Epigenetic Regulation of Myogenesis: Focus on the Histone Variants
Abstract
:1. Introduction
1.1. Establishment of the Skeletal Muscle Lineage
1.2. The Epigenetic Regulation of Gene Expression
2. The Role of the Non-Canonical h3 Histone Variant H3.3 in Myogenesis
2.1. The Histone H3 Family
2.2. H3.3 Function during Myogenesis
2.3. H3.3 Genetic Diversity
2.4. Role of the Histone Chaperone HIRA in Myogenic Cells
2.5. Primate-Specific H3 Variants H3.X and H3.Y
3. H2A Histone Variants and Myogenic Gene Expression
3.1. The H2A Family
3.2. The mH2A Family
3.3. H2A.X Histone Variant
4. The Linker Histone H1 Variants and Myoblast Differentiation
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ahmad, K.; Henikoff, S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 2002, 9, 1191–1200. [Google Scholar] [CrossRef]
- Asp, P.; Blum, R.; Vethantham, V.; Parisi, F.; Micsinai, M.; Cheng, J.; Bowman, C.; Kluger, Y.; Dynlacht, B.D. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. USA 2011, 108, E149–E158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ausió, J.; Abbott, D.W.; Wang, X.; Moore, S.C. Histone variants and histone modifications: A structural perspective. Biochem. Cell Biol. Biochim. Biol. Cell 2001, 79, 693–708. [Google Scholar] [CrossRef]
- Belotti, E.; Lacoste, N.; Simonet, T.; Papin, C.; Padmanabhan, K.; Scionti, I.; Gangloff, Y.-G.; Ramos, L.; Dalkara, D.; Hamiche, A.; et al. H2A.Z is dispensable for both basal and activated transcription in post-mitotic mouse muscles. Nucleic Acids Res. 2020, 48, 4601–4613. [Google Scholar] [CrossRef] [Green Version]
- Bernadotte, A.; Mikhelson, V.M.; Spivak, I.M. Markers of cellular senescence. Telomere shortening as a marker of cellular senescence. Aging 2016, 8, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boonsanay, V.; Zhang, T.; Georgieva, A.; Kostin, S.; Qi, H.; Yuan, X.; Zhou, Y.; Braun, T. Regulation of Skeletal Muscle Stem Cell Quiescence by Suv4-20h1-Dependent Facultative Heterochromatin Formation. Cell Stem Cell 2016, 18, 229–242. [Google Scholar] [CrossRef] [Green Version]
- Boulard, M.; Storck, S.; Cong, R.; Pinto, R.; Delage, H.; Bouvet, P. Histone variant macroH2A1 deletion in mice causes female-specific steatosis. Epigenet. Chromatin 2010, 3, 8. [Google Scholar] [CrossRef] [Green Version]
- Brush, D.; Dodgson, J.B.; Choi, O.R.; Stevens, P.W.; Engel, J.D. Replacement variant histone genes contain intervening sequences. Mol. Cell. Biol. 1985, 5, 1307–1317. [Google Scholar]
- Buckingham, M.; Rigby, P.W.J. Gene Regulatory Networks and Transcriptional Mechanisms that Control Myogenesis. Dev. Cell 2014, 28, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Buschbeck, M.; Uribesalgo, I.; Wibowo, I.; Rué, P.; Martin, D.; Gutierrez, A.; Morey, L.; Guigó, R.; López-Schier, H.; Di Croce, L. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol. 2009, 16, 1074–1079. [Google Scholar] [CrossRef]
