Sensitive Immunofluorescent Detection of the PRAME Antigen Using a Practical Antibody Conjugation Approach
Abstract
1. Introduction
2. Results and Discussion
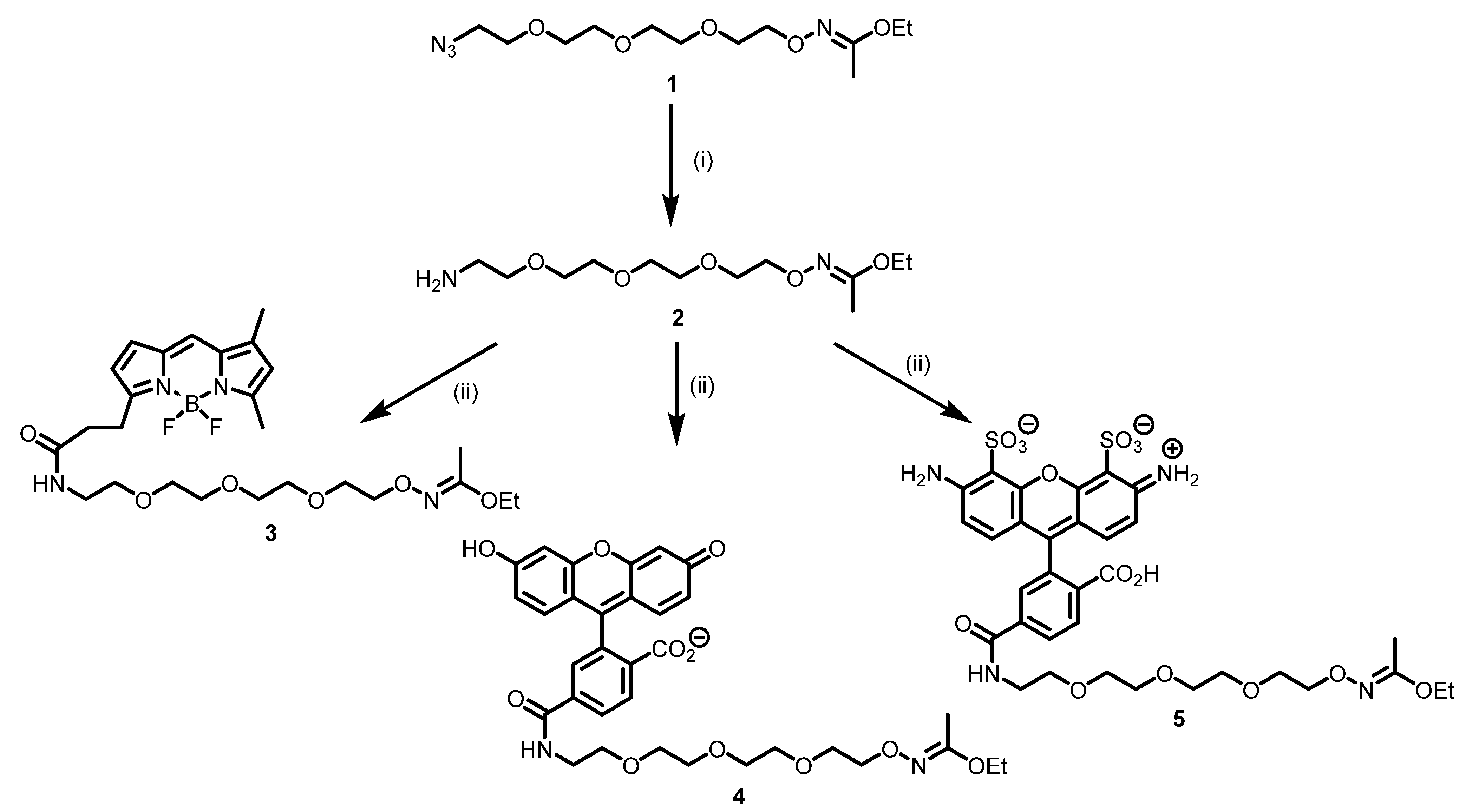
2.1. Synthesis of Modifying Reagents
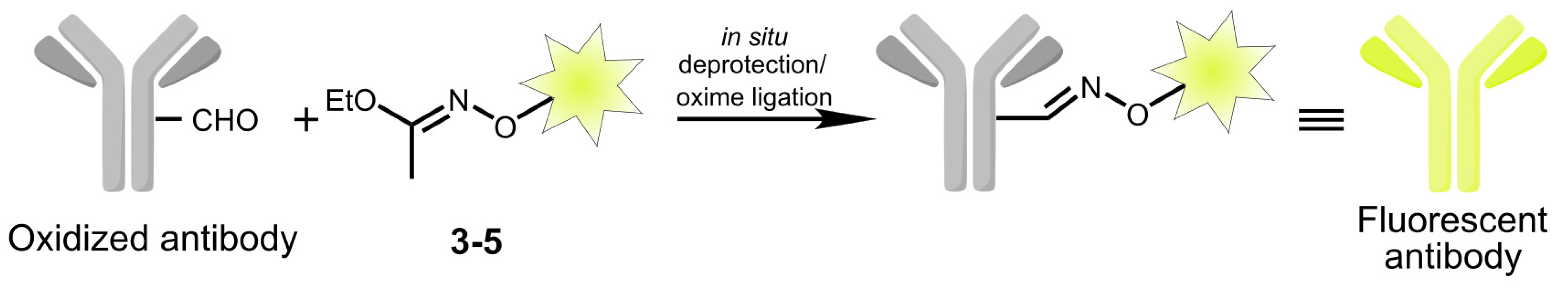
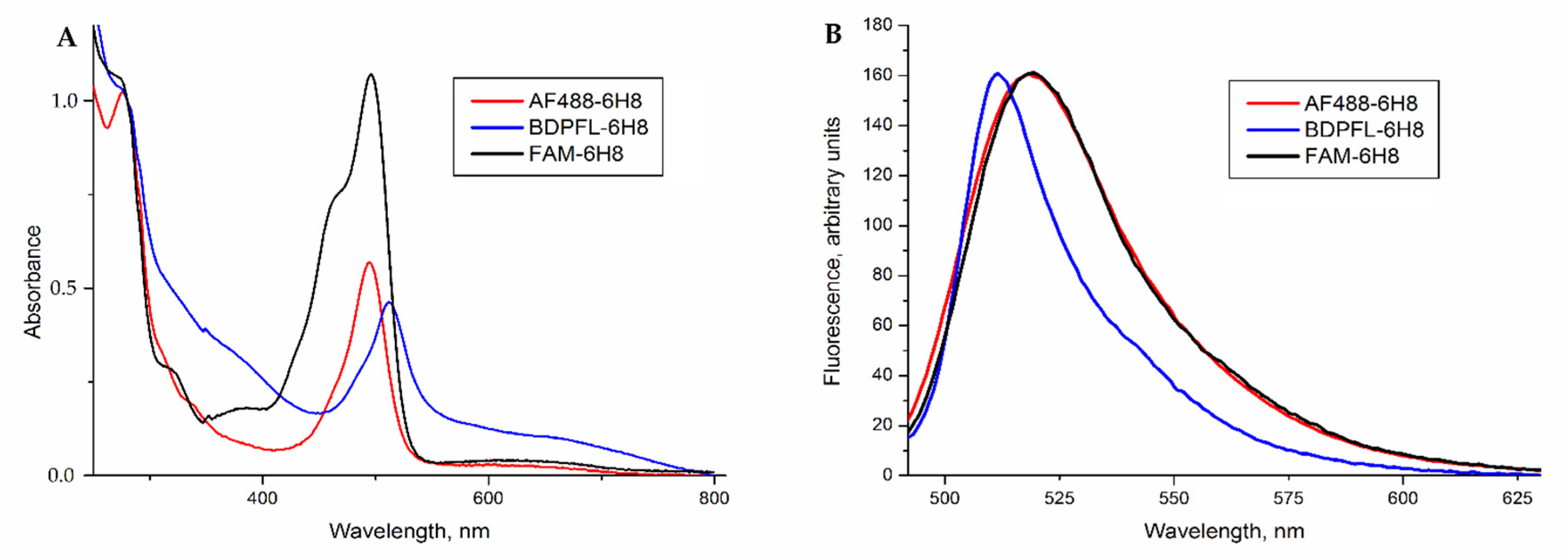
2.2. Modification of Monoclonal Antibody 6H8
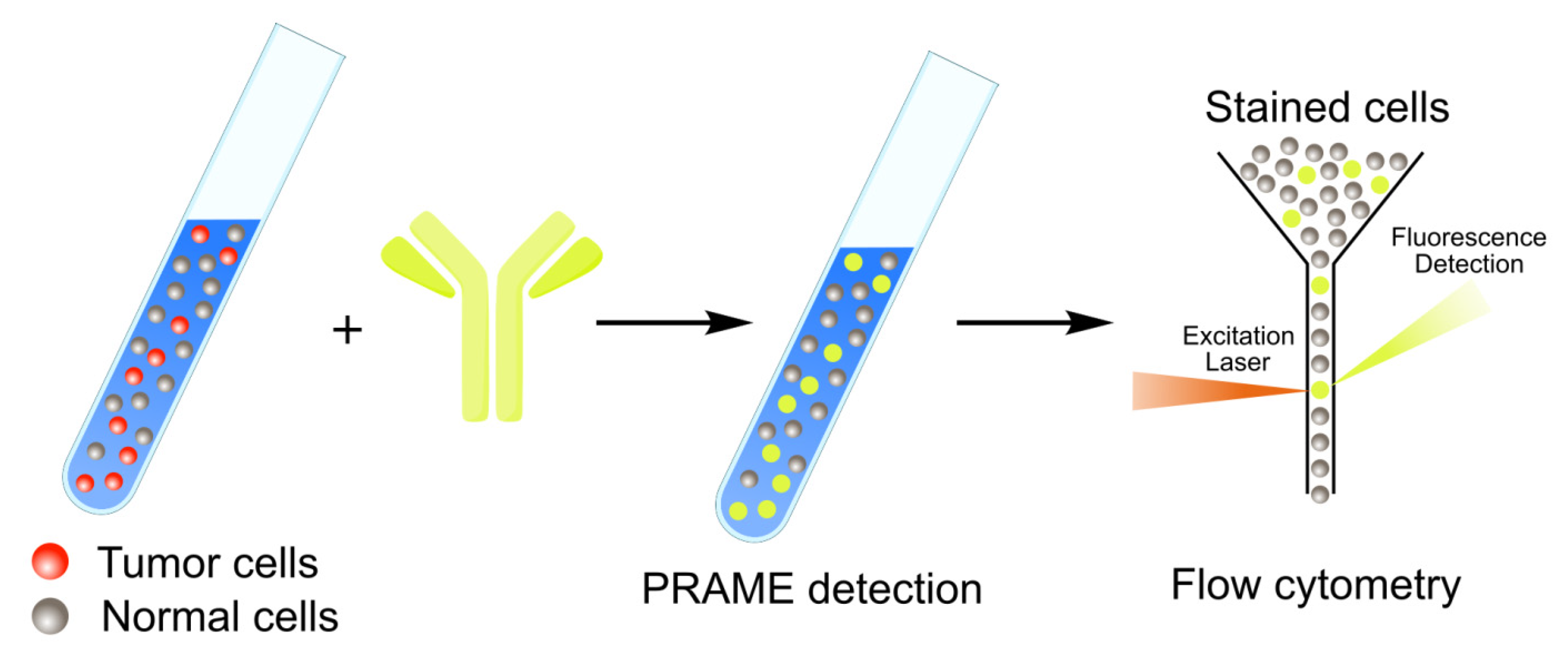
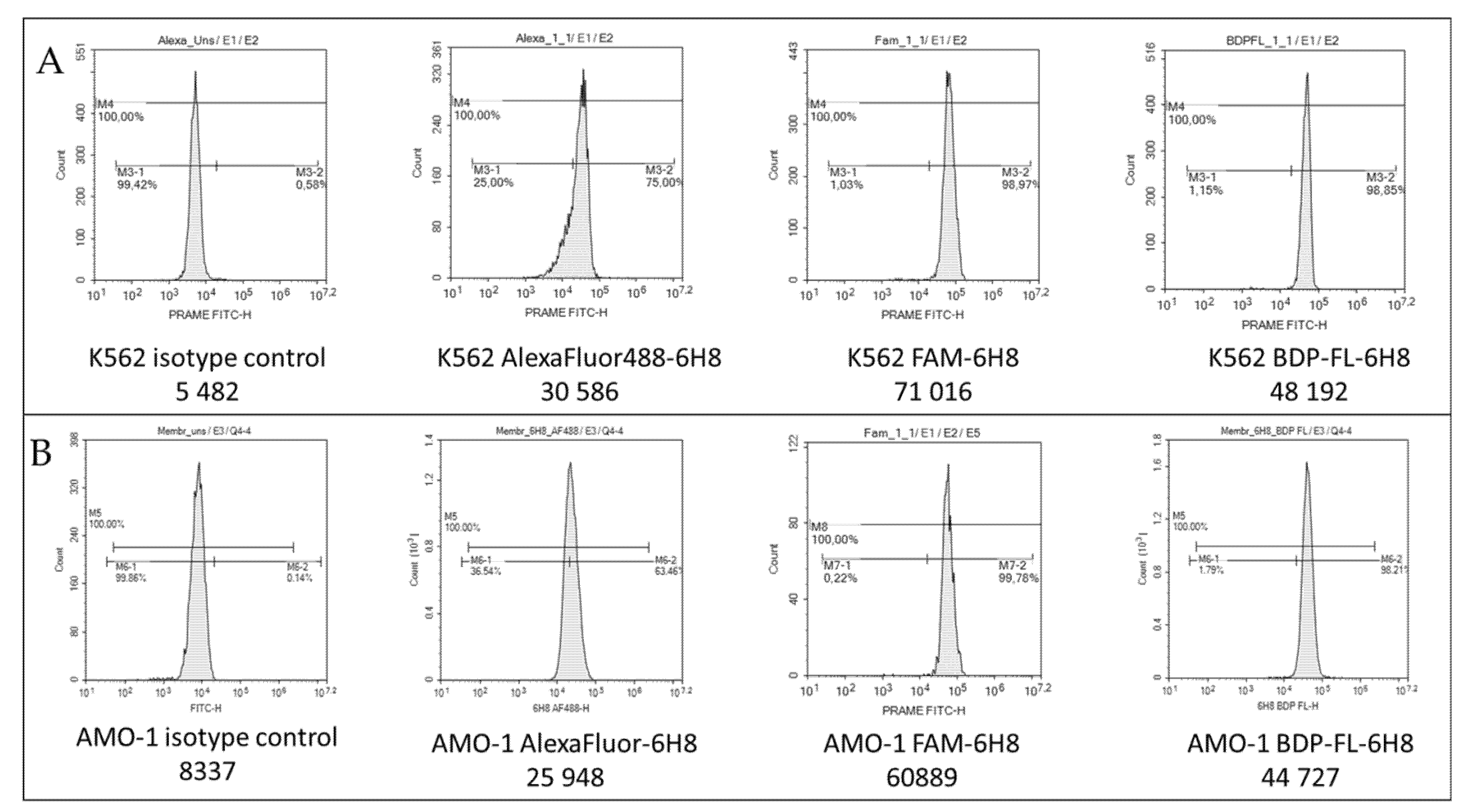
2.3. PRAME Detection by Flow Cytometry
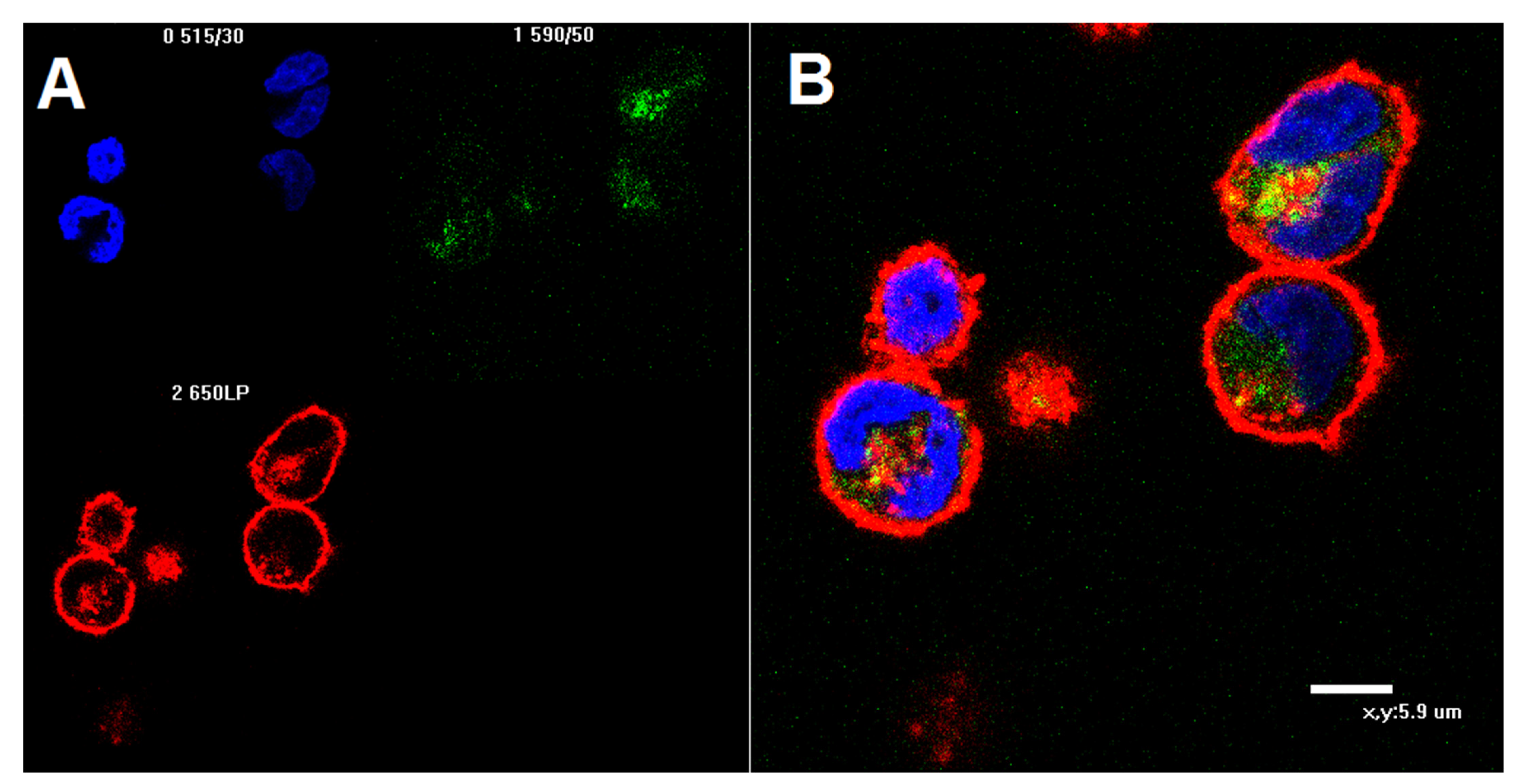
2.4. Confocal Microscopy of K562 Cells Stained with AF488-Labeled 6H8 Monoclonal Antibody
3. Materials and Methods
3.1. General Methods
3.2. Synthetic Procedures
3.3. Antibody Staining
3.4. Cell Line Handling
3.5. Bone Marrow Sample Collection
3.6. RNA Isolation and Real-Time PCR
3.7. Cells Staining and Flow Cytometry
3.8. Cell Staining for Confocal Microscopy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haugland, R.P. Coupling of monoclonal antibodies with fluorophores. In Monoclonal Antibody Protocols; Humana Press: Totowa, NJ, USA, 1995; pp. 205–222. ISBN 978-0-89603-308-5. [Google Scholar] [CrossRef]
- McCormack, T.; O’Keeffe, G.; Mac Craith, B.; O’Kennedy, R. Assessment of the effect of increased fluorophore labelling on the binding ability of an antibody. Anal. Lett. 1996, 29, 953–968. [Google Scholar] [CrossRef]
- Vira, S.; Mekhedov, E.; Humphrey, G.; Blank, P.S. Fluorescent-labeled antibodies: Balancing functionality and degree of labeling. Anal. Biochem. 2010, 402, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, S.; Dai, N.; Teo, Y.N.; Kool, E.T. Multispectral labeling of antibodies with polyfluorophores on a DNA backbone and application in cellular imaging. Proc. Natl. Acad. Sci. USA 2011, 108, 3493–3498. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, D.; Bagosi, A.; Szöllősi, J.; Jenei, A. Comparative study of the three different fluorophore antibody conjugation strategies. Anal. Bioanal. Chem. 2012, 404, 1449–1463. [Google Scholar] [CrossRef]
- Szabó, Á.; Szendi-Szatmári, T.; Ujlaky-Nagy, L.; Rádi, I.; Vereb, G.; Szöllősi, J.; Nagy, P. The effect of fluorophore conjugation on antibody affinity and the photophysical properties of dyes. Biophys. J. 2018, 114, 688–700. [Google Scholar] [CrossRef] [PubMed]
