum L. to salt-stress. J. Exp. Bot. 2003, 54, 2553–2562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, E.A.; Silva, M.d.G.; dos Santos Gomes, A.; Urakawa, A.H.; Barros, A.L. Carbon-nitrogen metabolism and oxidative stress in young plants of Dipteryx alata Vog. subjected to seasonal water regimes in the Cerrado. Environ. Exp. Bot. 2021, 183, 104361. [Google Scholar] [CrossRef]
- Pires, M.V.; Pereira, A.A., Jr.; Medeiros, D.B.; Daloso, D.M.; Pham, P.A.; Barros, K.A.; Fernie, A.R. The influence of alternative pathways of respiration that utilize branched-chain amino acids following water shortage in Arabidopsis. Plant Cell Environ. 2016, 39, 1304–1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukushima, A.; Kusano, M. A network perspective on nitrogen metabolism from model to crop plants using integrated ‘omics’ approaches. J. Exp. Bot. 2014, 65, 5619–5630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leberecht, M.; Dannenmann, M.; Tejedor, J.; Simon, J.; Rennenberg, H.; Polle, A. Segregation of nitrogen use between ammonium and nitrate of ectomycorrhizas and beech trees. Plant Cell Environ. 2016, 39, 2691–2700. [Google Scholar] [CrossRef]
- Casartelli, A.; Riewe, D.; Hubberten, H.M.; Altmann, T.; Hoefgen, R.; Heuer, S. Exploring traditional aus-type rice for metabolites conferring drought tolerance. Rice 2018, 11, 9. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.C.; Xu, T.; Li, X.; Tang, J.; Ma, D.F.; Li, Z.Y.; Sun, J. NaCl-induced changes of ion homeostasis and nitrogen metabolism in two sweet potato Ipomoea batatas L. cultivars exhibit different salt tolerance at adventitious root stage. Environ. Exp. Bot. 2016, 129, 23–36. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, M.S.; Guo, R.; Shi, D.C.; Liu, B.; Lin, X.Y.; Yang, C.W. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice Oryza sativa L. BMC Plant Biol. 2012, 12, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botella, M.A.; Martínez, V.; Nieves, M.; Cerdá, A. Effect of salinity on the growth and nitrogen uptake by wheat seedlings. J. Plant Nutr. 1997, 20, 793–804. [Google Scholar] [CrossRef]
- Zhang, C.X.; Meng, S.; Li, Y.M.; Zhao, Z. Net NH4+ and NO3− fluxes, and expression of NH4+ and NO3− transporter genes in roots of Populus simonii after acclimation to moderate salinity. Trees 2014, 28, 1813–1821. [Google Scholar] [CrossRef]
- Silveira, J.A.G.; Viégas, R.D.A.; Rocha, I.M.A.D.; Moreira, R.D.A.; Oliveira, J.T.A. Proline accumulation and glutamine synthetase activity are increased by salt-induced proteolysis in cashew leaves. J. Plant Physiol. 2003, 160, 115–123. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Yuan, Y.Z.; Ou, J.Q.; Lin, Q.H.; Zhang, C.F. Glutamine synthetase and glutamate dehydrogenase contribute differentially to proline accumulation in leaves of wheat Triticum aestivum seedlings exposed to different salinity. J. Plant Physiol. 2007, 164, 695–701. [Google Scholar] [CrossRef]
- Jabeen, N.; Ahmad, R. Growth response and nitrogen metabolism of sunflower Helianthus annuus L. to vermicompost and biogas slurry under salinity stress. J. Plant Nutr. 2017, 40, 104–114. [Google Scholar] [CrossRef]
- Surabhi, G.K.; Reddy, A.M.; Kumari, G.J.; Sudhakar, C. Modulations in key enzymes of nitrogen metabolism in two high yielding genotypes of mulberry Morus alba L. with differential sensitivity to salt stress. Environ. Exp. Bot. 2008, 64, 171–179. [Google Scholar] [CrossRef]
- Betti, M.; Udvardi, M.K.; Monza, J. Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol. 2010, 188, 1001–1013. [Google Scholar]
- Rong, Q.Q.; Liu, J.T.; Cai, Y.P.; Lu, Z.H.; Zhao, Z.Z.; Yue, W.C.; Xia, J.B. Leaf carbon, nitrogen and phosphorus stoichiometry of Tamarix chinensis Lour. in the Laizhou Bay coastal wetland, China. Ecol. Eng. 2015, 76, 57–65. [Google Scholar] [CrossRef]
