Implications of Gut Microbiota in Complex Human Diseases
Abstract
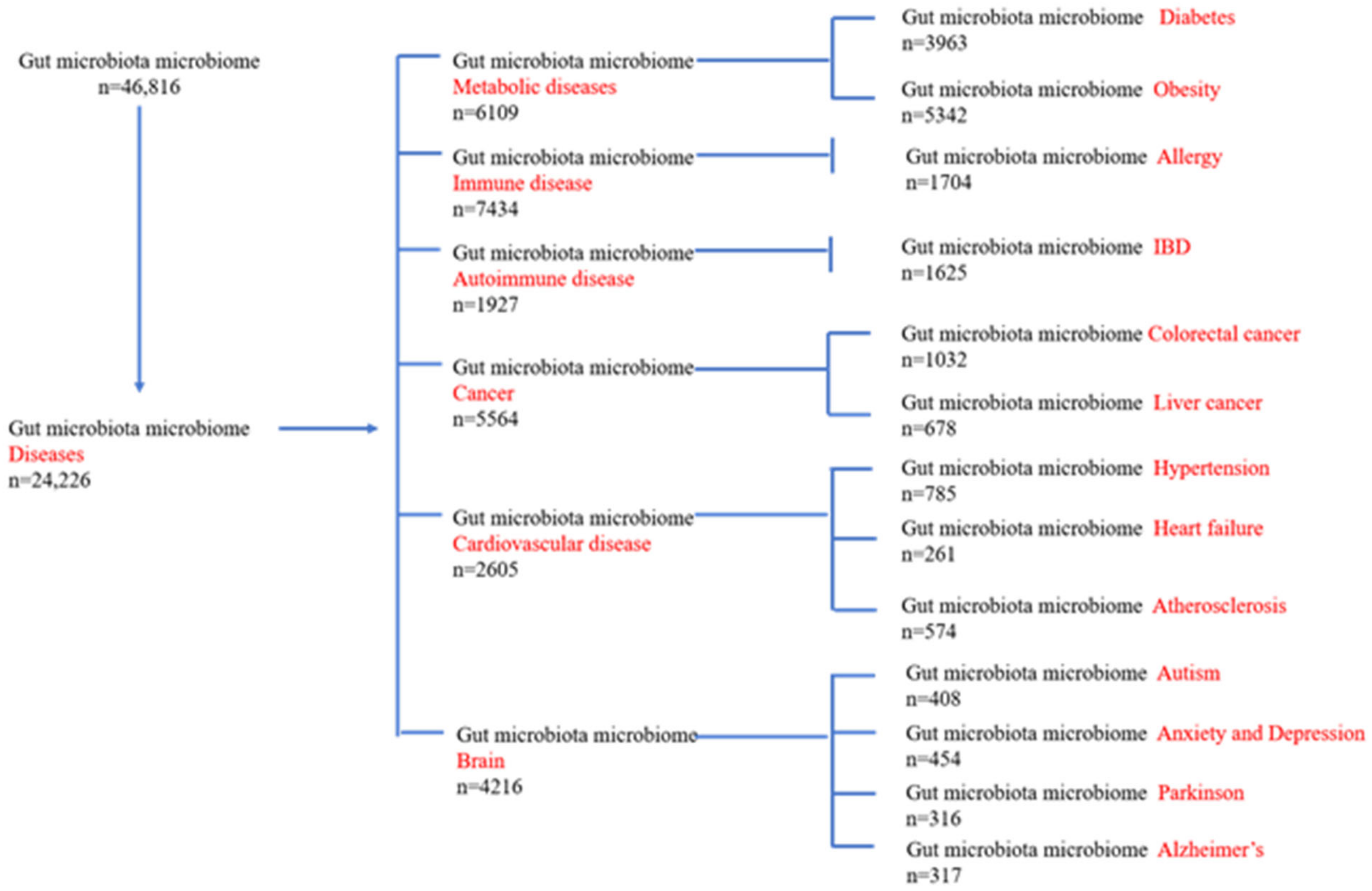
:1. Introduction
Methods of the Review
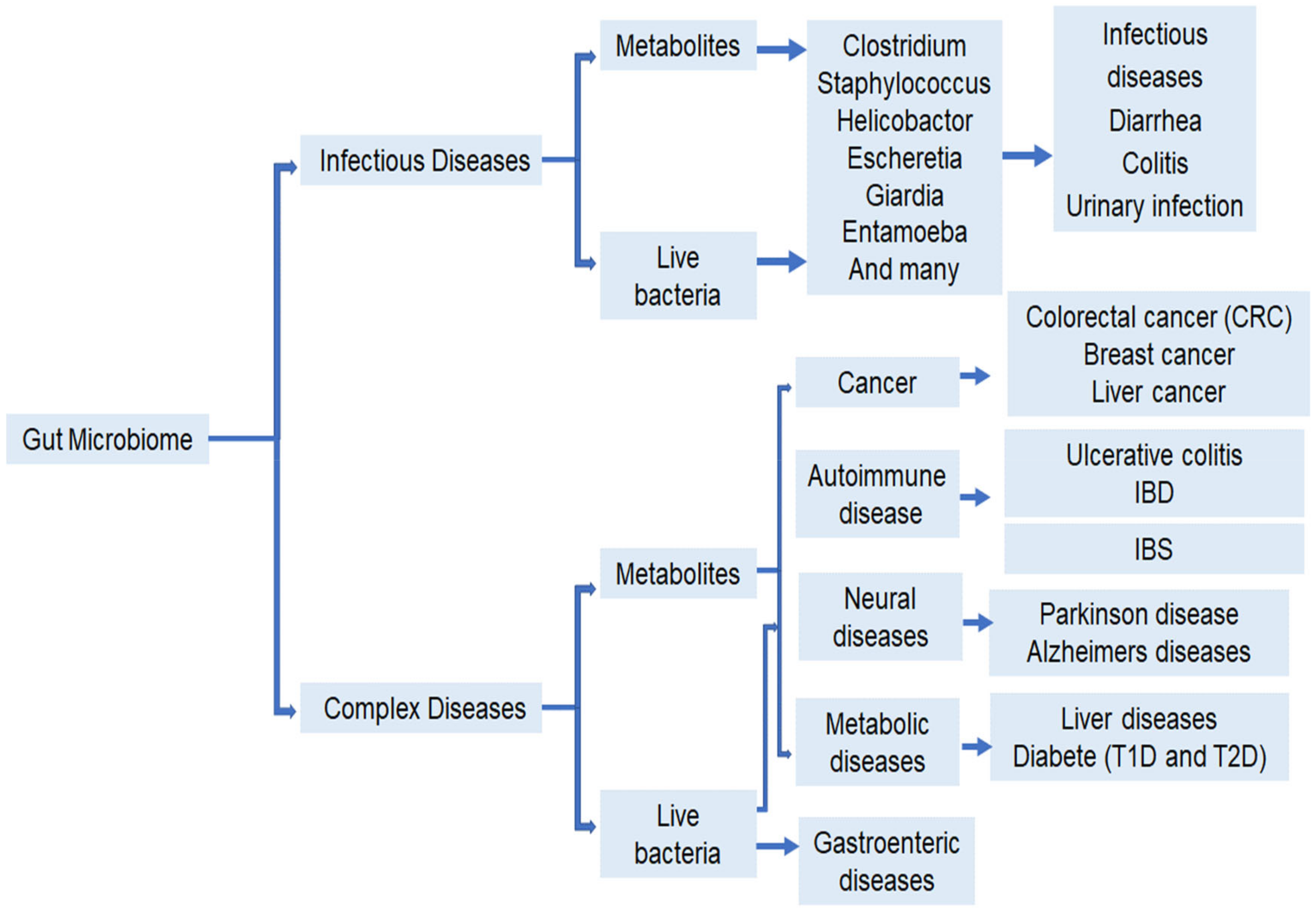
2. Gut Bacteria and Human Health
2.1. Digestive Tract Diseases
| Cells/Tissue/Other | Host | Disease | Methods | Bacteria Identified/Increased | References |
|---|---|---|---|---|---|
| Small intestine | Human | Digestive tract diseases | 16S RNA | Bacteroides, Lactobacillus, UC Erysipelotrichaceae | [28] |
| Intestinal tract cells | Mouse | Digestive tract diseases | IgA seq | Prevotellaceae, Helicobacter spp., Flexispira and SFB | [30]. |
| Ilium and rectum Biopsy | Human | IBD | 16S RNA | Proteobacteria | [32] |
| ileocecal biopsies | Human | PSC-IBD and UC | 16S RNA | Escherichia, Lachnospiraceae family, Veillonella and Megasphaera | [36] |
| Large intestine | Human | IBS | RT-QPCR | Lactobacillus, bifidobacteria, and Clostridium | [37] |
| Colon and caecum | Mice | Osmotic diarrhea | 16S RNA | Bacteroides | [38]. |
| Blood, stool, urine | Human | Anxiety and Depression in IBS | fMRI | Bifidobacterium longum NCC3001 | [39] |
| Fecal sample | Human | ASD | 16 S RNA | Faecalibacteriu, Ruminococcus,Sarcina and Clostridium | [40] |
| Fecal sample | Human | Schizophrenia | Magnetic Resonance Spectroscopy | Clostridium, Lactobacillus and Bacteroides | [41] |
| Frozen brain Biopsy | Human | Alzheimer’s | 16S RNA and Nextgen Sequencing | P. gingivalis, F. nucleatum and P. intermedia, Helicobacter pylori | [42] |
| Fecal sample | Human | Parkinson Disease | RT-QPCR | Enterobacteriaceae | [43] |
| Fecal sample | Human | Diabetes | 16S RNA | Bacteriodes, bifidobacteria, Clostridium | [44] |
| Cecum | Mice | Obesity | 16S RNA | Firmicutes and Bacteriodetes | [45] |
| Fecal sample | Human | Gaut | 16S RNA | Bacteroides, Porphyromonadaceae Rhodococcus, Erysipelatoclostridium and Anaerolineaceae | [46] |
| Fecal sample | Human | Hypertension | Metagenome shotgun sequencing | Klebsiella spp., Streptococcus spp., and Parabacteroides merdae | [47] |
| Fecal sample | Human | Atherosclerosis | Metagenome shotgun sequencing | Roseburia | [48]. |
| Fecal sample | Human | Colorectal cancer | RT-QPCR | F. nucleatum,Peptostreptococcus anaerobius, P. stomatis, Solobacterium moorei, Gemella morbillorum and Parvimonas micra | [49] |
| Fecal sample | Human | Colorectal Cancer | 16S RNA, NextGen sequencing | F. nucleatum | [50]. |
| Fecal sample | Human | Allergy | 16S RNA | Enterobacteriaceae and Parabacteroides | [51]. |
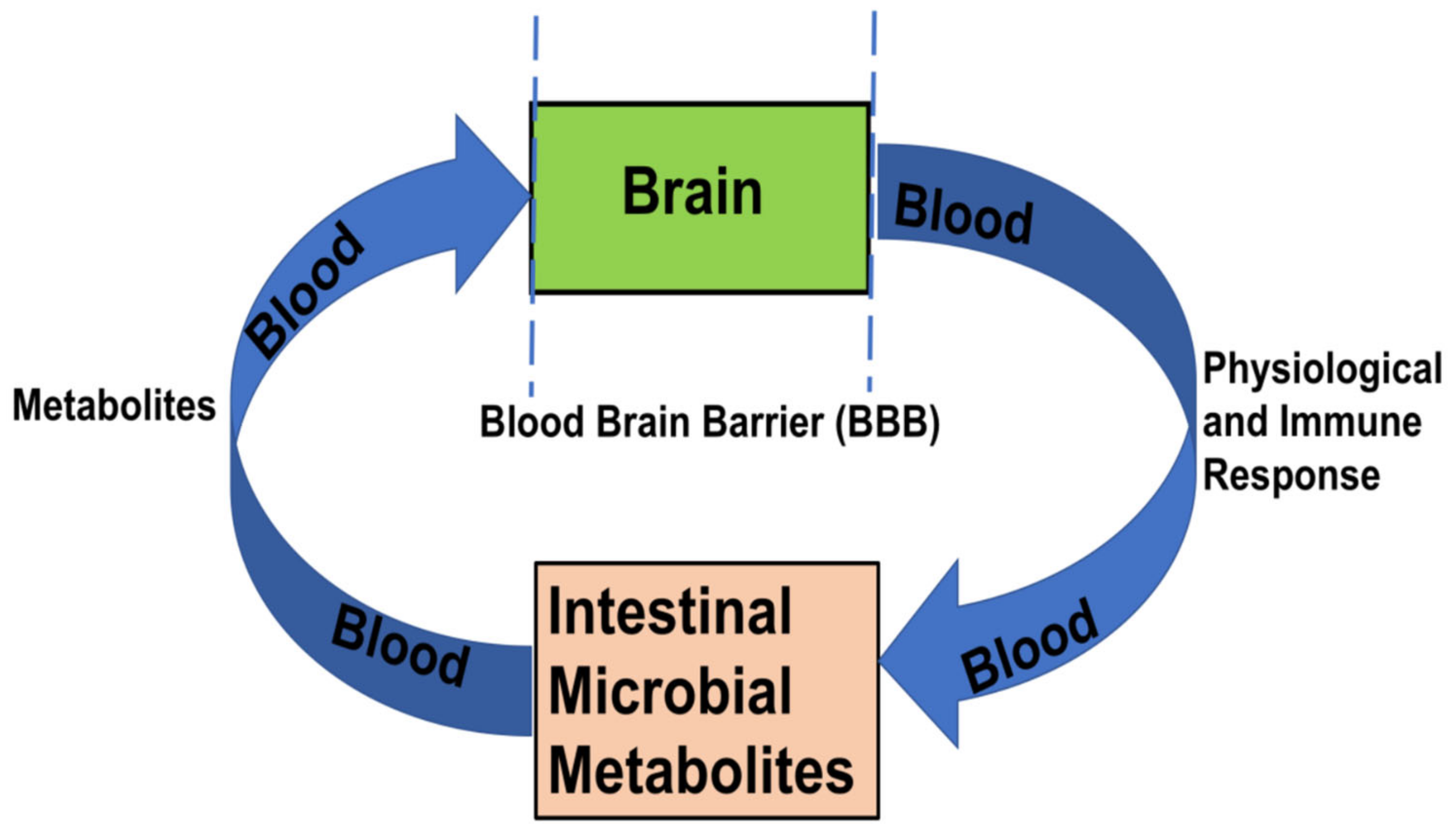
2.2. Intestinal Encephalopathy
2.2.1. Anxiety and Depression
2.2.2. Autism Spectrum Disorders (ASD)
2.2.3. Schizophrenia
2.2.4. Alzheimer’s Disease (AD)
2.2.5. Parkinson’s Disease (PD)
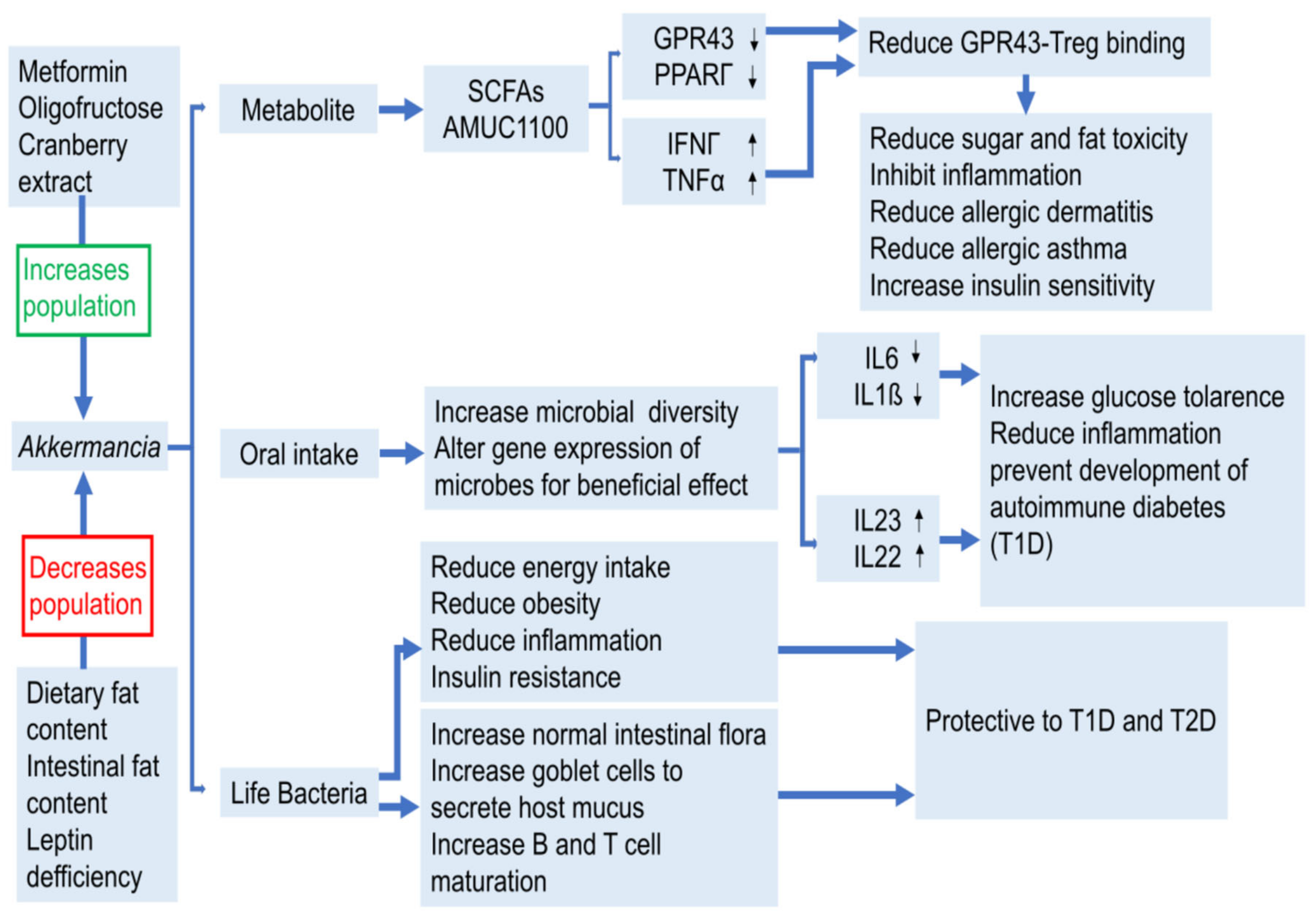
2.3. Metabolic Diseases
2.3.1. Diabetes & Obesity
2.3.2. Other Diseases with Abnormalities in Metabolism
Common Cardiovascular Diseases including Hypertension, Atherosclerosis and Chronic Hypertension
2.4. Cancer
2.5. Immune Dysfunction
3. The Mechanism of Action of Gut Bacteria in Health and Disease Development
3.1. Beneficial Bacteria and Harmful Bacteria
3.2. Metabolites
3.2.1. SCFAs
3.2.2. Small Molecules of the Microbial Enzymatic Pathways
3.2.3. Toxic Metabolites
3.2.4. Bile Acid Metabolism
4. Treatment and Adjustment
4.1. Fecal Microbial Transplantation (FMT) and Oral Administration
4.2. Dietary Interventions
4.2.1. Dietary Fiber
4.2.2. Probiotics and Prebiotics Products
4.3. Drug Adjustment
Drugs Have Therapeutic Effects on Some Diseases Caused by Microbial Disorders
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Banati, M.; Csecsei, P.; Koszegi, E.; Nielsen, H.H.; Suto, G.; Bors, L.; Trauninger, A.; Csepany, T.; Rozsa, C.; Jakab, G.; et al. Antibody response against gastrointestinal antigens in demyelinating diseases of the central nervous system. Eur. J. Neurol. 2013, 20, 1492–1495. [Google Scholar] [CrossRef] [PubMed]
- Barb, J.J.; Oler, A.J.; Kim, H.S.; Chalmers, N.; Wallen, G.R.; Cashion, A.; Munson, P.J.; Ames, N.J. Development of an Analysis Pipeline Characterizing Multiple Hypervariable Regions of 16S rRNA Using Mock Samples. PLoS ONE 2016, 11, 18. [Google Scholar]
- Buda, A.; Qualtrough, D.; Jepson, M.A.; Martines, D.; Paraskeva, C.; Pignatelli, M. Butyrate downregulates alpha(2)beta(1) integrin: A possible role in the induction of apoptosis in colorectal cancer cell lines. Gut 2003, 52, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Burn, J.; Gerdes, A.M.; Macrae, F.; Mecklin, J.P.; Moeslein, G.; Olschwang, S.; Eccles, D.; Evans, D.G.; Maher, E.R.; Bertario, L.; et al. Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: An analysis from the CAPP2 randomised controlled trial. Lancet 2011, 378, 2081–2087. [Google Scholar] [CrossRef] [Green Version]
- Zeng, M.Y.; Cisalpino, D.; Varadarajan, S.; Hellman, J.; Warren, H.S.; Cascalho, M.; Inohara, N.; Núñez, G. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity 2016, 44, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Qie, Y.; Park, J.; Kim, C.H. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host Microbe 2016, 20, 202–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clifford, M.N. Diet-derived Phenols in plasma and tissues and their implications for health. Planta Med. 2004, 70, 1103–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonnenburg, J.L.; Angenent, L.T.; Gordon, J.I. Getting a grip on things: How do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 2004, 5, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Lucas, C.; Barnich, N.; Nguyen, H.T.T. Microbiota, Inflammation and Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary intervention impact on gut microbial gene richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yu, J. The association of diet, gut microbiota and colorectal cancer: What we eat may imply what we get. Protein Cell 2018, 9, 474–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, H.C.; Pamp, S.J.; Hill, J.A.; Surana, N.K.; Edelman, S.M.; Troy, E.B.; Reading, N.C.; Villablanca, E.J.; Wang, S.; Mora, J.R.; et al. Gut Immune Maturation Depends on Colonization with a Host-Specific Microbiota. Cell 2012, 149, 1578–1593. [Google Scholar] [CrossRef] [Green Version]
- Circu, M.L.; Aw, T.Y. Intestinal redox biology and oxidative stress. Semin. Cell Dev. Biol. 2012, 23, 729–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marietta, E.; Horwath, I.; Taneja, V. Microbiome, Immunomodulation, and the Neuronal System. Neurotherapeutics 2018, 15, 23–30. [Google Scholar] [CrossRef] [Green Version]
- Seville, L.A.; Patterson, A.J.; Scott, K.P.; Mullany, P.; Quail, M.A.; Parkhill, J.; Ready, D.; Wilson, M.; Spratt, D.; Roberts, A.P. Distribution of Tetracycline and Erythromycin Resistance Genes Among Human Oral and Fecal Metagenomic DNA. Microb. Drug Resist. 2009, 15, 159–166. [Google Scholar] [CrossRef]
- Salyers, A.A.; Gupta, A.; Wang, Y.P. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004, 12, 412–416. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, X.; Lu, N.; Zhu, B. The abundance of antibiotic resistance genes in human guts has correlation to the consumption of antibiotics in animal. Gut Microbes 2014, 5, 245–249. [Google Scholar] [CrossRef] [Green Version]
- Forslund, K.; Sunagawa, S.; Kultima, J.R.; Mende, D.R.; Arumugam, M.; Typas, A.; Bork, P. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013, 23, 1163–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, N.; Hu, Y.; Zhu, L.; Yang, X.; Yin, Y.; Lei, F.; Zhu, Y.; Du, Q.; Wang, X.; Meng, Z.; et al. DNA microarray analysis reveals that antibiotic resistance-gene diversity in human gut microbiota is age related. Sci. Rep. 2014, 4, 4302. [Google Scholar] [CrossRef] [PubMed]
- Furness, J.B. The enteric nervous system and neurogastroenterology. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Ashton, P.M.; Nair, S.; Dallman, T.; Rubino, S.; Rabsch, W.; Mwaigwisya, S.; Wain, J.; O’Grady, J. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol. 2015, 33, 296–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berer, K.; Krishnamoorthy, G. Microbial view of central nervous system autoimmunity. FEBS Lett. 2014, 588, 4207–4213. [Google Scholar] [CrossRef]