- Calo, E.; Wysocka, J. Modification of enhancer chromatin: What, how, and why? Mol. Cell 2013, 49, 825–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, E.I.; Reinberg, D. New chaps in the histone chaperone arena. Genes Dev. 2010, 24, 1334–1338. [Google Scholar] [CrossRef] [Green Version]
- Caretti, G.; Di Padova, M.; Micales, B.; Lyons, G.E.; Sartorelli, V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004, 18, 2627–2638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celeste, A.; Petersen, S.; Romanienko, P.J.; Fernandez-Capetillo, O.; Chen, H.T.; Sedelnikova, O.A.; Reina-San-Martin, B.; Coppola, V.; Meffre, E.; Difilippantonio, M.J.; et al. Genomic instability in mice lacking histone H2AX. Science 2002, 296, 922–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chadwick, B.P.; Willard, H.F. Histone H2A variants and the inactive X chromosome: Identification of a second macroH2A variant. Hum. Mol. Genet. 2001, 10, 1101–1113. [Google Scholar] [CrossRef] [Green Version]
- Chakravarthy, S.; Gundimella, S.K.Y.; Caron, C.; Perche, P.-Y.; Pehrson, J.R.; Khochbin, S.; Luger, K. Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 2005, 25, 7616–7624. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.-W.; Chang, Y.-J.; Yeh, C.-M.; Lian, Y.-L.; Chan, M.W.Y.; Kao, C.-F.; Chen, L. SH2B1 modulates chromatin state and MyoD occupancy to enhance expressions of myogenic genes. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 270–281. [Google Scholar] [CrossRef]
- Costanzi, C.; Pehrson, J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 1998, 393, 599–601. [Google Scholar] [CrossRef]
- Costanzi, C.; Pehrson, J.R. MACROH2A2, a new member of the MARCOH2A core histone family. J. Biol. Chem. 2001, 276, 21776–21784. [Google Scholar] [CrossRef] [Green Version]
- Cuadrado, A.; Corrado, N.; Perdiguero, E.; Lafarga, V.; Muñoz-Canoves, P.; Nebreda, A.R. Essential role of p18Hamlet/SRCAP-mediated histone H2A.Z chromatin incorporation in muscle differentiation. EMBO J. 2010, 29, 2014–2025. [Google Scholar] [CrossRef] [Green Version]
- Dell’Orso, S.; Wang, A.H.; Shih, H.-Y.; Saso, K.; Berghella, L.; Gutierrez-Cruz, G.; Ladurner, A.G.; O’Shea, J.J.; Sartorelli, V.; Zare, H. The Histone Variant MacroH2A1.2 Is Necessary for the Activation of Muscle Enhancers and Recruitment of the Transcription Factor Pbx1. Cell Rep. 2016, 14, 1156–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Foggia, V.; Zhang, X.; Licastro, D.; Gerli, M.F.M.; Phadke, R.; Muntoni, F.; Mourikis, P.; Tajbakhsh, S.; Ellis, M.; Greaves, L.C.; et al. Bmi1 enhances skeletal muscle regeneration through MT1-mediated oxidative stress protection in a mouse model of dystrophinopathy. J. Exp. Med. 2014, 211, 2617–2633. [Google Scholar] [CrossRef] [Green Version]