- Chari, R.V.J.; Miller, M.L.; Widdison, W.C. Antibody-drug conjugates: An emerging concept in cancer therapy. Angew. Chem. Int. Ed. 2014, 53, 3796–3827. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, K.C.; Rigol, S. The role of organic synthesis in the emergence and development of antibody–drug conjugates as targeted cancer therapies. Angew. Chem. Int. Ed. 2019, 58, 11206–11241. [Google Scholar] [CrossRef]
- Bargh, J.D.; Isidro-Llobet, A.; Parker, J.S.; Spring, D.R. Cleavable linkers in antibody–drug conjugates. Chem. Soc. Rev. 2019, 48, 4361–4374. [Google Scholar] [CrossRef]
- Joubert, N.; Beck, A.; Dumontet, C.; Denevault-Sabourin, C. Antibody–drug conjugates: The last decade. Pharmaceuticals 2020, 13, 245. [Google Scholar] [CrossRef]
- Kostova, V.; Désos, P.; Starck, J.-B.; Kotschy, A. The chemistry behind ADCs. Pharmaceuticals 2021, 14, 442. [Google Scholar] [CrossRef]
- Walsh, S.J.; Bargh, J.D.; Dannheim, F.M.; Hanby, A.R.; Seki, H.; Counsell, A.J.; Ou, X.; Fowler, E.; Ashman, N.; Takada, Y.; et al. Site-selective modification strategies in antibody–drug conjugates. Chem. Soc. Rev. 2021, 50, 1305–1353. [Google Scholar] [CrossRef]
- Wang, T.; Liu, Y.D.; Cai, B.; Huang, G.; Flynn, G.C. Investigation of antibody disulfide reduction and re-oxidation and impact to biological activities. J. Pharm. Biomed. Anal. 2015, 102, 519–528. [Google Scholar] [CrossRef]
- Liu-Shin, L.P.-Y.; Fung, A.; Malhotra, A.; Ratnaswamy, G. Evidence of disulfide bond scrambling during production of an antibody-drug conjugate. mAbs 2018, 10, 1190–1199. [Google Scholar] [CrossRef] [PubMed]
- Szijj, P.; Chudasama, V. The renaissance of chemically generated bispecific antibodies. Nat. Rev. Chem. 2021, 5, 78–92. [Google Scholar] [CrossRef]
- Adumeau, P.; Sharma, S.K.; Brent, C.; Zeglis, B.M. Site-specifically labeled immunoconjugates for molecular imaging—Part 1: Cysteine residues and glycans. Mol. Imag. Biol. 2016, 18, 1–17. [Google Scholar] [CrossRef]