- Blount, Z.D. The unexhausted potential of E. coli. eLife 2015, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ 2009, 339, b2535. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Pabst, O.; Slack, E. IgA and the intestinal microbiota: The importance of being specific. Mucosal. Immunol. 2020, 13, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunker, J.J.; Erickson, S.A.; Flynn, T.M.; Henry, C.; Koval, J.C.; Meisel, M.; Jabri, B.; Antonopoulos, D.A.; Wilson, P.C.; Bendelac, A. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 2017, 358, eaan6619. [Google Scholar] [CrossRef] [Green Version]
- Palm, N.W.; de Zoete, M.R.; Cullen, T.W.; Barry, N.A.; Stefanowski, J.; Hao, L.; Degnan, P.H.; Hu, J.; Peter, I.; Zhang, W.; et al. Immunoglobulin A Coating Identifies Colitogenic Bacteria in Inflammatory Bowel Disease. Cell 2014, 158, 1000–1010. [Google Scholar] [CrossRef] [Green Version]
- Schultz, B.M.; Paduro, C.A.; Salazar, G.A.; Salazar-Echegarai, F.J.; Sebastian, V.P.; Riedel, C.A.; Kalergis, A.M.; Alvarez-Lobos, M.; Bueno, S.M. A Potential Role of Salmonella Infection in the Onset of Inflammatory Bowel Diseases. Front. Immunol. 2017, 8, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vazquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, H.; Takechi, M.; Kiyonari, H.; Shioi, G.; Tamura, A.; Tsukita, S. Intestinal deletion of Claudin-7 enhances paracellular organic solute flux and initiates colonic inflammation in mice. Gut 2015, 64, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Lu, P.; Burger-van Paassen, N.; van der Sluis, M.; Witte-Bouma, J.; Kerckaert, J.P.; van Goudoever, J.B.; Van Seuningen, I.; Renes, I.B. Colonic Gene Expression Patterns of Mucin Muc2 Knockout Mice Reveal Various Phases in Colitis Development. Inflamm. Bowel Dis. 2011, 17, 2047–2057. [Google Scholar] [CrossRef] [PubMed]
- Gewirtz, A.T.; Navas, T.A.; Lyons, S.; Godowski, P.J.; Madara, J.L. Cutting edge: Bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 2001, 167, 1882–1885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossen, N.G.; Fuentes, S.; Boonstra, K.; D’Haens, G.R.; Heilig, H.G.; Zoetendal, E.G.; de Vos, W.M.; Ponsioen, C.Y. The Mucosa-associated Microbiota of PSC Patients is Characterized by Low Diversity and Low Abundance of Uncultured Clostridiales II. J. Crohns Colitis 2015, 9, 342–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.N.; Wu, H.; Chen, Y.Z.; Chen, Y.J.; Shen, X.Z.; Liu, T.T. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Dig. Liver Dis. 2017, 49, 331–337. [Google Scholar] [CrossRef]
- Tropini, C.; Moss, E.L.; Merrill, B.D.; Ng, K.M.; Higginbottom, S.K.; Casavant, E.P.; Gonzalez, C.G.; Fremin, B.; Bouley, D.M.; Elias, J.E.; et al. Transient Osmotic Perturbation Causes Long-Term Alteration to the Gut Microbiota. Cell 2018, 173, 1742–1754. [Google Scholar] [CrossRef] [Green Version]
- Pinto-Sanchez, M.I.; Hall, G.B.; Ghajar, K.; Nardelli, A.; Bolino, C.; Lau, J.T.; Martin, F.-P.; Cominetti, O.; Welsh, C.; Rieder, A.; et al. Probiotic Bifidobacterium longum NCC3001 Reduces Depression Scores and Alters Brain Activity: A Pilot Study in Patients With Irritable Bowel Syndrome. Gastroenterology 2017, 153, 448–459. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal Microbiota and Metabolome of Children with Autism and Pervasive Developmental Disorder Not Otherwise Specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Kosciolek, T.; Tang, J.; Zhou, Y.; Li, Z.; Ma, X.; Zhu, Q.; Yuan, N.; Yuan, L.; Li, C.; et al. Gut microbiome and magnetic resonance spectroscopy study of subjects at ultra-high risk for psychosis may support the membrane hypothesis. Eur. Psychiatry 2018, 53, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Emery, D.C.; Shoemark, D.K.; Batstone, T.E.; Waterfall, C.M.; Coghill, J.A.; Cerajewska, T.L.; Davies, M.; West, N.X.; Allen, S.J. 16S rRNA Next Generation Sequencing Analysis Shows Bacteria in Alzheimer’s Post-Mortem Brain. Front. Aging Neurosci. 2017, 9, 195. [Google Scholar] [CrossRef]
- Unger, M.M.; Spiegel, J.; Dillmann, K.-U.; Grundmann, D.; Philippeit, H.; Buermann, J.; Fassbender, K.; Schwiertz, A.; Schaefer, K.-H. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyotylainen, T.; Hamalainen, A.-M.; Peet, A.; Tillmann, V.; Poho, P.; Mattila, I.; et al. The Dynamics of the Human Infant Gut Microbiome in Development and in Progression toward Type 1 Diabetes. Cell Host Microbe 2015, 17, 260–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, T.; Shao, L.; Li, H.; Xie, Z.; He, Z.; Wen, C. Combined Signature of the Fecal Microbiome and Metabolome in Patients with Gout. Front. Microbiol. 2017, 8, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Q.; Gu, Y.; Li, X.; Yang, W.; Jia, L.; Chen, C.; Han, X.; Huang, Y.; Zhao, L.; Li, P.; et al. Alterations of the Gut Microbiome in Hypertension. Front. Cell. Infect. Microbiol. 2017, 7, 381. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, F.H.; Fak, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Backhed, F.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Feng, Q.; Wong, S.H.; Zhang, D.; Liang, Q.Y.; Qin, Y.; Tang, L.; Zhao, H.; Stenvang, J.; Li, Y.; et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017, 66, 70–78. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Wopereis, H.; Sim, K.; Shaw, A.; Warner, J.O.; Knol, J.; Kroll, J.S. Intestinal microbiota in infants at high risk for allergy: Effects of prebiotics and role in eczema development. J. Allergy Clin. Immunol. 2018, 141, 1334–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donnachie, E.; Schneider, A.; Mehring, M.; Enck, P. Incidence of irritable bowel syndrome and chronic fatigue following GI infection: A population-level study using routinely collected claims data. Gut 2018, 67, 1078–1086. [Google Scholar] [CrossRef]
- Pimentel, M.; Lembo, A. Microbiome and Its Role in Irritable Bowel Syndrome. Dig. Dis. Sci. 2020, 65, 829–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tziatzios, G.; Giamarellos-Bourboulis, E.J.; Papanikolaou, I.S.; Pimentel, M.; Dimitriadis, G.D.; Triantafyllou, K. Is small intestinal bacterial overgrowth involved in the pathogenesis of functional dyspepsia? Med. Hypotheses 2017, 106, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Tziatzios, G.; Gkolfakis, P.; Papanikolaou, I.S.; Mathur, R.; Pimentel, M.; Giamarellos-Bourboulis, E.J.; Triantafyllou, K. Gut Microbiota Dysbiosis in Functional Dyspepsia. Microorganisms 2020, 8, 691. [Google Scholar] [CrossRef] [PubMed]
- Sugano, K.; Tack, J.; Kuipers, E.J.; Graham, D.Y.; El-Omar, E.M.; Miura, S.; Haruma, K.; Asaka, M.; Uemura, N.; Malfertheiner, P. Kyoto global consensus report on Helicobacter pylori gastritis. Gut 2015, 64, 1353–1367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, S.S.; Gensollen, T.; Gandhi, A.; Oh, S.F.; Neves, J.F.; Collin, F.; Lavin, R.; Serra, C.; Glickman, J.; de Silva, P.S.A.; et al. Dietary and Microbial Oxazoles Induce Intestinal Inflammation by Modulating Aryl Hydrocarbon Receptor Responses. Cell 2018, 173, 1123–1134. [Google Scholar] [CrossRef] [Green Version]
- Kundu, P.; Lee, H.U.; Garcia-Perez, I.; Tay, E.X.Y.; Kim, H.; Faylon, L.E.; Martin, K.A.; Purbojati, R.; Drautz-Moses, D.I.; Ghosh, S.; et al. Neurogenesis and prolongevity signaling in young germ-free mice transplanted with the gut microbiota of old mice. Sci. Transl. Med. 2019, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.Y.; Chua, A.S. Influence of gut microbes on the brain-gut axis (Gut 2011;60:307-317). J. Neurogastroenterol. Motil. 2011, 17, 427–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grenham, S.; Clarke, G.; Cryan, J.F.; Dinan, T.G. Brain-gut-microbe communication in health and disease. Front. Physiol. 2011, 2, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gareau, M.G.; Wine, E.; Rodrigues, D.M.; Cho, J.H.; Whary, M.T.; Philpott, D.J.; MacQueen, G.; Sherman, P.M. Bacterial infection causes stress-induced memory dysfunction in mice. Gut 2011, 60, 307–317. [Google Scholar] [CrossRef]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bercik, P.; Park, A.J.; Sinclair, D.; Khoshdel, A.; Lu, J.; Huang, X.; Deng, Y.; Blennerhassett, P.A.; Fahnestock, M.; Moine, D.; et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol. Motil. 2011, 23, 1132–1139. [Google Scholar] [CrossRef] [Green Version]