- Drane, P.; Ouararhni, K.; Depaux, A.; Shuaib, M.; Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010, 24, 1253–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dungan, C.M.; Peck, B.D.; Walton, R.G.; Huang, Z.; Bamman, M.M.; Kern, P.A.; Peterson, C.A. In vivo analysis of γH2AX+ cells in skeletal muscle from aged and obese humans. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 7018–7035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteves de Lima, J.; Bou Akar, R.; Machado, L.; Li, Y.; Drayton-Libotte, B.; Dilworth, F.J.; Relaix, F. HIRA stabilizes skeletal muscle lineage identity. Nat. Commun. 2021, 12, 3450. [Google Scholar] [CrossRef] [PubMed]
- Evano, B.; Khalilian, S.; Le Carrou, G.; Almouzni, G.; Tajbakhsh, S. Dynamics of Asymmetric and Symmetric Divisions of Muscle Stem Cells In Vivo and on Artificial Niches. Cell Rep. 2020, 30, 3195–3206. [Google Scholar] [CrossRef] [Green Version]
- Faast, R.; Thonglairoam, V.; Schulz, T.C.; Beall, J.; Wells, J.R.E.; Taylor, H.; Matthaei, K.; Rathjen, P.D.; Tremethick, D.J.; Lyons, I. Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 2001, 11, 1183–1187. [Google Scholar] [CrossRef]
- Fan, Y.; Nikitina, T.; Morin-Kensicki, E.M.; Zhao, J.; Magnuson, T.R.; Woodcock, C.L.; Skoultchi, A.I. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 2003, 23, 4559–4572. [Google Scholar] [CrossRef] [Green Version]
- Gaspar-Maia, A.; Qadeer, Z.A.; Hasson, D.; Ratnakumar, K.; Leu, N.A.; Leroy, G.; Liu, S.; Costanzi, C.; Valle-Garcia, D.; Schaniel, C.; et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat. Commun. 2013, 4, 1565. [Google Scholar] [CrossRef] [Green Version]
- Giaimo, B.D.; Ferrante, F.; Herchenröther, A.; Hake, S.B.; Borggrefe, T. The histone variant H2A.Z in gene regulation. Epigenet. Chromatin 2019, 12, 37. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.-M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010, 140, 678–691. [Google Scholar] [CrossRef] [Green Version]
- Gros, J.; Manceau, M.; Thomé, V.; Marcelle, C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 2005, 435, 954–958. [Google Scholar] [CrossRef]
- Hammond, C.M.; Strømme, C.B.; Huang, H.; Patel, D.J.; Groth, A. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 2017, 18, 141–158. [Google Scholar] [CrossRef] [Green Version]
- Harada, A.; Okada, S.; Konno, D.; Odawara, J.; Yoshimi, T.; Yoshimura, S.; Kumamaru, H.; Saiwai, H.; Tsubota, T.; Kurumizaka, H.; et al. Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J. 2012, 31, 2994–3007. [Google Scholar] [CrossRef] [Green Version]
- Harada, A.; Maehara, K.; Sato, Y.; Konno, D.; Tachibana, T.; Kimura, H.; Ohkawa, Y. Incorporation of histone H3.1 suppresses the lineage potential of skeletal muscle. Nucleic Acids Res. 2015, 43, 775–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, A.; Maehara, K.; Ono, Y.; Taguchi, H.; Yoshioka, K.; Kitajima, Y.; Xie, Y.; Sato, Y.; Iwasaki, T.; Nogami, J.; et al. Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration. Nat. Commun. 2018, 9, 1400. [Google Scholar] [CrossRef] [Green Version]