- Song, M.L.; Chai, Q.; Li, X.Z.; Yao, X.; Li, C.J.; Christensen, M.J.; Nan, Z.B. An asexual Epichloë endophyte modifies the nutrient stoichiometry of wild barley Hordeum brevisubulatum under salt stress. Plant Soil 2015, 387, 153–165. [Google Scholar] [CrossRef]
- Cabot, C.; Sibole, J.V.; Barceló, J.; Poschenrieder, C. Lessons from crop plants struggling with salinity. Plant Sci. 2014, 226, 2–13. [Google Scholar] [CrossRef]
- Khan, M.H.; Meghvansi, M.K.; Gupta, R.; Veer, V.; Singh, L.; Kalita, M.C. Foliar spray with vermiwash modifies the Arbuscular mycorrhizal dependency and nutrient stoichiometry of Bhut Jolokia Capsicum assamicum. PLoS ONE 2014, 9, e92318. [Google Scholar]
- Kiani-Pouya, A.; Roessner, U.; Jayasinghe, N.S.; Lutz, A.; Rupasinghe, T.; Bazihizina, N.; Bohm, J.; Alharbi, S.; Hedrich, R.; Shabala, S. Epidermal bladder cells confer salinity stress tolerance in the Halophyte quinoa and Atriplex species. Plant Cell Environ. 2017, 409, 1900–1915. [Google Scholar] [CrossRef] [Green Version]
- Gong, Q.; Li, P.; Ma, S.; Rupassara, S.I.; Bohnert, H.J. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005, 445, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, H.; Ji, C.; Wei, L.; Zhao, J.; Yu, J. An integrated proteomic and metabolomic study on the chronic effects of mercury in Suaeda salsa under an environmentally relevant salinity. PLoS ONE 2013, 85, e64041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patterson, J.H.; Newbigin, E.; Tester, M.; Bacic, A.; Roessner, U. Metabolic responses to salt stress of barley Hordeum vulgare L. cultivars, Sahara and Clipper, which differ in salinity tolerance. J. Exp. Bot. 2009, 60, 4089–4103. [Google Scholar]
- Shu, L.; Lou, Q.; Ma, C.; Ding, W.; Zhou, J.; Wu, J.; Feng, F.; Lu, X.; Luo, L.; Xu, G.; et al. Genetic, proteomic and metabolic analysis of the regulation of energy storage in rice seedlings in response to drought. Proteomics 2011, 11, 4122–4138. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xing, W.W.; Shao, Q.S.; Shu, S.; Sun, J.; Guo, S.R. The effects of grafting on glycolysis and the tricarboxylic acid cycle in Ca(NO3)2-stressed cucumber seedlings with pumpkin as rootstock. Acta Physiol. Plant 2015, 37, 259. [Google Scholar] [CrossRef]
- Rangani, J.; Parida, A.K.; Panda, A.; Kumari, A. Coordinated changes in antioxidative enzymes protect the photosynthetic machinery from salinity induced oxidative damage and confer salt tolerance in an extreme halophyte Salvadora persica L. Front. Plant Sci. 2016, 7, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Caballero, B.; Trugo, L.; Finglas, P. Encyclopedia of food sciences and nutrition. In Encyclopedia of Food Sciences and Nutrition, 2nd ed.; Academic Press: Cambridge, MA, USA, 2003; Volume 1–10. [Google Scholar]
- Hodgesm, D.M.; DeLong, J.M.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Yang, L.; Wang, X.; Chang, N.; Nan, W.; Wang, S.; Ruan, M.; Sun, L.; Li, S.; Bi, Y.R. Cytosolic glucose-6-phosphate dehydrogenase is involved in seed germination and root growth under salinity in Arabidopsis. Front. Plant Sci. 2019, 10, 182. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Yuan, G.; Zhang, Q.; Xuan, L.; Li, J.; Zhou, L.; Shi, H.; Wang, X.; Wang, C. Transcriptome and metabolome analyses reveal the pivotal role of hydrogen sulfide in promoting submergence tolerance in Arabidopsis. Environ. Exp. Bot. 2021, 183, 104365. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelenal, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, D.J.; Halsall, A.; Haselden, N.J.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 7, 1060–1083. [Google Scholar] [CrossRef]
- Hou, W.; Wang, J.; Christensen, M.J.; Liu, J.; Zhang, Y.; Liu, Y.; Cheng, C. Metabolomics insights into the mechanism by which Epichloë gansuensis endophyte increased Achnatherum inebrians tolerance to low nitrogen stress. Plant Soil 2021, 463, 487–508. [Google Scholar] [CrossRef]