- Krajmalnik-Brown, R.; Lozupone, C.; Kang, D.-W.; Adams, J.B. Gut bacteria in children with autism spectrum disorders: Challenges and promise of studying how a complex community influences a complex disease. Microb. Ecol. Health Dis. 2015, 26, 26914. [Google Scholar] [CrossRef] [PubMed]
- Severance, E.G.; Yolken, R.H.; Eaton, W.W. Autoimmune diseases, gastrointestinal disorders and the microbiome in schizophrenia: More than a gut feeling. Schizophr. Res. 2016, 176, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Keshavarzian, A.; Green, S.J.; Engen, P.A.; Voigt, R.M.; Naqib, A.; Forsyth, C.B.; Mutlu, E.; Shannon, K.M. Colonic Bacterial Composition in Parkinson’s Disease. Mov. Disord. 2015, 30, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Schachter, J.; Martel, J.; Lin, C.-S.; Chang, C.-J.; Wu, T.-R.; Lu, C.-C.; Ko, Y.-F.; Lai, H.-C.; Ojcius, D.M.; Young, J.D. Effects of obesity on depression: A role for inflammation and the gut microbiota. Brain Behav. Immun. 2018, 69, 1–8. [Google Scholar] [CrossRef]
- Huang, F.; Wu, X. Brain Neurotransmitter Modulation by Gut Microbiota in Anxiety and Depression. Front. Cell Dev. Biol. 2021, 9, 649103. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.A.; Diaz-Arteche, C.; Eliby, D.; Schwartz, O.S.; Simmons, J.G.; Cowan, C.S.M. The gut microbiota in anxiety and depression—A systematic review. Clin. Psychol. Rev. 2021, 83, 101943. [Google Scholar] [CrossRef] [PubMed]
- Clapp, M.; Aurora, N.; Herrera, L.; Bhatia, M.; Wilen, E.; Wakefield, S. Gut microbiota’s effect on mental health: The gut-brain axis. Clin Pract. 2017, 7, 987. [Google Scholar] [CrossRef]
- Coretti, L.; Cristiano, C.; Florio, E.; Scala, G.; Lama, A.; Keller, S.; Cuomo, M.; Russo, R.; Pero, R.; Paciello, O.; et al. Sex-related alterations of gut microbiota composition in the BTBR mouse model of autism spectrum disorder. Sci. Rep. 2017, 7, srep45356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roussin, L.; Prince, N.; Perez-Pardo, P.; Kraneveld, A.D.; Rabot, S.; Naudon, L. Role of the Gut Microbiota in the Pathophysiology of Autism Spectrum Disorder: Clinical and Preclinical Evidence. Microorganisms 2020, 8, 1369. [Google Scholar] [CrossRef]
- Li, Q.; Han, Y.; Dy, A.B.C.; Hagerman, R.J. The Gut Microbiota and Autism Spectrum Disorders. Front Cell Neurosci 2017, 11, 120. [Google Scholar] [CrossRef]
- Kang, D.-W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; LaBaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced Incidence of Prevotella and Other Fermenters in Intestinal Microflora of Autistic Children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [Green Version]
- Mannion, A.; Leader, G.; Healy, O. An investigation of comorbid psychological disorders, sleep problems, gastrointestinal symptoms and epilepsy in children and adolescents with Autism Spectrum Disorder. Res. Autism Spectr. Disord. 2013, 7, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism 2013, 4, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segata, N.; Waldron, L.; Ballarini, A.; Narasimhan, V.; Jousson, O.; Huttenhower, C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 2012, 9, 811–814. [Google Scholar] [CrossRef]
- Loohuis, L.M.O.; Mangul, S.; Ori, A.P.S.; Jospin, G.; Koslicki, D.; Yang, H.T.; Wu, T.; Boks, M.P.; Lomen-Hoerth, C.; Wiedau-Pazos, M.; et al. Transcriptome analysis in whole blood reveals increased microbial diversity in schizophrenia. Transl. Psychiatry 2018, 8, 96. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Xu, J.; Li, Z.; Huang, Y.; Yuan, Y.; Wang, J.; Zhang, M.; Hu, S.; Liang, Y. Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: A cross-sectional study. Schizophr. Res. 2018, 197, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Blennow, K.; Dubois, B.; Fagan, A.M.; Lewczuk, P.; de Leon, M.J.; Hampel, H. Clinical utility of cerebrospinal fluid biomarkers in the diagnosis of early Alzheimer’s disease. Alzheimers Dement 2015, 11, 58–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, E.A.; Duffy, L.C.; Schanbacher, F.L.; Qiao, H.P.; Dryja, D.; Leavens, A.; Rossman, J.; Rich, G.; Dirienzo, D.; Ogra, P.L. In vivo effects of bifidobacteria and lactoferrin on gut endotoxin concentration and mucosal immunity in balb/c mice. Dig. Dis. Sci. 2004, 49, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Hellwig, K.; Kvartsberg, H.; Portelius, E.; Andreasson, U.; Oberstein, T.J.; Lewczuk, P.; Blennow, K.; Kornhuber, J.; Maler, J.M.; Zetterberg, H.; et al. Neurogranin and YKL-40: Independent markers of synaptic degeneration and neuroinflammation in Alzheimer’s disease. Alzheimers Res. Ther. 2015, 7, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doulberis, M.; Kotronis, G.; Thomann, R.; Polyzos, S.A.; Boziki, M.; Gialamprinou, D.; Deretzi, G.; Katsinelos, P.; Kountouras, J. Review: Impact of Helicobacter pylori on Alzheimer’s disease: What do we know so far? Helicobacter 2018, 23, e12454. [Google Scholar] [CrossRef]
- Albaret, G.; Sifré, E.; Floch, P.; Laye, S.; Aubert, A.; Dubus, P.; Azzi-Martin, L.; Giese, A.; Salles, N.; Mégraud, F.; et al. Alzheimer’s Disease and Helicobacter pylori Infection: Inflammation from Stomach to Brain? J. Alzheimers Dis. 2020, 73, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, C.; Santangelo, R. Alzheimer’s disease and gut microbiota modifications: The long way between preclinical studies and clinical evidence. Pharmacol. Res. 2018, 129, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Arora, K.; Green, M.; Prakash, S. The Microbiome and Alzheimer’s Disease: Potential and Limitations of Prebiotic, Synbiotic, and Probiotic Formulations. Front. Bioeng Biotechnol. 2020, 8, 537847. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jaber, V.; Lukiw, W.J. Secretory Products of the Human GI Tract Microbiome and Their Potential Impact on Alzheimer’s Disease (AD): Detection of Lipopolysaccharide (LPS) in AD Hippocampus. Front. Cell. Infect. Microbiol. 2017, 7, 318. [Google Scholar] [CrossRef] [PubMed]
- Cui, B.; Su, D.; Li, W.; She, X.; Zhang, M.; Wang, R.; Zhai, Q. Effects of chronic noise exposure on the microbiome-gut-brain axis in senescence-accelerated prone mice: Implications for Alzheimer’s disease. J. Neuroinflammation 2018, 15, 190. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.-K.; Wang, J.-H.; Lei, W.-Y.; Chen, C.-L.; Chang, C.-Y.; Liou, L.-S. Helicobacter pylori infection is associated with an increased risk of Parkinson’s disease: A population-based retrospective cohort study. Parkinsonism. Relat. Disord. 2018, 47, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qian, Y.; Xu, S.; Song, Y.; Xiao, Q. Longitudinal Analysis of Fecal Microbiome and Pathologic Processes in a Rotenone Induced Mice Model of Parkinson’s Disease. Front. Aging Neurosci. 2018, 9, 441. [Google Scholar] [CrossRef] [Green Version]
- Crusell, M.K.W.; Hansen, T.H.; Nielsen, T.; Allin, K.H.; Ruehlemann, M.C.; Damm, P.; Vestergaard, H.; Rorbye, C.; Jorgensen, N.R.; Christiansen, O.B.; et al. Gestational diabetes is associated with change in the gut microbiota composition in third trimester of pregnancy and postpartum. Microbiome 2018, 6, 89. [Google Scholar] [CrossRef]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; Vlieg, J.E.T.V.-H.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015, 9, 1–15. [Google Scholar] [CrossRef]
- Giongo, A.; Gano, K.A.; Crabb, D.B.; Mukherjee, N.; Novelo, L.L.; Casella, G.; Drew, J.C.; Ilonen, J.; Knip, M.; Hyoty, H.; et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011, 5, 82–91. [Google Scholar] [CrossRef]