- Adumeau, P.; Sharma, S.K.; Brent, C.; Zeglis, B.M. Site-specifically labeled immunoconjugates for molecular imaging—Part 2: Peptide tags and unnatural amino acids. Mol. Imag. Biol. 2016, 18, 153–165. [Google Scholar] [CrossRef]
- Alam, M.K.; El-Sayed, A.; Barreto, K.; Bernhard, W.; Fonge, H.; Geyer, C.R. Site-specific fluorescent labeling of antibodies and diabodies using SpyTag/SpyCatcher system for in vivo optical imaging. Mol. Imag. Biol. 2019, 21, 54–66. [Google Scholar] [CrossRef]
- Hoyt, E.A.; Cal, P.M.S.D.; Oliveira, B.L.; Bernardes, G.J.L. Contemporary approaches to site-selective protein modification. Nat. Rev. Chem. 2019, 3, 147–171. [Google Scholar] [CrossRef]
- Stieger, C.E.; Franz, L.; Körlin, F.; Hackenberger, C.P.R. Diethynyl phosphinates for cysteine-selective protein labeling and disulfide rebridging. Angew. Chem. Int. Ed. 2021, 60, 15359–15364. [Google Scholar] [CrossRef]
- Kölmel, D.K.; Kool, E.T. Oximes and hydrazones in bioconjugation: Mechanism and catalysis. Chem. Rev. 2017, 117, 10358–10376. [Google Scholar] [CrossRef] [PubMed]
- Kalia, J.; Raines, R.T. Hydrolytic stability of hydrazones and oximes. Angew. Chem. Int. Ed. 2008, 47, 7523–7526. [Google Scholar] [CrossRef]
- Wolfe, C.A.C.; Hage, D.S. Studies on the rate and control of antibody oxidation by periodate. Anal. Biochem. 1995, 231, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Zuberbühler, K.; Casi, G.; Bernardes, G.J.L.; Neri, D. Fucose-specific conjugation of hydrazide derivatives to a vascular-targeting monoclonal antibody in IgG format. Chem. Commun. 2012, 48, 7100. [Google Scholar] [CrossRef]
- Zhou, Q.; Stefano, J.E.; Manning, C.; Kyazike, J.; Chen, B.; Gianolio, D.A.; Park, A.; Busch, M.; Bird, J.; Zheng, X.; et al. Site-specific antibody–drug conjugation through glycoengineering. Bioconjug. Chem. 2014, 25, 510–520. [Google Scholar] [CrossRef]
- Zhu, Z.; Ramakrishnan, B.; Li, J.; Wang, Y.; Feng, Y.; Prabakaran, P.; Colantonio, S.; Dyba, M.A.; Qasba, P.K.; Dimitrov, D.S. Site-specific antibody-drug conjugation through an engineered glycotransferase and a chemically reactive sugar. mAbs 2014, 6, 1190–1200. [Google Scholar] [CrossRef]