- Wang, J.F.; Nan, Z.B.; Christensen, M.J.; Zhang, X.X.; Tian, P.; Zhang, Z.X.; Niu, X.L.; Gao, P.; Chen, T.; Ma, L.X. Effect of Epichloë gansuensis endophyte on the nitrogen metabolism, nitrogen use efficiency, and stoichiometry of Achnatherum inebrians under nitrogen limitation. J. Agric. Food Chem. 2018, 66, 4022–4031. [Google Scholar] [CrossRef]
- Li, B.; Qiu, J.; Jayakannan, M.; Xu, B.; Li, Y.; Mayo, G.M.; Tester, M.; Gilliham, M.; Roy, S.J. AtNPF2.5 modulates chloride (Cl−) efflux from roots of Arabidopsis thaliana. Front. Plant Sci. 2017, 7, 2013. [Google Scholar] [CrossRef] [Green Version]
- Coelho, E.M.; da Silva Padilha, C.V.; Miskinis, G.A.; de Sá, A.G.B.; Pereira, G.E.; de Azevêdo, L.C.; dos Santos Lima, M. Simultaneous analysis of sugars and organic acids in wine and grape juices by HPLC: Method validation and characterization of products from northeast Brazil. J. Food Compos. Anal. 2018, 66, 160–167. [Google Scholar] [CrossRef] [Green Version]
- Du, S.; Zhang, Y.; Lin, X.Y.; Wang, Y.; Tang, C.X. Regulation of nitrate reductase by nitric oxide in Chinese cabbage pakchoi Brassica chinensis L. Plant Cell Environ. 2008, 31, 195–204. [Google Scholar] [CrossRef]
- Singh, R.P.; Srivastava, H.S. Increase in glutamate synthase NADH activity in maize seedlings in response to nitrate and ammonium nitrogen. Physiol. Plant 1986, 66, 413–416. [Google Scholar] [CrossRef]
- Wang, F.; Wang, X.; Zhao, C.; Wang, J.; Li, P.; Dou, Y.; Bi, Y. Alternative pathway is involved in the tolerance of highland barley to the low-nitrogen stress by maintaining the cellular redox homeostasis. Plant Cell Rep. 2016, 352, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.H.; Knorpp, C.; Leaver, C.J.; Hill, S.A. Plant mitochondrial pyruvate dehydrogenase complex: Purification and identification of catalytic components in potato. Biochem. J. 1998, 334, 571–576. [Google Scholar] [CrossRef] [Green Version]
- Jenner, H.L.; Winning, B.M.; Millar, A.H.; Tomlinson, K.L.; Leaver, C.J.; Hill, S.A. NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol. 2001, 126, 1139–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pekovich, S.R.; Martin, P.R.; Singleton, C.K. Thiamine pyrophosphate-requiring enzymes are altered during pyrithiamine-induced thiamine deficiency in cultured human lymphoblasts. J. Nutri. 1996, 126, 1791–1798. [Google Scholar]
- Schirawski, J.; Unden, G. Anaerobic respiration of Bacillus macerans with fumarate, TMAO, nitrate and nitrite and regulation of the pathways by oxygen and nitrate. Arch. Microbiol. 1995, 163, 148–154. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, J.; Wang, J.; Li, K.; Wang, S.; Qin, J.; Zhang, G.; Na, X.; Wang, X.; Bi, Y. Integrated Physiological, Transcriptomic, and Metabolomic Analyses Revealed Molecular Mechanism for Salt Resistance in Soybean Roots. Int. J. Mol. Sci. 2021, 22, 12848. https://doi.org/10.3390/ijms222312848
Jin J, Wang J, Li K, Wang S, Qin J, Zhang G, Na X, Wang X, Bi Y. Integrated Physiological, Transcriptomic, and Metabolomic Analyses Revealed Molecular Mechanism for Salt Resistance in Soybean Roots. International Journal of Molecular Sciences. 2021; 22(23):12848. https://doi.org/10.3390/ijms222312848
Chicago/Turabian StyleJin, Jie, Jianfeng Wang, Keke Li, Shengwang Wang, Juan Qin, Guohong Zhang, Xiaofan Na, Xiaomin Wang, and Yurong Bi. 2021. "Integrated Physiological, Transcriptomic, and Metabolomic Analyses Revealed Molecular Mechanism for Salt Resistance in Soybean Roots" International Journal of Molecular Sciences 22, no. 23: 12848. https://doi.org/10.3390/ijms222312848
APA StyleJin, J., Wang, J., Li, K., Wang, S., Qin, J., Zhang, G., Na, X., Wang, X., & Bi, Y. (2021). Integrated Physiological, Transcriptomic, and Metabolomic Analyses Revealed Molecular Mechanism for Salt Resistance in Soybean Roots. International Journal of Molecular Sciences, 22(23), 12848. https://doi.org/10.3390/ijms222312848