- Brown, C.T.; Davis-Richardson, A.G.; Giongo, A.; Gano, K.A.; Crabb, D.B.; Mukherjee, N.; Casella, G.; Drew, J.C.; Ilonen, J.; Knip, M.; et al. Gut Microbiome Metagenomics Analysis Suggests a Functional Model for the Development of Autoimmunity for Type 1 Diabetes. PLoS ONE 2011, 6, e25792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leiva-Gea, I.; Sanchez-Alcoholado, L.; Martin-Tejedor, B.; Castellano-Castillo, D.; Moreno-Indias, I.; Urda-Cardona, A.; Tinahones, F.J.; Carlos Fernandez-Garcia, J.; Isabel Queipo-Ortuno, M. Gut Microbiota Differs in Composition and Functionality Between Children With Type 1 Diabetes and MODY2 and Healthy Control Subjects: A Case-Control Study. Diabetes Care 2018, 41, 2385–2395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markle, J.G.M.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef] [Green Version]
- Gavin, P.G.; Mullaney, J.A.; Loo, D.; Cao, K.-A.L.; Gottlieb, P.A.; Hill, M.M.; Zipris, D.; Hamilton-Williams, E.E. Intestinal Metaproteomics Reveals Host-Microbiota Interactions in Subjects at Risk for Type 1 Diabetes. Diabetes Care 2018, 41, 2178–2186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Neyrinck, A.M.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Qin, Q.; Liu, M.; Zhang, X.; He, F.; Wang, G. Akkermansia muciniphila can reduce the damage of gluco/lipotoxicity, oxidative stress and inflammation, and normalize intestine microbiota in streptozotocin-induced diabetic rats. Pathog. Dis 2018, 76, fty028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miani, M.; Le Naour, J.; Waeckel-Enee, E.; Verma, S.C.; Straube, M.; Emond, P.; Ryffel, B.; Van Endert, P.; Sokol, H.; Diana, J. Gut Microbiota-Stimulated Innate Lymphoid Cells Support beta-Defensin 14 Expression in Pancreatic Endocrine Cells, Preventing Autoimmune Diabetes. Cell Metab. 2018, 28, 557–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.-S.; Li, J.; Krautkramer, K.A.; Badri, M.; Battaglia, T.; Borbet, T.C.; Koh, H.; Ng, S.; Sibley, R.A.; Li, Y.; et al. Antibiotic-induced acceleration of type 1 diabetes alters maturation of innate intestinal immunity. eLife 2018, 7, e22153. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; Molinaro, A.; Stahlman, M.; Khan, M.T.; Schmidt, C.; Manneras-Holm, L.; Wu, H.; Carreras, A.; Jeong, H.; Olofsson, L.E.; et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell 2018, 175, 947–961. [Google Scholar] [CrossRef] [Green Version]
- Moon, J.-Y.; Zolnik, C.P.; Wang, Z.; Qiu, Y.; Usyk, M.; Wang, T.; Kizer, J.R.; Landay, A.L.; Kurland, I.J.; Anastos, K.; et al. Gut microbiota and plasma metabolites associated with diabetes in women with, or at high risk for, HIV infection. Ebiomedicine 2018, 37, 392–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Cui, L.; Yan, X.; Zhao, X.; Cheng, J.; Zhou, L.; Gao, J.; Cao, Z.; Ye, X.; Hu, S. Analysis of Oral Microbiota Revealed High Abundance of Prevotella Intermedia in Gout Patients. Cell. Physiol. Biochem. 2018, 49, 1804–1812. [Google Scholar] [CrossRef]
- Guo, Z.; Zhang, J.; Wang, Z.; Ang, K.Y.; Huang, S.; Hou, Q.; Su, X.; Qiao, J.; Zheng, Y.; Wang, L.; et al. Intestinal Microbiota Distinguish Gout Patients from Healthy Humans. Sci. Rep. 2016, 6, 20602. [Google Scholar] [CrossRef] [Green Version]
- Jubair, W.K.; Hendrickson, J.D.; Severs, E.L.; Schulz, H.M.; Adhikari, S.; Ir, D.; Pagan, J.D.; Anthony, R.M.; Robertson, C.E.; Frank, D.N.; et al. Modulation of Inflammatory Arthritis in Mice by Gut Microbiota Through Mucosal Inflammation and Autoantibody Generation. Arthritis Rheumatol. 2018, 70, 1220–1233. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zeng, B.; Zhang, J.; Li, W.; Mou, F.; Wang, H.; Zou, Q.; Zhong, B.; Wu, L.; Wei, H.; et al. Role of the Gut Microbiome in Modulating Arthritis Progression in Mice. Sci. Rep. 2016, 6, 30594. [Google Scholar] [CrossRef]
- Schmidt, T.S.B.; Hayward, M.R.; Coelho, L.P.; Li, S.S.; Costea, P.I.; Voigt, A.Y.; Wirbel, J.; Maistrenko, O.M.; Alves, R.J.C.; Bergsten, E.; et al. Extensive transmission of microbes along the gastrointestinal tract. Elife 2019, 8, e42693. [Google Scholar] [CrossRef]
- Breban, M.; Tap, J.; Leboime, A.; Said-Nahal, R.; Langella, P.; Chiocchia, G.; Furet, J.-P.; Sokol, H. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann. Rheum. Dis. 2017, 76, 1614–1622. [Google Scholar] [CrossRef]
- van Dijkhuizen, E.H.P.; Del Chierico, F.; Malattia, C.; Russo, A.; Pires Marafon, D.; Ter Haar, N.M.; Magni-Manzoni, S.; Vastert, S.J.; Dallapiccola, B.; Prakken, B.; et al. Microbiome Analytics of the Gut Microbiota in Patients With Juvenile Idiopathic Arthritis: A Longitudinal Observational Cohort Study. Arthritis Rheumatol. 2018, 71, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
- Mell, B.; Jala, V.R.; Mathew, A.V.; Byun, J.; Waghulde, H.; Zhang, Y.; Haribabu, B.; Vijay-Kumar, M.; Pennathur, S.; Joe, B. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol. Genom. 2015, 47, 187–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walejko, J.M.; Kim, S.; Goel, R.; Handberg, E.M.; Richards, E.M.; Pepine, C.J.; Raizada, M.K. Gut microbiota and serum metabolite differences in African Americans and White Americans with high blood pressure. Int. J. Cardiol. 2018, 271, 336–339. [Google Scholar] [CrossRef] [PubMed]
- Karbach, S.H.; Schonfelder, T.; Brandao, I.; Wilms, E.; Hormann, N.; Jackel, S.; Schuler, R.; Finger, S.; Knorr, M.; Lagrange, J.; et al. Gut Microbiota Promote Angiotensin II-Induced Arterial Hypertension and Vascular Dysfunction. J. Am. Heart Assoc. 2016, 5, e003698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Y.; Aranda, J.M.; Rodriguez, V.; Raizada, M.K.; Pepine, C.J. Impact of antibiotics on arterial blood pressure in a patient with resistant hypertension—A case report. Int. J. Cardiol. 2015, 201, 157–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Arango, L.F.; Barrett, H.L.; McIntyre, H.D.; Callaway, L.K.; Morrison, M.; Nitert, M.D.; Grp, S.T. Increased Systolic and Diastolic Blood Pressure Is Associated With Altered Gut Microbiota Composition and Butyrate Production in Early Pregnancy. Hypertension 2016, 68, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Goel, R.; Kumar, A.; Qi, Y.; Lobaton, G.; Hosaka, K.; Mohammed, M.; Handberg, E.M.; Richards, E.M.; Pepine, C.J.; et al. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin. Sci. 2018, 132, 701–718. [Google Scholar] [CrossRef] [Green Version]
- Pasini, E.; Aquilani, R.; Testa, C.; Baiardi, P.; Angioletti, S.; Boschi, F.; Verri, M.; Dioguardi, F. Pathogenic Gut Flora in Patients With Chronic Heart Failure. Jacc-Heart Fail. 2016, 4, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.N.; Bazzano, L.A.; Ajami, N.J.; He, H.; Zhao, J.; Petrosino, J.F.; Correa, A.; He, J. Gut Microbiome Associates With Lifetime Cardiovascular Disease Risk Profile Among Bogalusa Heart Study Participants. Circ. Res. 2016, 119, 956–964. [Google Scholar] [CrossRef] [Green Version]
- Koren, O.; Spor, A.; Felin, J.; Fak, F.; Stombaugh, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108, 4592–4598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuevas-Ramos, G.; Petit, C.R.; Marcq, I.; Boury, M.; Oswald, E.; Nougayrede, J.-P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11537–11542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/beta-Catenin Signaling via its FadA Adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Tsoi, H.; Chu, E.S.H.; Zhang, X.; Sheng, J.; Nakatsu, G.; Ng, S.C.; Chan, A.W.H.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Peptostreptococcus anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology 2017, 152, 1419–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, S.H.; Zhao, L.; Zhang, X.; Nakatsu, G.; Han, J.; Xu, W.; Xiao, X.; Kwong, T.N.Y.; Tsoi, H.; Wu, W.K.K.; et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 2017, 153, 1621–1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grivennikov, S.I.; Wang, K.; Mucida, D.; Stewart, C.A.; Schnabl, B.; Jauch, D.; Taniguchi, K.; Yu, G.-Y.; Oesterreicher, C.H.; Hung, K.E.; et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012, 491, 254–1633. [Google Scholar] [CrossRef] [Green Version]