- Hurtado-Bagès, S.; Posavec Marjanovic, M.; Valero, V.; Malinverni, R.; Corujo, D.; Bouvet, P.; Lavigne, A.-C.; Bystricky, K.; Buschbeck, M. The Histone Variant MacroH2A1 Regulates Key Genes for Myogenic Cell Fusion in a Splice-Isoform Dependent Manner. Cells 2020, 9, 1109. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.-W.; Shibata, Y.; Starmer, J.; Yee, D.; Magnuson, T. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 2015, 29, 1377–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, W.; Shang, Y.; Peng, J.; Jiang, S. Histone H3 Methyltransferase Suv39h1 Prevents Myogenic Terminal Differentiation by Repressing MEF2 Activity in Muscle Cells. Int. J. Mol. Sci. 2016, 17, 1908. [Google Scholar] [CrossRef] [Green Version]
- Juan, A.H.; Derfoul, A.; Feng, X.; Ryall, J.G.; Dell’Orso, S.; Pasut, A.; Zare, H.; Simone, J.M.; Rudnicki, M.A.; Sartorelli, V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011, 25, 789–794. [Google Scholar] [CrossRef] [Green Version]
- Karthik, N.; Taneja, R. Histone variants in skeletal myogenesis. Epigenetics 2021, 16, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Kassar-Duchossoy, L. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005, 19, 1426–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krogan, N.J.; Keogh, M.-C.; Datta, N.; Sawa, C.; Ryan, O.W.; Ding, H.; Haw, R.A.; Pootoolal, J.; Tong, A.; Canadien, V.; et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 2003, 12, 1565–1576. [Google Scholar] [CrossRef] [Green Version]
- Kujirai, T.; Horikoshi, N.; Sato, K.; Maehara, K.; Machida, S.; Osakabe, A.; Kimura, H.; Ohkawa, Y.; Kurumizaka, H. Structure and function of human histone H3.Y nucleosome. Nucleic Acids Res. 2016, 44, 6127–6141. [Google Scholar] [CrossRef] [Green Version]
- Law, C.; Cheung, P. Expression of Non-acetylatable H2A.Z in Myoblast Cells Blocks Myoblast Differentiation through Disruption of MyoD Expression. J. Biol. Chem. 2015, 290, 13234–13249. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Habas, R.; Abate-Shen, C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 2004, 304, 1675–1678. [Google Scholar] [CrossRef]
- Loyola, A.; Almouzni, G. Marking histone H3 variants: How, when and why? Trends Biochem. Sci. 2007, 32, 425–433. [Google Scholar] [CrossRef]
- Maehara, K.; Harada, A.; Sato, Y.; Matsumoto, M.; Nakayama, K.I.; Kimura, H.; Ohkawa, Y. Tissue-specific expression of histone H3 variants diversified after species separation. Epigenet. Chromatin 2015, 8, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mal, A.K. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006, 25, 3323–3334. [Google Scholar] [CrossRef]
- Matsuda, R.; Hori, T.; Kitamura, H.; Takeuchi, K.; Fukagawa, T.; Harata, M. Identification and characterization of the two isoforms of the vertebrate H2A.Z histone variant. Nucleic Acids Res. 2010, 38, 4263–4273. [Google Scholar] [CrossRef] [Green Version]
- Meneghini, M.D.; Wu, M.; Madhani, H.D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 2003, 112, 725–736. [Google Scholar] [CrossRef] [Green Version]
