Antiseptic 9-Meric Peptide with Potency against Carbapenem-Resistant Acinetobacter baumannii Infection
Abstract
:1. Introduction
2. Results
2.1. Peptide Design
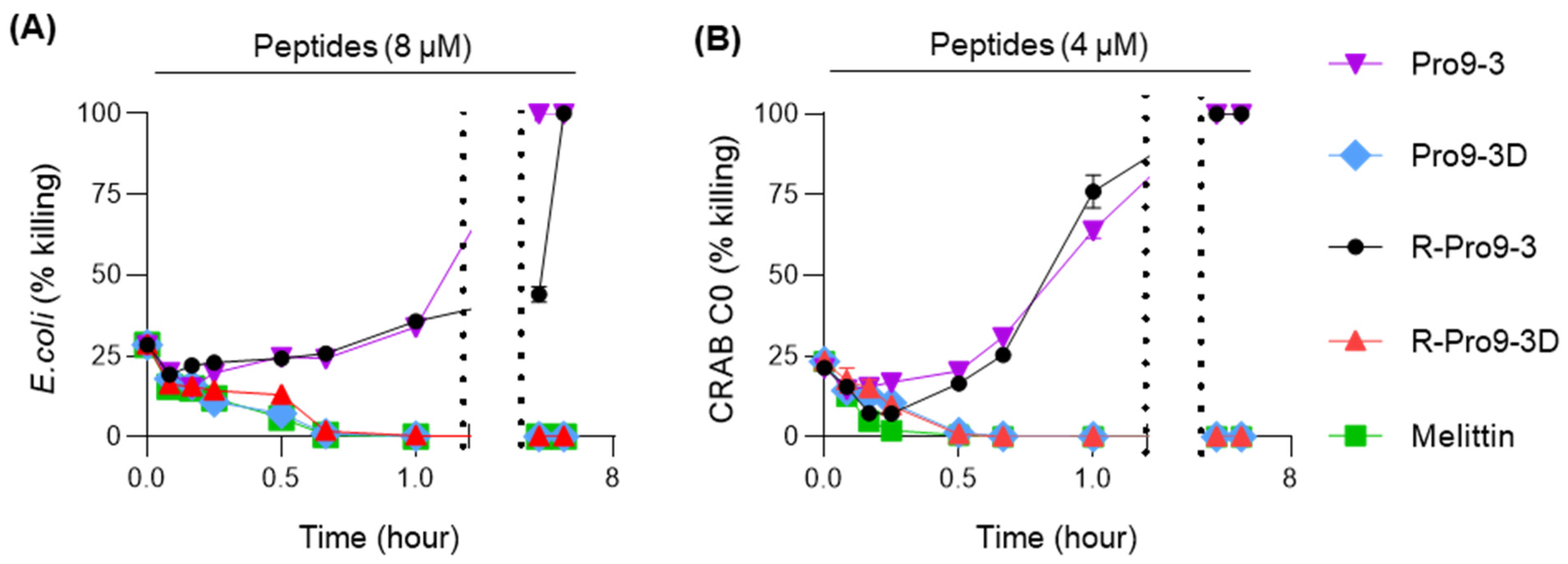
2.2. Antibacterial Activity of Peptides against Carbapenem-Resistant A. baumannii (CRAB)