- Huber, S.; Gagliani, N.; Zenewicz, L.A.; Huber, F.J.; Bosurgi, L.; Hu, B.; Hedl, M.; Zhang, W.; O’Connor, W., Jr.; Murphy, A.J.; et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 2012, 491, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Flynn, K.J.; Baxter, N.T.; Schloss, P.D. Metabolic and Community Synergy of Oral Bacteria in Colorectal Cancer. Msphere 2016, 1, e00102. [Google Scholar] [CrossRef] [Green Version]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Claney, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef] [Green Version]
- Mima, K.; Cao, Y.; Chan, A.T.; Qian, Z.R.; Nowak, J.A.; Masugi, Y.; Shi, Y.; Song, M.; da Silva, A.; Gu, M.C.; et al. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin. Transl. Gastroenterol. 2016, 7, e200. [Google Scholar] [CrossRef] [PubMed]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 Protein of Fusobacterium nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors from Immune Cell Attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abed, J.; Emgard, J.E.M.; Zamir, G.; Faroja, M.; Almogy, G.; Grenov, A.; Sol, A.; Naor, R.; Pikarsky, E.; Atlan, K.A.; et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016, 20, 215–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomkovich, S.; Yang, Y.; Winglee, K.; Gauthier, J.; Muhlbauer, M.; Sun, X.; Mohamadzadeh, M.; Liu, X.; Martin, P.; Wang, G.P.; et al. Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis. Cancer Res. 2017, 77, 2620–2632. [Google Scholar] [CrossRef] [Green Version]
- Santaolalla, R.; Sussman, D.A.; Ruiz, J.R.; Davies, J.M.; Pastorini, C.; Espana, C.L.; Sotolongo, J.; Burlingame, O.; Bejarano, P.A.; Philip, S.; et al. TLR4 Activates the beta-catenin Pathway to Cause Intestinal Neoplasia. PLoS ONE 2013, 8, e63298. [Google Scholar]
- Shi, C.; Yang, Y.; Xia, Y.; Okugawa, Y.; Yang, J.; Liang, Y.; Chen, H.; Zhang, P.; Wang, F.; Han, H.; et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut 2016, 65, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Peng, Y.; Yu, J.; Chen, T.; Wu, Y.; Shi, L.; Li, Q.; Wu, J.; Fu, X. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget 2017, 8, 31705–31717. [Google Scholar] [CrossRef]
- Dejea, C.M.; Wick, E.C.; Hechenbleikner, E.M.; White, J.R.; Welch, J.L.M.; Rossetti, B.J.; Peterson, S.N.; Snesrud, E.C.; Borisy, G.G.; Lazarev, M.; et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 18321–18326. [Google Scholar] [CrossRef] [Green Version]
- Drewes, J.L.; White, J.R.; Dejea, C.M.; Fathi, P.; Iyadorai, T.; Vadivelu, J.; Roslani, A.C.; Wick, E.C.; Mongodin, E.F.; Loke, M.F.; et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. Npj Biofilms Microbiomes 2017, 3, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, D.J. Acute appendicitis and dietary fibre: An alternative hypothesis. Br. Med. J. (Clin. Res. Ed.) 1985, 290, 1125–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strachan, D.P. Hay fever, hygiene, and household size. BMJ 1989, 299, 1259–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endesfelder, D.; Castell, W.Z.; Ardissone, A.; Davis-Richardson, A.G.; Achenbach, P.; Hagen, M.; Pflueger, M.; Gano, K.A.; Fagen, J.R.; Drew, J.C.; et al. Compromised Gut Microbiota Networks in Children With Anti-Islet Cell Autoimmunity. Diabetes 2014, 63, 2006–2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dzidic, M.; Abrahamsson, T.R.; Artacho, A.; Collado, M.C.; Mira, A.; Jenmalm, M.C. Oral microbiota maturation during the first 7 years of life in relation to allergy development. Allergy 2018, 73, 2000–2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wypych, T.P.; Marsland, B. Antibiotics as Instigators of Microbial Dysbiosis: Implications for Asthma and Allergy. Trends Immunol. 2018, 39, 697–711. [Google Scholar] [CrossRef] [PubMed]
- Jenmalm, M.C. The mother-offspring dyad: Microbial transmission, immune interactions and allergy development. J. Intern. Med. 2017, 282, 484–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitre, E.; Susi, A.; Kropp, L.E.; Schwartz, D.J.; Gorman, G.H.; Nylund, C.M. Association Between Use of Acid-Suppressive Medications and Antibiotics During Infancy and Allergic Diseases in Early Childhood. JAMA Pediatr. 2018, 172, e180315. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.T.; Whyatt, R.; Hoepner, L.; Oberfield, S.; Dominguez-Bello, M.G.; Widen, E.M.; Hassoun, A.; Perera, F.; Rundle, A. Prenatal exposure to antibiotics, cesarean section and risk of childhood obesity. Int. J. Obes. 2015, 39, 665–670. [Google Scholar] [CrossRef] [Green Version]
- Lapin, B.; Piorkowski, J.; Ownby, D.; Freels, S.; Chavez, N.; Hernandez, E.; Wagner-Cassanova, C.; Pelzel, D.; Vergara, C.; Persky, V. Relationship between prenatal antibiotic use and asthma in at-risk children. Ann. Allergy Asthma Immunol. 2015, 114, 203–207. [Google Scholar] [CrossRef] [Green Version]
- Zachariassen, L.F.; Krych, L.; Engkilde, K.; Nielsen, D.S.; Kot, W.; Hansen, C.H.F.; Hansen, A.K. Sensitivity to oxazolone induced dermatitis is transferable with gut microbiota in mice. Sci. Rep. 2017, 7, srep44385. [Google Scholar] [CrossRef] [Green Version]
- Plantamura, E.; Dzutsev, A.; Chamaillard, M.; Djebali, S.; Moudombi, L.; Boucinha, L.; Grau, M.; Macari, C.; Bauche, D.; Dumitrescu, O.; et al. MAVS deficiency induces gut dysbiotic microbiota conferring a proallergic phenotype. Proc. Natl. Acad. Sci. USA 2018, 115, 10404–10409. [Google Scholar] [CrossRef] [Green Version]
- Teo, S.M.; Tang, H.H.F.; Mok, D.; Judd, L.M.; Watts, S.C.; Pham, K.; Holt, B.J.; Kusel, M.; Serralha, M.; Troy, N.; et al. Airway Microbiota Dynamics Uncover a Critical Window for Interplay of Pathogenic Bacteria and Allergy in Childhood Respiratory Disease. Cell Host Microbe 2018, 24, 341–352. [Google Scholar] [CrossRef] [Green Version]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, I.S.; Broere, F.; Manurung, S.; Lambers, T.T.; van der Zee, R.; van Eden, W. Lactobacillus rhamnosus GG-Derived Soluble Mediators Modulate Adaptive Immune Cells. Front. Immunol. 2018, 9, 1546. [Google Scholar] [CrossRef]
- Rodriguez-Nogales, A.; Algieri, F.; Garrido-Mesa, J.; Vezza, T.; Pilar Utrilla, M.; Chueca, N.; Garcia, F.; Elena Rodriguez-Cabezas, M.; Galvez, J. Intestinal anti-inflammatory effect of the probiotic Saccharomyces boulardii in DSS-induced colitis in mice: Impact on microRNAs expression and gut microbiota composition. J. Nutr. Biochem. 2018, 61, 129–139. [Google Scholar] [CrossRef]
- Kwon, M.-S.; Lim, S.K.; Jang, J.-Y.; Lee, J.; Park, H.K.; Kim, N.; Yun, M.; Shin, M.-Y.; Jo, H.E.; Oh, Y.J.; et al. Lactobacillus sakei WIKIM30 Ameliorates Atopic Dermatitis-Like Skin Lesions by Inducing Regulatory T cells and Altering Gut Microbiota Structure in Mice. Front. Immunol. 2018, 9, 1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saglani, S.; Gregory, L.G.; Manghera, A.K.; Branchett, W.J.; Uwadiae, F.; Entwistle, L.J.; Oliver, R.A.; Vasiliou, J.E.; Sherburn, R.; Lui, S.; et al. Inception of early-life allergen-induced airway hyperresponsiveness is reliant on IL-13(+)CD4(+) T cells. Sci. Immunol. 2018, 3, eaan4128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lew, L.-C.; Hor, Y.-Y.; Yusoff, N.A.A.; Choi, S.-B.; Yusoff, M.S.B.; Roslan, N.S.; Ahmad, A.; Mohammad, J.A.M.; Abdullah, M.F.I.L.; Zakaria, N.; et al. Probiotic Lactobacillus plantarum P8 alleviated stress and anxiety while enhancing memory and cognition in stressed adults: A randomised, double-blind, placebo-controlled study. Clin. Nutr. 2018, 38, 2053–2064. [Google Scholar] [CrossRef] [PubMed]
- Buffington, S.A.; Di Prisco, G.V.; Auchtung, T.A.; Ajami, N.J.; Petrosino, J.F.; Costa-Mattioli, M. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell 2016, 165, 1762–1775. [Google Scholar] [CrossRef] [Green Version]
- Majeed, M.; Nagabhushanam, K.; Arumugam, S.; Majeed, S.; Ali, F. Bacillus coagulans MTCC 5856 for the management of major depression with irritable bowel syndrome: A randomised, double-blind, placebo controlled, multi-centre, pilot clinical study. Food Nutr. Res. 2018, 62, 1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araya, R.E.; Goldszmid, R.S. Two Bugs a NOD Away from Improving Cancer Therapy Efficacy. Immunity 2016, 45, 714–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konishi, H.; Fujiya, M.; Tanaka, H.; Ueno, N.; Moriichi, K.; Sasajima, J.; Ikuta, K.; Akutsu, H.; Tanabe, H.; Kohgo, Y. Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nat. Commun. 2016, 7, 12365. [Google Scholar] [CrossRef]