- Michod, D.; Bartesaghi, S.; Khelifi, A.; Bellodi, C.; Berliocchi, L.; Nicotera, P.; Salomoni, P. Calcium-Dependent Dephosphorylation of the Histone Chaperone DAXX Regulates H3.3 Loading and Transcription upon Neuronal Activation. Neuron 2012, 74, 122–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, R.K.; Gurdon, J.B. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 2008, 10, 102–109. [Google Scholar] [CrossRef]
- Palacios, D.; Mozzetta, C.; Consalvi, S.; Caretti, G.; Saccone, V.; Proserpio, V.; Marquez, V.E.; Valente, S.; Mai, A.; Forcales, S.V.; et al. TNF/p38α/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 2010, 7, 455–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, C.; Fan, Y. Role of H1 Linker Histones in Mammalian Development and Stem Cell Differentiation. Biochim. Biophys. Acta 2016, 1859, 496–509. [Google Scholar]
- Pehrson, J.R.; Fried, V.A. MacroH2A, a core histone containing a large nonhistone region. Science 1992, 257, 1398–1400. [Google Scholar] [CrossRef]
- Pehrson, J.R.; Changolkar, L.N.; Costanzi, C.; Leu, N.A. Mice without macroH2A histone variants. Mol. Cell. Biol. 2014, 34, 4523–4533. [Google Scholar] [CrossRef] [Green Version]
- Posavec Marjanović, M.; Hurtado-Bagès, S.; Lassi, M.; Valero, V.; Malinverni, R.; Delage, H.; Navarro, M.; Corujo, D.; Guberovic, I.; Douet, J.; et al. MacroH2A1.1 regulates mitochondrial respiration by limiting nuclear NAD+ consumption. Nat. Struct. Mol. Biol. 2017, 24, 902–910. [Google Scholar] [CrossRef] [Green Version]
- Relaix, F.; Rocancourt, D.; Mansouri, A.; Buckingham, M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 2005, 435, 948–953. [Google Scholar] [CrossRef] [Green Version]
- Relaix, F.; Bencze, M.; Borok, M.J.; Der Vartanian, A.; Gattazzo, F.; Mademtzoglou, D.; Perez-Diaz, S.; Prola, A.; Reyes-Fernandez, P.C.; Rotini, A.; et al. Perspectives on skeletal muscle stem cells. Nat. Commun. 2021, 12, 692. [Google Scholar] [CrossRef]
- Resnick, R.; Wong, C.-J.; Hamm, D.C.; Bennett, S.R.; Skene, P.J.; Hake, S.B.; Henikoff, S.; van der Maarel, S.M.; Tapscott, S.J. DUX4-Induced Histone Variants H3.X and H3.Y Mark DUX4 Target Genes for Expression. Cell Rep. 2019, 29, 1812–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, C.; Sutherland, H.F.; Farmer, H.; Kimber, W.; Halford, S.; Carey, A.; Brickman, J.M.; Wynshaw-Boris, A.; Scambler, P.J. Targeted Mutagenesis of the Hira Gene Results in Gastrulation Defects and Patterning Abnormalities of Mesoendodermal Derivatives Prior to Early Embryonic Lethality. Mol. Cell. Biol. 2002, 22, 2318–2328. [Google Scholar] [CrossRef] [Green Version]
- Semba, Y.; Harada, A.; Maehara, K.; Oki, S.; Meno, C.; Ueda, J.; Yamagata, K.; Suzuki, A.; Onimaru, M.; Nogami, J.; et al. Chd2 regulates chromatin for proper gene expression toward differentiation in mouse embryonic stem cells. Nucleic Acids Res. 2017, 45, 8758–8772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef]