- Axup, J.Y.; Bajjuri, K.M.; Ritland, M.; Hutchins, B.M.; Kim, C.H.; Kazane, S.A.; Halder, R.; Forsyth, J.S.; Santidrian, A.F.; Stafin, K.; et al. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc. Natl. Acad. Sci. USA 2012, 109, 16101–16106. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.A.; Mozhdehi, D.; Dzuricky, M.J.; Isaacs, F.J.; Brustad, E.M.; Chilkoti, A. Active targeting of cancer cells by nanobody decorated polypeptide micelle with bio-orthogonally conjugated drug. Nano Lett. 2019, 19, 247–254. [Google Scholar] [CrossRef]
- Field, M.G.; Decatur, C.L.; Kurtenbach, S.; Gezgin, G.; van der Velden, P.A.; Jager, M.J.; Kozak, K.N.; Harbour, J.W. PRAME as an independent biomarker for metastasis in uveal melanoma. Clin. Cancer Res. 2016, 22, 1234–1242. [Google Scholar] [CrossRef]
- Field, M.G.; Durante, M.A.; Decatur, C.L.; Tarlan, B.; Oelschlager, K.M.; Stone, J.F.; Kuznetsov, J.; Bowcock, A.M.; Kurtenbach, S.; Harbour, J.W. Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in Class 1 and Class 2 uveal melanomas. Oncotarget 2016, 7, 59209–59219. [Google Scholar] [CrossRef]
- Gerami, P.; Yao, Z.; Polsky, D.; Jansen, B.; Busam, K.; Ho, J.; Martini, M.; Ferris, L.K. Development and validation of a noninvasive 2-gene molecular assay for cutaneous melanoma. J. Am. Acad. Dermatol. 2017, 76, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Pankov, D.; Sjöström, L.; Kalidindi, T.; Lee, S.-G.; Sjöström, K.; Gardner, R.; McDevitt, M.R.; O’Reilly, R.; Thorek, D.L.J.; Larson, S.M.; et al. In vivo immuno-targeting of an extracellular epitope of membrane bound preferentially expressed antigen in melanoma (PRAME). Oncotarget 2017, 8, 65917–65931. [Google Scholar] [CrossRef] [PubMed]
- Ferris, L.K.; Gerami, P.; Skelsey, M.K.; Peck, G.; Hren, C.; Gorman, C.; Frumento, T.; Siegel, D.M. Real-world performance and utility of a noninvasive gene expression assay to evaluate melanoma risk in pigmented lesions. Melanoma Res. 2018, 28, 478–482. [Google Scholar] [CrossRef]
- Wang, W.-L.; Gokgoz, N.; Samman, B.; Andrulis, I.L.; Wunder, J.S.; Demicco, E.G. RNA expression profiling reveals PRAME, a potential immunotherapy target, is frequently expressed in solitary fibrous tumors. Mod. Pathol. 2021, 34, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, W.; Lai, X.; Zhou, Y.; Zhou, X.; Li, J.; Liang, Y.; Zhu, X.; Ren, X.; Ding, Y.; et al. CD24 and PRAME are novel grading and prognostic indicators for pineal parenchymal tumors of intermediate differentiation. Am. J. Surg. Pathol. 2020, 44, 11–20. [Google Scholar] [CrossRef]