- Balcazar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wake, N.; Asahi, Y.; Noiri, Y.; Hayashi, M.; Motooka, D.; Nakamura, S.; Gotoh, K.; Miura, J.; Machi, H.; Iida, T.; et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. Npj Biofilms Microbiomes 2016, 2, 16018. [Google Scholar] [CrossRef] [PubMed]
- Licht, T.R.; Christensen, B.B.; Krogfelt, K.A.; Molin, S. Plasmid transfer in the animal intestine and other dynamic bacterial populations: The role of community structure and environment. Microbiol.-Sgm 1999, 145, 2615–2622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, A.; Jose Salas-Jara, M.; Herrera, C.; Gonzalez, C. Biofilm and Helicobacter pylori: From environment to human host. World J. Gastroenterol. 2014, 20, 5632–5638. [Google Scholar] [CrossRef]
- Zarco, M.F.; Vess, T.J.; Ginsburg, G.S. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012, 18, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Weber, J.; Loening-Baucke, V.; Hale, L.P.; Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005, 43, 3380–3389. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.E.V.; Gustafsson, J.K.; Holmen-Larsson, J.; Jabbar, K.S.; Xia, L.; Xu, H.; Ghishan, F.K.; Carvalho, F.A.; Gewirtz, A.T.; Sjovall, H.; et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 2014, 63, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Hoarau, G.; Mukherjee, P.K.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. Mbio 2016, 7, e01250-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scales, B.S.; Huffnagle, G.B. The microbiome in wound repair and tissue fibrosis. J. Pathol. 2013, 229, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Dejea, C.M.; Fathi, P.; Craig, J.M.; Boleij, A.; Taddese, R.; Geis, A.L.; Wu, X.; Shields, C.E.D.; Hechenbleikner, E.M.; Huso, D.L.; et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018, 359, 592–597. [Google Scholar] [CrossRef] [Green Version]
- Arthur, J.C.; Gharaibeh, R.Z.; Muehlbauer, M.; Perez-Chanona, E.; Uronis, J.M.; McCafferty, J.; Fodor, A.A.; Jobin, C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun. 2014, 5, 4724. [Google Scholar] [CrossRef] [Green Version]
- Reti, K.L.; Tymensen, L.D.; Davis, S.P.; Amrein, M.W.; Buret, A.G. Campylobacter jejuni Increases Flagellar Expression and Adhesion of Noninvasive Escherichia coli: Effects on Enterocytic Toll-Like Receptor 4 and CXCL-8 Expression. Infect. Immun. 2015, 83, 4571–4581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeBlanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact 2017, 16, 79. [Google Scholar] [CrossRef] [Green Version]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ (Clin. Res. Ed.) 2018, 361, k2179. [Google Scholar] [CrossRef] [Green Version]
- Sleeth, M.L.; Thompson, E.L.; Ford, H.E.; Zac-Varghese, S.E.K.; Frost, G. Free fatty acid receptor 2 and nutrient sensing: A proposed role for fibre, fermentable carbohydrates and short-chain fatty acids in appetite regulation. Nutr. Res. Rev. 2010, 23, 135–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic T-reg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [Green Version]
- Belcheva, A.; Irrazabal, T.; Robertson, S.J.; Streutker, C.; Maughan, H.; Rubino, S.; Moriyama, E.H.; Copeland, J.K.; Kumar, S.; Green, B.; et al. Gut Microbial Metabolism Drives Transformation of Msh2-Deficient Colon Epithelial Cells. Cell 2014, 158, 288–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Thangaraju, M.; Cresci, G.A.; Liu, K.; Ananth, S.; Gnanaprakasam, J.P.; Browning, D.D.; Mellinger, J.D.; Smith, S.B.; Digby, G.J.; Lambert, N.A.; et al. GPR109A Is a G-protein-Coupled Receptor for the Bacterial Fermentation Product Butyrate and Functions as a Tumor Suppressor in Colon. Cancer Res. 2009, 69, 2826–2832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Chen, F.; Wu, W.; Sun, M.; Bilotta, A.J.; Yao, S.; Xiao, Y.; Huang, X.; Eaves-Pyles, T.D.; Golovko, G.; et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018, 11, 752–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef] [Green Version]
- Etxeberria, U.; Fernandez-Quintela, A.; Milagro, F.I.; Aguirre, L.; Alfredo Martinez, J.; Portillo, M.P. Impact of Polyphenols and Polyphenol-Rich Dietary Sources on Gut Microbiota Composition. J. Agric. Food Chem. 2013, 61, 9517–9533. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.B.; Gu, X.; Buffa, J.A.; Hurd, A.G.; Wang, Z.; Zhu, W.; Gupta, N.; Skye, S.M.; Cody, D.B.; Levison, B.S.; et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 2018, 24, 1407–1417. [Google Scholar] [CrossRef]
- Li, H.; Goh, B.N.; Teh, W.K.; Jiang, Z.; Goh, J.P.Z.; Goh, A.; Wu, G.; Hoon, S.S.; Raida, M.; Camattari, A.; et al. Skin Commensal Malassezia globosa Secreted Protease Attenuates Staphylococcus aureus Biofilm Formation. J. Investig. Dermatol. 2018, 138, 1137–1145. [Google Scholar] [CrossRef]
- Dinić, M.; Pecikoza, U.; Djokić, J.; Stepanović-Petrović, R.; Milenković, M.; Stevanović, M.; Filipović, N.; Begović, J.; Golić, N.; Lukić, J. Exopolysaccharide Produced by Probiotic Strain. Front Pharm. 2018, 9, 1. [Google Scholar] [CrossRef]
- Rolig, A.S.; Sweeney, E.G.; Kaye, L.E.; DeSantis, M.D.; Perkins, A.; Banse, A.V.; Hamilton, M.K.; Guillemin, K. A bacterial immunomodulatory protein with lipocalin-like domains facilitates host-bacteria mutualism in larval zebrafish. eLife 2018, 7, e37172. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.D.; Myers, C.J.; Harris, S.C.; Kakiyama, G.; Lee, I.-K.; Yun, B.-S.; Matsuzaki, K.; Furukawa, M.; Min, H.-K.; Bajaj, J.S.; et al. Bile Acid 7alpha-Dehydroxylating Gut Bacteria Secrete Antibiotics that Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell Chem. Biol. 2018, 26, 27–34.e4. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Wei, Y.; Liu, X.; Zhou, Y.; Jiang, L.; Yin, J.; Wang, F.; Hu, Y.; Urs, A.N.N.; Liu, Y.; et al. Indoleacetate decarboxylase is a glycyl radical enzyme catalysing the formation of malodorant skatole. Nat. Commun. 2018, 9, 4224. [Google Scholar] [CrossRef] [PubMed]
- Koppel, N.; Bisanz, J.E.; Pandelia, M.-E.; Turnbaugh, P.J.; Balskus, E.P. Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. eLife 2018, 7, e33953. [Google Scholar] [CrossRef]
- Russell, W.R.; Duncan, S.H.; Scobbie, L.; Duncan, G.; Cantlay, L.; Calder, A.G.; Anderson, S.E.; Flint, H.J. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 2013, 57, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Loh, Y.H.; Jakszyn, P.; Luben, R.N.; Mulligan, A.A.; Mitrou, P.N.; Khaw, K.-T. N-nitroso compounds and cancer incidence: The European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Am. J. Clin. Nutr. 2011, 93, 1053–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Windey, K.; De Preter, V.; Verbeke, K. Relevance of protein fermentation to gut health. Mol. Nutr. Food Res. 2012, 56, 184–196. [Google Scholar] [CrossRef]
- Hanfrey, C.C.; Pearson, B.M.; Hazeldine, S.; Lee, J.; Gaskin, D.J.; Woster, P.M.; Phillips, M.A.; Michael, A.J. Alternative Spermidine Biosynthetic Route Is Critical for Growth of Campylobacter jejuni and Is the Dominant Polyamine Pathway in Human Gut Microbiota. J. Biol. Chem. 2011, 286, 43301–43312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Martino, M.L.; Campilongo, R.; Casalino, M.; Micheli, G.; Colonna, B.; Prosseda, G. Polyamines: Emerging players in bacteria-host interactions. Int. J. Med Microbiol. 2013, 303, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Marquet, P.; Duncan, S.H.; Chassard, C.; Bernalier-Donadille, A.; Flint, H.J. Lactate has the potential to promote hydrogen sulphide formation in the human colon. Fems Microbiol. Lett. 2009, 299, 128–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roediger, W.E.; Moore, J.; Babidge, W. Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig. Dis. Sci. 1997, 42, 1571–1579. [Google Scholar] [CrossRef]