- Song, K.; Wang, Y.; Sassoon, D. Expression of Hox-7.1 in myoblasts inhibits terminal differentiation and induces cell transformation. Nature 1992, 360, 477–481. [Google Scholar] [CrossRef]
- Song, T.-Y.; Yang, J.-H.; Park, J.Y.; Song, Y.; Han, J.-W.; Youn, H.-D.; Cho, E.-J. The role of histone chaperones in osteoblastic differentiation of C2C12 myoblasts. Biochem. Biophys. Res. Commun. 2012, 423, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Sousa-Victor, P.; Gutarra, S.; García-Prat, L.; Rodriguez-Ubreva, J.; Ortet, L.; Ruiz-Bonilla, V.; Jardí, M.; Ballestar, E.; González, S.; Serrano, A.L.; et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 2014, 506, 316–321. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Histone variants at a glance. J. Cell Sci. 2021, 134, jcs244749. [Google Scholar] [CrossRef]
- Tanasijevic, B.; Rasmussen, T.P. X chromosome inactivation and differentiation occur readily in ES cells doubly-deficient for macroH2A1 and macroH2A2. PLoS ONE 2011, 6, e21512. [Google Scholar] [CrossRef] [Green Version]
- Tang, M.C.W.; Jacobs, S.A.; Mattiske, D.M.; Soh, Y.M.; Graham, A.N.; Tran, A.; Lim, S.L.; Hudson, D.F.; Kalitsis, P.; O’Bryan, M.K.; et al. Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice. PLoS Genet. 2015, 11, e1004964. [Google Scholar]
- Valenzuela, N.; Fan, Q.; Fa’ak, F.; Soibam, B.; Nagandla, H.; Liu, Y.; Schwartz, R.J.; McConnell, B.K.; Stewart, M.D. Cardiomyocyte-specific conditional knockout of the histone chaperone HIRA in mice results in hypertrophy, sarcolemmal damage and focal replacement fibrosis. Dis. Model. Mech. 2016, 9, 335–345. [Google Scholar] [CrossRef] [Green Version]
- Valenzuela, N.; Soibam, B.; Li, L.; Wang, J.; Byers, L.A.; Liu, Y.; Schwartz, R.J.; Stewart, M.D. HIRA deficiency in muscle fibers causes hypertrophy and susceptibility to oxidative stress. J. Cell Sci. 2017, 130, 2551–2563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Kumar, R.M.; Biggs, V.J.; Lee, H.; Chen, Y.; Kagey, M.H.; Young, R.A.; Abate-Shen, C. The Msx1 Homeoprotein Recruits Polycomb to the Nuclear Periphery during Development. Dev. Cell 2011, 21, 575–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiedemann, S.M.; Mildner, S.N.; Bönisch, C.; Israel, L.; Maiser, A.; Matheisl, S.; Straub, T.; Merkl, R.; Leonhardt, H.; Kremmer, E.; et al. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J. Cell Biol. 2010, 190, 777–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wunsch, A.M.; Lough, J. Modulation of histone H3 variant synthesis during the myoblast-myotube transition of chicken myogenesis. Dev. Biol. 1987, 119, 94–99. [Google Scholar] [CrossRef]
- Yang, J.-H.; Song, Y.; Seol, J.-H.; Park, J.Y.; Yang, Y.-J.; Han, J.-W.; Youn, H.-D.; Cho, E.-J. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc. Natl. Acad. Sci. USA 2011, 108, 85–90. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.-H.; Choi, J.-H.; Jang, H.; Park, J.-Y.; Han, J.-W.; Youn, H.-D.; Cho, E.-J. Histone chaperones cooperate to mediate Mef2-targeted transcriptional regulation during skeletal myogenesis. Biochem. Biophys. Res. Commun. 2011, 407, 541–547. [Google Scholar] [CrossRef]