- Lee, K.H.; Gowrishankar, K.; Street, J.; McGuire, H.M.; Luciani, F.; Hughes, B.; Singh, M.; Clancy, L.E.; Gottlieb, D.J.; Micklethwaite, K.P.; et al. Ex vivo enrichment of PRAME antigen-specific T cells for adoptive immunotherapy using CD137 activation marker selection. Clin. Transl. Immunol. 2020, 9, e1200. [Google Scholar] [CrossRef] [PubMed]
- Lezcano, C.; Pulitzer, M.; Moy, A.P.; Hollmann, T.J.; Jungbluth, A.A.; Busam, K.J. Immunohistochemistry for PRAME in the distinction of nodal nevi from metastatic melanoma. Am. J. Surg. Pathol. 2020, 44, 503–508. [Google Scholar] [CrossRef]
- Taniguchi, Y.; Ishida, M.; Saito, T.; Ryota, H.; Utsumi, T.; Maru, N.; Matsui, H.; Hino, H.; Tsuta, K.; Murakawa, T. Preferentially expressed antigen in melanoma as a novel diagnostic marker differentiating thymic squamous cell carcinoma from thymoma. Sci. Rep. 2020, 10, 12286. [Google Scholar] [CrossRef]
- See, S.H.C.; Finkelman, B.S.; Yeldandi, A.V. The diagnostic utility of PRAME and p16 in distinguishing nodal nevi from nodal metastatic melanoma. Pathol. Res. Pract. 2020, 216, 153105. [Google Scholar] [CrossRef] [PubMed]
- Gradecki, S.E.; Slingluff, C.L.; Gru, A.A. PRAME expression in 155 cases of metastatic melanoma. J. Cutan. Pathol. 2021, 48, 479–485. [Google Scholar] [CrossRef]
- Gassenmaier, M.; Hahn, M.; Metzler, G.; Bauer, J.; Yazdi, A.S.; Keim, U.; Garbe, C.; Wagner, N.B.; Forchhammer, S. Diffuse PRAME expression is highly specific for thin melanomas in the distinction from severely dysplastic nevi but does not distinguish metastasizing from non-metastasizing thin melanomas. Cancers 2021, 13, 3864. [Google Scholar] [CrossRef]
- Sapozhnikova, K.A.; Misyurin, A.V.; Pestov, N.B.; Meleshkina, E.G.; Oreshkov, S.D.; Ganzha, E.P.; Mikhailova, A.V.; Korshun, V.A.; Misyurin, V.A.; Brylev, V.A. Detection of the PRAME protein on the surface of melanoma cells using a fluorescently labeled monoclonal antibody. Russ. J. Bioorg. Chem. 2021, 47, 1077–1085. [Google Scholar] [CrossRef]