- Barrasa, J.I.; Olmo, N.; Antonia Lizarbe, M.; Turnay, J. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol. Vitr. 2013, 27, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.M.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef]
- Islam, K.B.M.S.; Fukiya, S.; Hagio, M.; Fujii, N.; Ishizuka, S.; Ooka, T.; Ogura, Y.; Hayashi, T.; Yokota, A. Bile Acid Is a Host Factor That Regulates the Composition of the Cecal Microbiota in Rats. Gastroenterology 2011, 141, 1773–1781. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, N.; Kronenberg, M. Cancer immunity thwarted by the microbiome. Science 2018, 360, 858–859. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, Y.; Wang, S.; Bian, X.; Jiang, X.; Wu, J.; Wang, K.; Wang, Q.; Xia, J.; Jiang, S.; et al. Western Diet Aggravated Carbon Tetrachloride-Induced Chronic Liver Injury by Disturbing Gut Microbiota and Bile Acid Metabolism. Mol. Nutr. Food Res. 2021, 65, e2000811. [Google Scholar] [CrossRef] [PubMed]
- Majer, F.; Sharma, R.; Mullins, C.; Keogh, L.; Phipps, S.; Duggan, S.; Kelleher, D.; Keely, S.; Long, A.; Radics, G.; et al. New highly toxic bile acids derived from deoxycholic acid, chenodeoxycholic acid and lithocholic acid. Bioorganic Med. Chem. 2014, 22, 256–268. [Google Scholar] [CrossRef]
- Cui, B.; Feng, Q.; Wang, H.; Wang, M.; Peng, Z.; Li, P.; Huang, G.; Liu, Z.; Wu, P.; Fan, Z.; et al. Fecal microbiota transplantation through mid-gut for refractory Crohn’s disease: Safety, feasibility, and efficacy trial results. J. Gastroenterol. Hepatol. 2015, 30, 51–58. [Google Scholar] [CrossRef] [PubMed]
- van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; de Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.W.M.; Tijssen, J.G.P.; et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Angelberger, S.; Reinisch, W.; Makristathis, A.; Lichtenberger, C.; Dejaco, C.; Papay, P.; Novacek, G.; Trauner, M.; Loy, A.; Erry, D.B. Temporal Bacterial Community Dynamics Vary Among Ulcerative Colitis Patients After Fecal Microbiota Transplantation. Am. J. Gastroenterol. 2013, 108, 1620–1630. [Google Scholar] [CrossRef] [PubMed]
- Tariq, R.; Pardi, D.S.; Tosh, P.K.; Walker, R.C.; Razonable, R.R.; Khanna, S. Fecal Microbiota Transplantation for Recurrent Clostridium difficile Infection Reduces Recurrent Urinary Tract Infection Frequency. Clin. Infect. Dis. 2017, 65, 1745–1747. [Google Scholar] [CrossRef]
- Oppfeldt, A.M.; Dahlerup, J.F.; Christensen, L.A.; Hvas, C.L. Faecal microbiota transplantation for recurring Clostridium difficile infection in a patient with Crohn’s disease and ileorectal anastomosis. BMJ Case Rep. 2016, 2016, bcr2016217209. [Google Scholar] [CrossRef] [PubMed]
- Rebello, D.; Wang, E.; Yen, E.; Lio, P.A.; Kelly, C.R. Hair Growth in Two Alopecia Patients after Fecal Microbiota Transplant. Acg Case Rep. J. 2017, 4, e107. [Google Scholar] [CrossRef]
- Holvoet, T.; Joossens, M.; Wang, J.; Boelens, J.; Verhasselt, B.; Laukens, D.; van Vlierberghe, H.; Hindryckx, P.; De Vos, M.; De Looze, D.; et al. Assessment of faecal microbial transfer in irritable bowel syndrome with severe bloating. Gut 2017, 66, 980–982. [Google Scholar] [CrossRef] [Green Version]
- Keshteli, A.H.; Millan, B.; Madsen, K.L. Pretreatment with antibiotics may enhance the efficacy of fecal microbiota transplantation in ulcerative colitis: A meta-analysis. Mucosal Immunol. 2017, 10, 565–566. [Google Scholar] [CrossRef] [Green Version]
- van Beurden, Y.H.; van Gills, T.; van Gils, N.A.; Kassam, Z.; Mulder, C.J.J.; Aparicio-Pages, N. Serendipity in Refractory Celiac Disease: Full Recovery of Duodenal Villi and Clinical Symptoms after Fecal Microbiota Transfer. J. Gastrointest. Liver Dis. 2016, 25, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Kassam, Z.; Fagan, A.; Gavis, E.A.; Liu, E.; Cox, I.J.; Kheradman, R.; Heuman, D.; Wang, J.; Gurry, T.; et al. Fecal Microbiota Transplant From a Rational Stool Donor Improves Hepatic Encephalopathy: A Randomized Clinical Trial. Hepatology 2017, 66, 1727–1738. [Google Scholar] [CrossRef]
- Tian, H.; Ge, X.; Nie, Y.; Yang, L.; Ding, C.; McFarland, L.V.; Zhang, X.; Chen, Q.; Gong, J.; Li, N. Fecal microbiota transplantation in patients with slow-transit constipation: A randomized, clinical trial. PLoS ONE 2017, 12, e0171308. [Google Scholar] [CrossRef] [PubMed]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, D.; Fang, Z.; Jie, Z.; Qiu, X.; Zhang, C.; Chen, Y.; Ji, L. Human Gut Microbiota Changes Reveal the Progression of Glucose Intolerance. PLoS ONE 2013, 8, e71108. [Google Scholar] [CrossRef] [PubMed]
- Jakobsdottir, G.; Xu, J.; Molin, G.; Ahrne, S.; Nyman, M. High-Fat Diet Reduces the Formation of Butyrate, but Increases Succinate, Inflammation, Liver Fat and Cholesterol in Rats, while Dietary Fibre Counteracts These Effects. PLoS ONE 2013, 8, e80476. [Google Scholar] [CrossRef] [Green Version]
- Ganesh, B.P.; Klopfleisch, R.; Loh, G.; Blaut, M. Commensal Akkermansia muciniphila Exacerbates Gut Inflammation in Salmonella Typhimurium-Infected Gnotobiotic Mice. PLoS ONE 2013, 8, e74963. [Google Scholar] [CrossRef] [PubMed]
- Mithieux, G. Does Akkermansia muciniphila play a role in type 1 diabetes? Gut 2018, 67, 1373–1374. [Google Scholar] [CrossRef]
- Grander, C.; Adolph, T.E.; Wieser, V.; Lowe, P.; Wrzosek, L.; Gyongyosi, B.; Ward, D.V.; Grabherr, F.; Gerner, R.R.; Pfister, A.; et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut 2018, 67, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillere, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, J.; Wang, X.; Liu, K.; He, D.; Niu, P.; Cao, R.; Jin, L.; Wu, J. Oral delivery of staphylococcal nuclease by Lactococcus lactis prevents type 1 diabetes mellitus in NOD mice. Appl. Microbiol. Biotechnol. 2017, 101, 7653–7662. [Google Scholar] [CrossRef] [PubMed]
- Bharwani, A.; Mian, M.F.; Surette, M.G.; Bienenstock, J.; Forsythe, P. Oral treatment with Lactobacillus rhamnosus attenuates behavioural deficits and immune changes in chronic social stress. Bmc Med. 2017, 15, 7. [Google Scholar] [CrossRef] [Green Version]
- Secher, T.; Kassem, S.; Benamar, M.; Bernard, I.; Boury, M.; Barreau, F.; Oswald, E.; Saoudi, A. Oral Administration of the Probiotic Strain Escherichia coli Nissle 1917 Reduces Susceptibility to Neuroinflammation and Repairs Experimental Autoimmune Encephalomyelitis-Induced Intestinal Barrier Dysfunction. Front. Immunol. 2017, 8, 1096. [Google Scholar] [CrossRef] [PubMed]
- Strus, M.; Helwich, E.; Lauterbach, R.; Rzepecka-Weglarz, B.; Nowicka, K.; Wilinska, M.; Szczapa, J.; Rudnicka, M.; Slawska, H.; Szczepanski, M.; et al. Effects of oral probiotic supplementation on gut Lactobacillus and Bifidobacterium populations and the clinical status of low-birth-weight preterm neonates: A multicenter randomized, double-blind, placebo-controlled trial. Infect. Drug Resist. 2018, 11, 1557–1571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, R.; Edu, A.; De Seta, F. Study on the effects of an oral lactobacilli and lactoferrin complex in women with intermediate vaginal microbiota. Arch. Gynecol. Obstet. 2018, 298, 139–145. [Google Scholar] [CrossRef]
- Spacova, I.; Petrova, M.I.; Fremau, A.; Pollaris, L.; Vanoirbeek, J.; Ceuppens, J.L.; Seys, S.; Lebeer, S. Intranasal administration of probiotic Lactobacillus rhamnosus GG prevents birch pollen-induced allergic asthma in a murine model. Allergy 2018, 74, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Verspreet, J.; Damen, B.; Broekaert, W.F.; Verbeke, K.; Delcour, J.A.; Courtin, C.M. A Critical Look at Prebiotics Within the Dietary Fiber Concept. Annu. Rev. Food Sci. Technol. 2016, 7, 167–190. [Google Scholar]
- Staller, K.; Song, M.; Grodstein, F.; Whitehead, W.E.; Matthews, C.A.; Kuo, B.; Chan, A.T. Increased Long-term Dietary Fiber Intake Is Associated With a Decreased Risk of Fecal Incontinence in Older Women. Gastroenterology 2018, 155, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Berer, K.; Martinez, I.; Walker, A.; Kunkel, B.; Schmitt-Kopplin, P.; Walter, J.; Krishnamoorthy, G. Dietary non-fermentable fiber prevents autoimmune neurological disease by changing gut metabolic and immune status. Sci. Rep. 2018, 8, 10431. [Google Scholar] [CrossRef] [PubMed]