- Yang, J.-H.; Song, T.-Y.; Jo, C.; Park, J.; Lee, H.-Y.; Song, I.; Hong, S.; Jung, K.Y.; Kim, J.; Han, J.-W.; et al. Differential regulation of the histone chaperone HIRA during muscle cell differentiation by a phosphorylation switch. Exp. Mol. Med. 2016, 48, e252. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Tan, J.; Liu, D.; Loreni, F.; Peng, X.; Yang, Q.; He, W.; Yao, Z.; Zhang, X.; Dal Prà, I.; et al. Eukaryotic initiation factor 6 modulates myofibroblast differentiation at transforming growth factor-β1 transcription level via H2A.Z occupancy and Sp1 recruitment. J. Cell Sci. 2015, 128, 3977–3989. [Google Scholar] [CrossRef]
- Yao, F.; Yu, P.; Li, Y.; Yuan, X.; Li, Z.; Zhang, T.; Liu, F.; Wang, Y.; Wang, Y.; Li, D.; et al. Histone Variant H2A.Z Is Required for the Maintenance of Smooth Muscle Cell Identity as Revealed by Single-Cell Transcriptomics. Circulation 2018, 138, 2274–2288. [Google Scholar] [CrossRef]
- Zernov, N.; Skoblov, M. Genotype-phenotype correlations in FSHD. BMC Med. Genom. 2019, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ryu, D.; Wu, Y.; Gariani, K.; Wang, X.; Luan, P.; D’Amico, D.; Ropelle, E.R.; Lutolf, M.P.; Aebersold, R.; et al. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 2016, 352, 1436–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Cooke, M.; Panjwani, S.; Cao, K.; Krauth, B.; Ho, P.-Y.; Medrzycki, M.; Berhe, D.T.; Pan, C.; McDevitt, T.C.; et al. Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 2012, 8, e1002691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zink, L.-M.; Delbarre, E.; Eberl, H.C.; Keilhauer, E.C.; Bönisch, C.; Pünzeler, S.; Bartkuhn, M.; Collas, P.; Mann, M.; Hake, S.B. H3.Y discriminates between HIRA and DAXX chaperone complexes and reveals unexpected insights into human DAXX-H3.3-H4 binding and deposition requirements. Nucleic Acids Res. 2017, 45, 5691–5706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
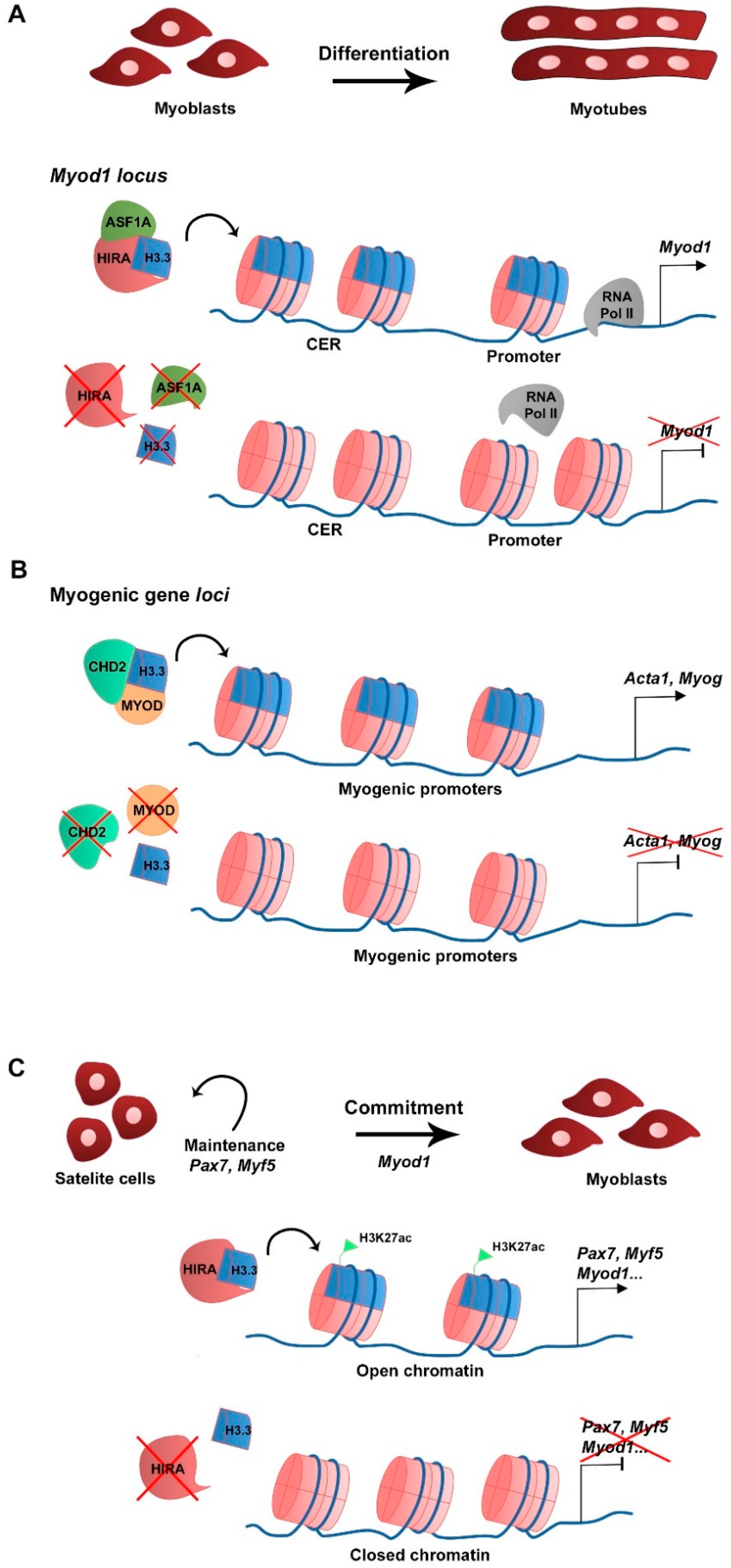
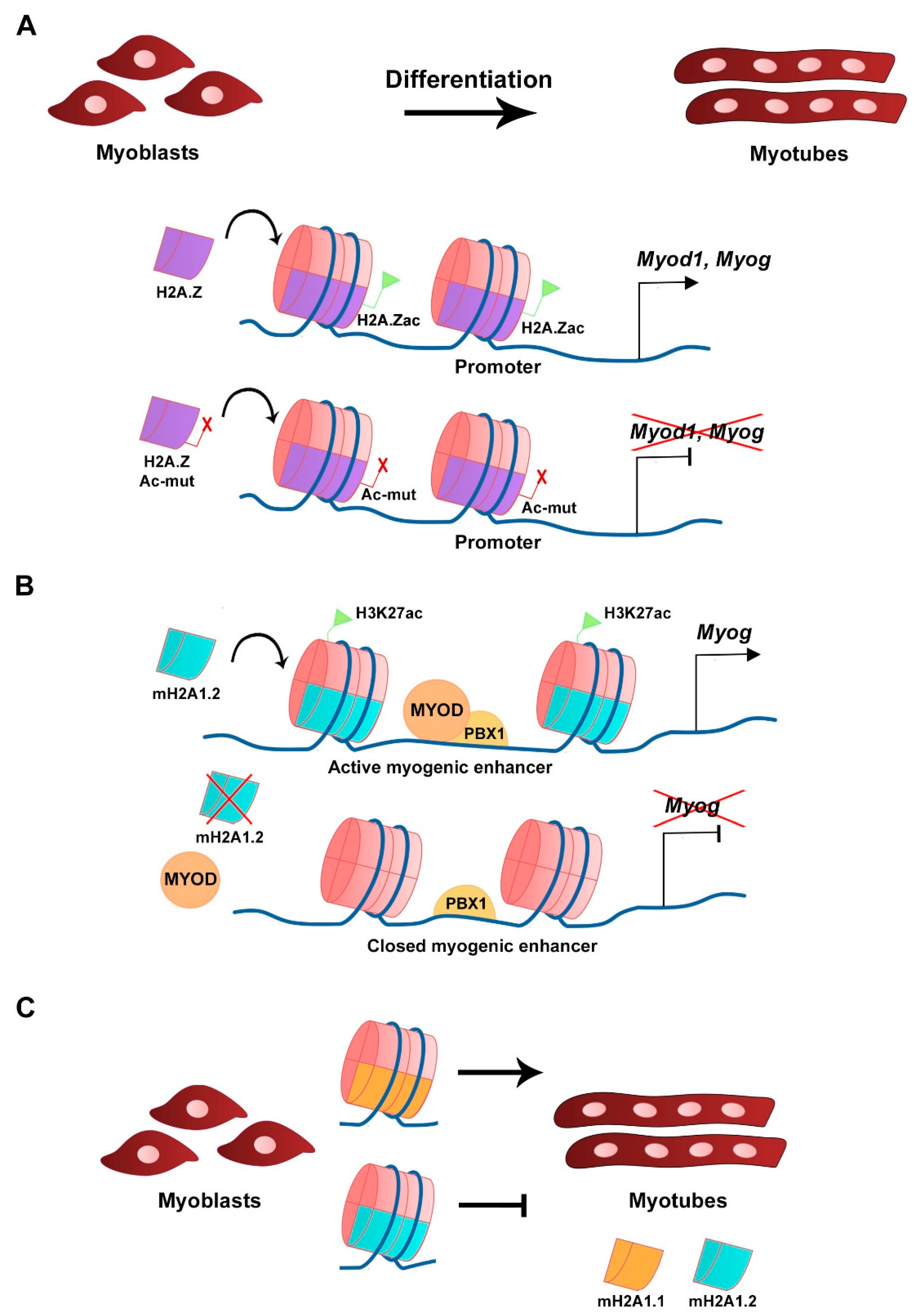
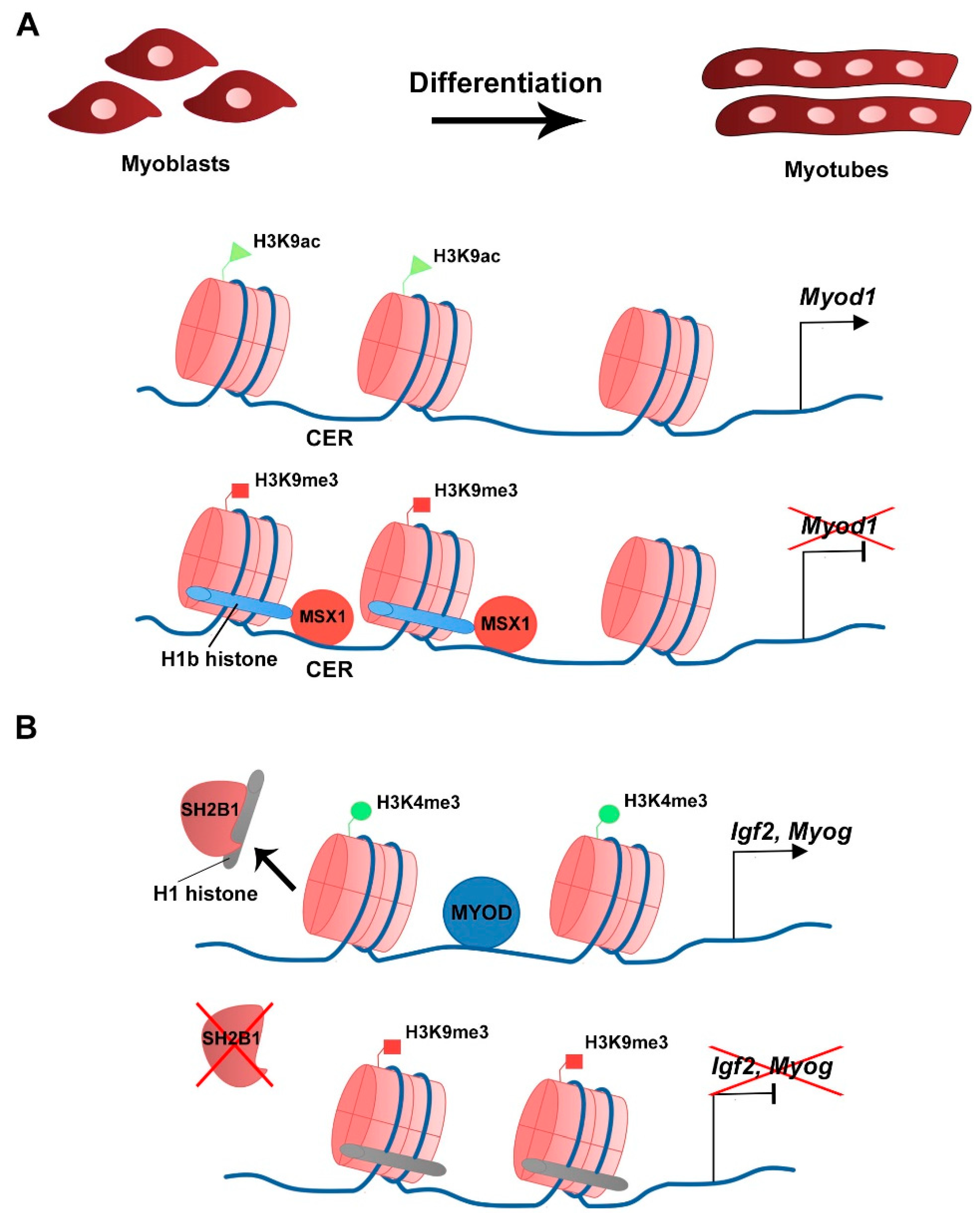
| Histone Variant | Expression/Deposition/Function | Reference |
|---|---|---|
| H3.3 | Synthesis (protein) increases during chicken primary myoblast differentiation. | Wunsch and Lough, 1987 |
| Expression (mRNA) in C2C12 cells during growth and differentiation. Deposition by HIRA/ASF1A in Myod1 CER in C2C12 cells. Required for Myod1 expression and myoblast differentiation. | Yang et al., 2011 | |
| Deposition by CHD2/MYOD complex in myogenic gene loci in C2C12 cells. Required for Myog expression and myoblast differentiation. | Harada et al., 2012 | |
| Deposition in myogenic gene loci is associated with the histone mark H3K4me3. | Harada et al., 2015 | |