- Foillard, S.; Rasmussen, M.O.; Razkin, J.; Boturyn, D.; Dumy, P. 1-Ethoxyethylidene, a new group for the stepwise SPPS of aminooxyacetic acid containing peptides. J. Org. Chem. 2008, 73, 983–991. [Google Scholar] [CrossRef]
- Khomutov, M.A.; Mandal, S.; Weisell, J.; Saxena, N.; Simonian, A.R.; Vepsalainen, J.; Madhubala, R.; Kochetkov, S.N. Novel convenient synthesis of biologically active esters of hydroxylamine. Amino Acids 2010, 38, 509–517. [Google Scholar] [CrossRef]
- Ikeda, H.; Lethé, B.; Lehmann, F.; Van Baren, N.; Baurain, J.-F.; De Smet, C.; Chambost, H.; Vitale, M.; Moretta, A.; Boon, T.; et al. Characterization of an antigen that Is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity 1997, 6, 199–208. [Google Scholar] [CrossRef]
- Ringhoffer, M.; Müller, C.R.; Schenk, A.; Kirsche, H.; Schmitt, M.; Greiner, J.; Gschwend, J.E. Simultaneous expression of T-cell activating antigens in renal cell carcinoma: Implications for specific immunotherapy. J. Urol. 2004, 171, 2456–2460. [Google Scholar] [CrossRef]
- Li, L.; Reinhardt, P.; Schmitt, A.; Barth, T.F.E.; Greiner, J.; Ringhoffer, M.; Döhner, H.; Wiesneth, M.; Schmitt, M. Dendritic cells generated from acute myeloid leukemia (AML) blasts maintain the expression of immunogenic leukemia associated antigens. Cancer Immunol. Immunother. 2005, 54, 685–693. [Google Scholar] [CrossRef]
- Tajeddine, N.; Millard, I.; Gailly, P.; Gala, J.-L. Real-time RT-PCR quantification of PRAME gene expression for monitoring minimal residual disease in acute myeloblastic leukaemia. Clin. Chem. Lab. Med. 2006, 44. [Google Scholar] [CrossRef] [PubMed]
- Griffioen, M.; Kessler, J.H.; Borghi, M.; van Soest, R.A.; van der Minne, C.E.; Nouta, J.; van der Burg, S.H.; Medema, J.P.; Schrier, P.I.; Falkenburg, J.H.F.; et al. Detection and functional analysis of CD8+ T cells specific for PRAME: A target for T-cell therapy. Clin. Cancer Res. 2006, 12, 3130. [Google Scholar] [CrossRef]