- Shively, C.A.; Register, T.C.; Appt, S.E.; Clarkson, T.B.; Uberseder, B.; Clear, K.Y.J.; Wilson, A.S.; Chiba, A.; Tooze, J.A.; Cook, K.L. Consumption of Mediterranean versus Western Diet Leads to Distinct Mammary Gland Microbiome Populations. Cell Rep. 2018, 25, 47–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roager, H.M.; Vogt, J.K.; Kristensen, M.; Hansen, L.B.S.; Ibrugger, S.; Markedahl, R.B.; Bahl, M.I.; Lind, M.V.; Nielsen, R.L.; Frokiar, H.; et al. Whole grain-rich diet reduces body weight and systemic low-grade inflammation without inducing major changes of the gut microbiome: A randomised cross-over trial. Gut 2019, 68, 83–93. [Google Scholar] [CrossRef] [Green Version]
- Whisner, C.M.; Martin, B.R.; Nakatsu, C.H.; Story, J.A.; MacDonald-Clarke, C.J.; McCabe, L.D.; McCabe, G.P.; Weaver, C.M. Soluble Corn Fiber Increases Calcium Absorption Associated with Shifts in the Gut Microbiome: A Randomized Dose-Response Trial in Free-Living Pubertal Females. J. Nutr. 2016, 146, 1298–1306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peñalver, R.; Lorenzo, J.M.; Ros, G.; Amarowicz, R.; Pateiro, M.; Nieto, G. Seaweeds as a Functional Ingredient for a Healthy Diet. Mar Drugs 2020, 18, 18. [Google Scholar] [CrossRef] [PubMed]
- Kearney, S.M.; Gibbons, S.M.; Erdman, S.E.; Alm, E.J. Orthogonal Dietary Niche Enables Reversible Engraftment of a Gut Bacterial Commensal. Cell Rep. 2018, 24, 1842–1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- So, D.; Whelan, K.; Rossi, M.; Morrison, M.; Holtmann, G.; Kelly, J.T.; Shanahan, E.R.; Staudacher, H.M.; Campbell, K.L. Dietary fiber intervention on gut microbiota composition in healthy adults: A systematic review and meta-analysis. Am. J. Clin. Nutr. 2018, 107, 965–983. [Google Scholar] [CrossRef] [Green Version]
- Katsnelson, A. Prebiotics gain prominence but remain poorly defined. Proc. Natl. Acad. Sci. USA 2016, 113, 14168–14169. [Google Scholar] [CrossRef] [Green Version]
- Sohn, K.; Underwood, M.A. Prenatal and postnatal administration of prebiotics and probiotics. Semin. Fetal Neonatal Med. 2017, 22, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Chua, M.C.; Ben-Amor, K.; Lay, C.; Neo, A.G.E.; Chiang, W.C.; Rao, R.; Chew, C.; Chaithongwongwatthana, S.; Khemapech, N.; Knol, J.; et al. Effect of Synbiotic on the Gut Microbiota of Cesarean Delivered Infants: A Randomized, Double-blind, Multicenter Study. J. Pediatric Gastroenterol. Nutr. 2017, 65, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Tunapong, W.; Apaijai, N.; Yasom, S.; Tanajak, P.; Wanchai, K.; Chunchai, T.; Kerdphoo, S.; Eaimworawuthikul, S.; Thiennimitr, P.; Pongchaidecha, A.; et al. Chronic treatment with prebiotics, probiotics and synbiotics attenuated cardiac dysfunction by improving cardiac mitochondrial dysfunction in male obese insulin-resistant rats. Eur. J. Nutr. 2018, 57, 2091–2104. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, P.; Wu, Y.; Guo, P.; Liu, L.; Ma, N.; Levesque, C.; Chen, Y.; Zhao, J.; Zhang, J.; et al. Dietary Fiber Increases Butyrate-Producing Bacteria and Improves the Growth Performance of Weaned Piglets. J. Agric. Food Chem. 2018, 66, 7995–8004. [Google Scholar] [CrossRef]
- Qian, L.; Gao, R.; Huang, J.; Qin, H. Supplementation of triple viable probiotics combined with dietary intervention is associated with gut microbial improvement in humans on a high-fat diet. Exp. Med. 2019, 18, 2262–2270. [Google Scholar] [CrossRef]
- Grimaldi, R.; Gibson, G.R.; Vulevic, J.; Giallourou, N.; Castro-Mejia, J.L.; Hansen, L.H.; Gibson, E.L.; Nielsen, D.S.; Costabile, A. A prebiotic intervention study in children with autism spectrum disorders (ASDs). Microbiome 2018, 6, 133. [Google Scholar] [CrossRef]
- Le Bourgot, C.; Apper, E.; Blat, S.; Respondek, F. Fructo-oligosaccharides and glucose homeostasis: A systematic review and meta-analysis in animal models. Nutr. Metab. 2018, 15, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinborn, V.; Valenzuela, C.; Olivares, M.; Arredondo, M.; Weill, R.; Pizarro, F. Prebiotics increase heme iron bioavailability and do not affect non-heme iron bioavailability in humans. Food Funct. 2017, 8, 1994–1999. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Chassaing, B.; Singh, V.; Pellizzon, M.; Ricci, M.; Fythe, M.D.; Kumar, M.V.; Gewirtz, A.T. Fiber-Mediated Nourishment of Gut Microbiota Protects against Diet-Induced Obesity by Restoring IL-22-Mediated Colonic Health. Cell Host Microbe 2018, 23, 41–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hryckowian, A.J.; Van Treuren, W.; Smits, S.A.; Davis, N.M.; Gardner, J.O.; Bouley, D.M.; Sonnenburg, J.L. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat. Microbiol. 2018, 3, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Bindels, L.B.; Neyrinck, A.M.; Claus, S.P.; Le Roy, C.I.; Grangette, C.; Pot, B.; Martinez, I.; Walter, J.; Cani, P.D.; Delzenne, N.M. Synbiotic approach restores intestinal homeostasis and prolongs survival in leukaemic mice with cachexia. Isme J. 2016, 10, 1456–1470. [Google Scholar] [CrossRef] [PubMed]
- Francavilla, R.; Piccolo, M.; Francavilla, A.; Polimeno, L.; Semeraro, F.; Cristofori, F.; Castellaneta, S.; Barone, M.; Indrio, F.; Gobbetti, M.; et al. Clinical and Microbiological Effect of a Multispecies Probiotic Supplementation in Celiac Patients With Persistent IBS-type Symptoms: A Randomized, Double-Blind, Placebo-controlled, Multicenter Trial. J. Clin. Gastroenterol. 2018, 53, e117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, X.; Tian, H.; Ding, C.; Gu, L.; Wei, Y.; Gong, J.; Zhu, W.; Li, N.; Li, J. Fecal Microbiota Transplantation in Combination with Soluble Dietary Fiber for Treatment of Slow Transit Constipation: A Pilot Study. Arch. Med Res. 2016, 47, 236–242. [Google Scholar] [CrossRef]
- Bohula, E.A.; Scirica, B.M.; Inzucchi, S.E.; McGuire, D.K.; Keech, A.C.; Smith, S.R.; Kanevsky, E.; Murphy, S.A.; Leiter, L.A.; Dwyer, J.P.; et al. Effect of lorcaserin on prevention and remission of type 2 diabetes in overweight and obese patients (CAMELLIA-TIMI 61): A randomised, placebo-controlled trial. Lancet 2018, 392, 2269–2279. [Google Scholar] [CrossRef]
- Kanarek, N.; Keys, H.R.; Cantor, J.R.; Lewis, C.A.; Chan, S.H.; Kunchok, T.; Abu-Remaileh, M.; Freinkman, E.; Schweitzer, L.D.; Sabatini, D.M. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature 2018, 559, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Seddon, M.M.; Bookstaver, P.B.; Justo, J.A.; Kohn, J.; Rac, H.; Haggard, E.; Mediwala, K.N.; Dash, S.; Al-Hasan, M.N. Role of Early De-escalation of Antimicrobial Therapy on Risk of Clostridioides difficile Infection following Enterobacteriaceae Bloodstream Infections. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018, 69, 414–420. [Google Scholar] [CrossRef]
- Ferrario, C.; Statello, R.; Carnevali, L.; Mancabelli, L.; Milani, C.; Mangifesta, M.; Duranti, S.; Lugli, G.A.; Jimenez, B.; Lodge, S.; et al. How to Feed the Mammalian Gut Microbiota: Bacterial and Metabolic Modulation by Dietary Fibers. Front. Microbiol. 2017, 8, 1749. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, D.; Meng, X.; de Vos, W.M.; Wu, H.; Fang, X.; Maiti, A.K. Implications of Gut Microbiota in Complex Human Diseases. Int. J. Mol. Sci. 2021, 22, 12661. https://doi.org/10.3390/ijms222312661
Yu D, Meng X, de Vos WM, Wu H, Fang X, Maiti AK. Implications of Gut Microbiota in Complex Human Diseases. International Journal of Molecular Sciences. 2021; 22(23):12661. https://doi.org/10.3390/ijms222312661
Chicago/Turabian StyleYu, Dahai, Xin Meng, Willem M. de Vos, Hao Wu, Xuexun Fang, and Amit K. Maiti. 2021. "Implications of Gut Microbiota in Complex Human Diseases" International Journal of Molecular Sciences 22, no. 23: 12661. https://doi.org/10.3390/ijms222312661
APA StyleYu, D., Meng, X., de Vos, W. M., Wu, H., Fang, X., & Maiti, A. K. (2021). Implications of Gut Microbiota in Complex Human Diseases. International Journal of Molecular Sciences, 22(23), 12661. https://doi.org/10.3390/ijms222312661