| Hira KO in satellite cells leads to decreased H3.3 at myogenic gene loci. Association with the histone mark H3K27ac and accessible chromatin. Required for myogenic gene expression, regeneration and satellite cell identity. | Esteves de Lima et al., 2021 | |
| H3mm7 | Expression (mRNA) in satellite cells in vivo and in C2C12 cells. Promotion of myogenic gene expression and differentiation. Required for normal regeneration. | Harada et al., 2018 |
| H3.X, H3.Y | Deposition in regulatory regions of DUX4 target genes in FSHD. Activation of DUX4 target gene expression. | Resnick et al., 2019 |
| H2A.Z | Deposited at myogenic genes promoters in primary myoblasts and C2C12 cells. Deposition is p38 MAPK-dependent and enriched during differentiation. Required for myogenic gene expression and myoblast differentiation. | Cuadrado et al., 2010 |
| Acetylation of H2A.Z is required for Myod1 expression and C2C12 myoblast differentiation. Acetylation of H2A.Z is required for RNA Pol II recruitment to myogenic gene loci. | Law and Cheung, 2015 | |
| Enrichment in actively transcribed myogenic genes in vivo. Dispensable for gene expression, homeostasis and regeneration in post-mitotic fibers. | Belotti et al., 2020 | |
| mH2A1.1 | Expression (mRNA) in C2C12 cells during growth and differentiation. | Dell’Orso et al., 2016 Posavec Marjanović et al., 2017 |
| Promotion of myoblast fusion in C2C12 cells. | Hurtado-Bagès et al., 2020 | |
| mH2A1.2 | Expression (mRNA) in C2C12 cells during growth and differentiation. | Dell’Orso et al., 2016 Posavec Marjanović et al., 2017 |
| Deposition is required for H3K27 acetylation and activation of muscle enhancers. Required for myogenic gene expression and myoblast differentiation. | Dell’Orso et al., 2016 | |
| Inhibition of C2C12 myoblast fusion. | Hurtado-Bagès et al., 2020 | |
| H1b | Inhibition of C2C12 myoblast differentiation. Binding to Myod1 locus and inhibition of Myod1 transcription. Direct interaction with MSX1 at Myod1 locus. | Lee et al., 2004 |
| H1 variants are associated with the histone mark H3K9me3 in myogenic gene loci. Inhibition of C2C12 myoblast differentiation. | Chen et al., 2017 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esteves de Lima, J.; Relaix, F. Epigenetic Regulation of Myogenesis: Focus on the Histone Variants. Int. J. Mol. Sci. 2021, 22, 12727. https://doi.org/10.3390/ijms222312727
Esteves de Lima J, Relaix F. Epigenetic Regulation of Myogenesis: Focus on the Histone Variants. International Journal of Molecular Sciences. 2021; 22(23):12727. https://doi.org/10.3390/ijms222312727
Chicago/Turabian StyleEsteves de Lima, Joana, and Frédéric Relaix. 2021. "Epigenetic Regulation of Myogenesis: Focus on the Histone Variants" International Journal of Molecular Sciences 22, no. 23: 12727. https://doi.org/10.3390/ijms222312727
APA StyleEsteves de Lima, J., & Relaix, F. (2021). Epigenetic Regulation of Myogenesis: Focus on the Histone Variants. International Journal of Molecular Sciences, 22(23), 12727. https://doi.org/10.3390/ijms222312727