- Grünebach, F.; Mirakaj, V.; Mirakaj, V.; Müller, M.R.; Brümmendorf, T.; Brossart, P. BCR-ABL is not an immunodominant antigen in chronic myelogenous leukemia. Cancer Res. 2006, 66, 5892. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Epping, M.T.; Bernards, R. A causal role for the human tumor antigen preferentially expressed antigen of melanoma in cancer. Cancer Res. 2006, 66, 10639. [Google Scholar] [CrossRef] [PubMed]
- Schenk, T.; Stengel, S.; Goellner, S.; Steinbach, D.; Saluz, H.P. Hypomethylation of PRAME is responsible for its aberrant overexpression in human malignancies. Genes Chromosom. Cancer 2007, 46, 796–804. [Google Scholar] [CrossRef]
- Quintarelli, C.; Dotti, G.; De Angelis, B.; Hoyos, V.; Mims, M.; Luciano, L.; Heslop, H.E.; Rooney, C.M.; Pane, F.; Savoldo, B. Cytotoxic T lymphocytes directed to the preferentially expressed antigen of melanoma (PRAME) target chronic myeloid leukemia. Blood 2008, 112, 1876–1885. [Google Scholar] [CrossRef][Green Version]
- Oehler, V.G.; Guthrie, K.A.; Cummings, C.L.; Sabo, K.; Wood, B.L.; Gooley, T.; Yang, T.; Epping, M.T.; Shou, Y.; Pogosova-Agadjanyan, E.; et al. The preferentially expressed antigen in melanoma (PRAME) inhibits myeloid differentiation in normal hematopoietic and leukemic progenitor cells. Blood 2009, 114, 3299–3308. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, E.; Holyoake, T. The chronic myeloid leukemia stem cell. Clin. Lymphoma Myeloma 2009, 9, S376–S381. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Lu, J.; Bao, L.; Zhu, H.; Li, J.; Li, L.; Lai, Y.; Shi, H.; Wang, Y.; Liu, Y.; et al. Bortezomib improves progression-free survival in multiple myeloma patients overexpressing preferentially expressed antigen of melanoma. Chin. Med. J. 2014, 127, 1666–1671. [Google Scholar] [PubMed]
- Wadelin, F.R.; Fulton, J.; Collins, H.M.; Tertipis, N.; Bottley, A.; Spriggs, K.A.; Falcone, F.H.; Heery, D.M. PRAME Is a golgi-targeted protein that associates with the elongin BC complex and is upregulated by interferon-gamma and bacterial PAMPs. PLoS ONE 2013, 8, e58052. [Google Scholar] [CrossRef] [PubMed]
- Misyurin, V.A.; Finashutina, Y.P.; Turba, A.A.; Larina, M.V.; Solopova, O.N.; Lyzhko, N.A.; Kesaeva, L.A.; Kasatkina, N.N.; Aliev, T.K.; Misyurin, A.V.; et al. Epitope analysis of murine and chimeric monoclonal antibodies recognizing the cancer testis antigen PRAME. Dokl. Biochem. Biophys. 2020, 492, 135–138. [Google Scholar] [CrossRef]
| Patient | PRAME Gene Expression Level (% Relative to ABL) | Blast Cell Fluorescence | Lymphocyte Fluorescence | ||
|---|---|---|---|---|---|
| Isotypic Control | BDP-FL-6H8 | Isotypic Control | BDP-FL-6H8 | ||
| M2 | 255 | 324 | 6872 | 113 | 124 |
| M4-53 | 13 | 352 | 580 | 1453 | 1428 |
| M4-40 | 378 | 295 | 8656 | 1135 | 1156 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sapozhnikova, K.A.; Misyurin, V.A.; Ryazantsev, D.Y.; Kokin, E.A.; Finashutina, Y.P.; Alexeeva, A.V.; Ivanov, I.A.; Kocharovskaya, M.V.; Tikhonova, N.A.; Popova, G.P.; et al. Sensitive Immunofluorescent Detection of the PRAME Antigen Using a Practical Antibody Conjugation Approach. Int. J. Mol. Sci. 2021, 22, 12845. https://doi.org/10.3390/ijms222312845
Sapozhnikova KA, Misyurin VA, Ryazantsev DY, Kokin EA, Finashutina YP, Alexeeva AV, Ivanov IA, Kocharovskaya MV, Tikhonova NA, Popova GP, et al. Sensitive Immunofluorescent Detection of the PRAME Antigen Using a Practical Antibody Conjugation Approach. International Journal of Molecular Sciences. 2021; 22(23):12845. https://doi.org/10.3390/ijms222312845
Chicago/Turabian StyleSapozhnikova, Ksenia A., Vsevolod A. Misyurin, Dmitry Y. Ryazantsev, Egor A. Kokin, Yulia P. Finashutina, Anastasiya V. Alexeeva, Igor A. Ivanov, Milita V. Kocharovskaya, Nataliya A. Tikhonova, Galina P. Popova, and et al. 2021. "Sensitive Immunofluorescent Detection of the PRAME Antigen Using a Practical Antibody Conjugation Approach" International Journal of Molecular Sciences 22, no. 23: 12845. https://doi.org/10.3390/ijms222312845
APA StyleSapozhnikova, K. A., Misyurin, V. A., Ryazantsev, D. Y., Kokin, E. A., Finashutina, Y. P., Alexeeva, A. V., Ivanov, I. A., Kocharovskaya, M. V., Tikhonova, N. A., Popova, G. P., Alferova, V. A., Ustinov, A. V., Korshun, V. A., & Brylev, V. A. (2021). Sensitive Immunofluorescent Detection of the PRAME Antigen Using a Practical Antibody Conjugation Approach. International Journal of Molecular Sciences, 22(23), 12845. https://doi.org/10.3390/ijms222312845