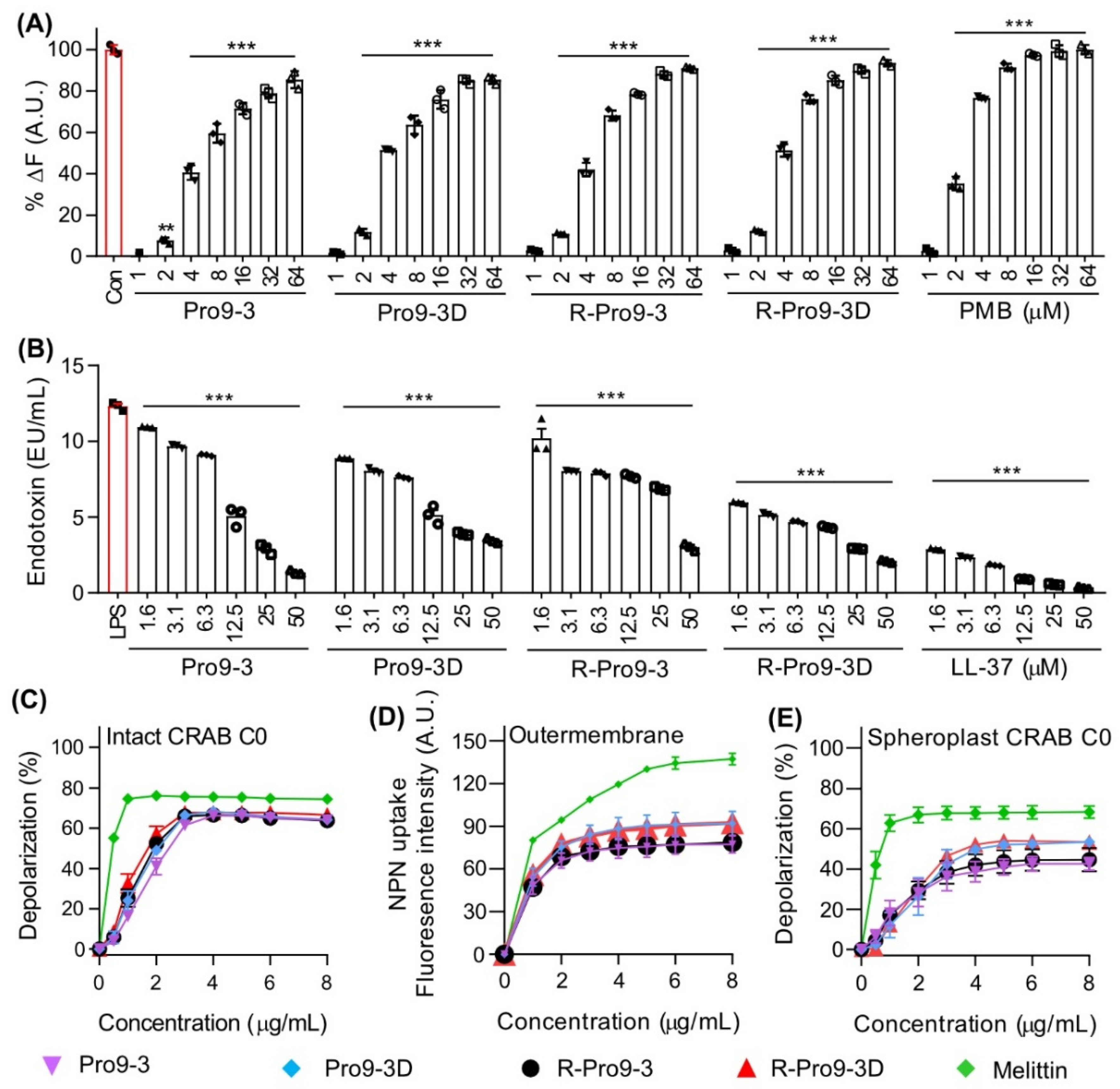
2.3. Mechanism of Antibacterial Activity against CRAB
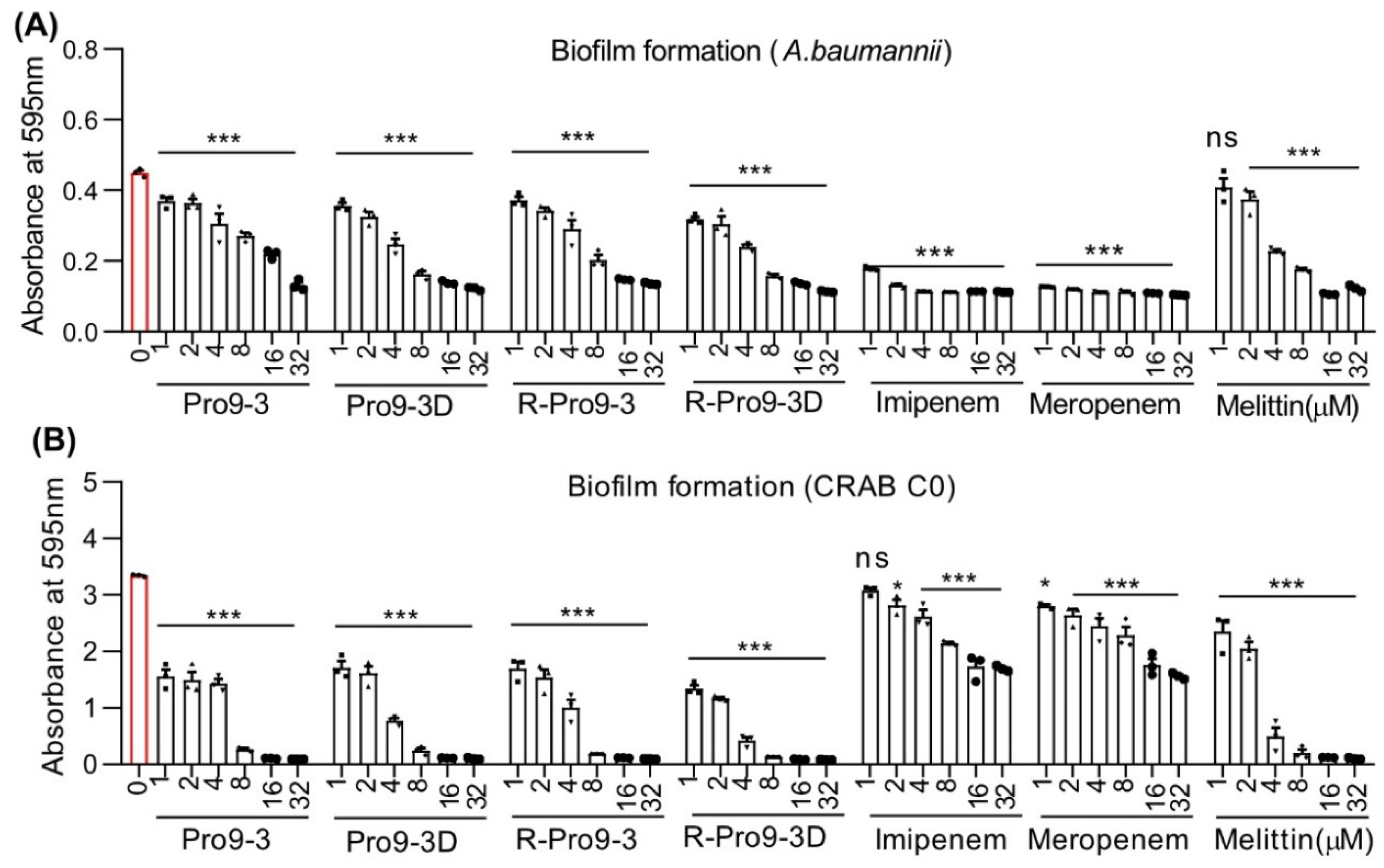
2.4. Effect of Peptides on Killing Biofilm Forming Bacteria
2.5. Resistance of Peptides against Protease Digestion
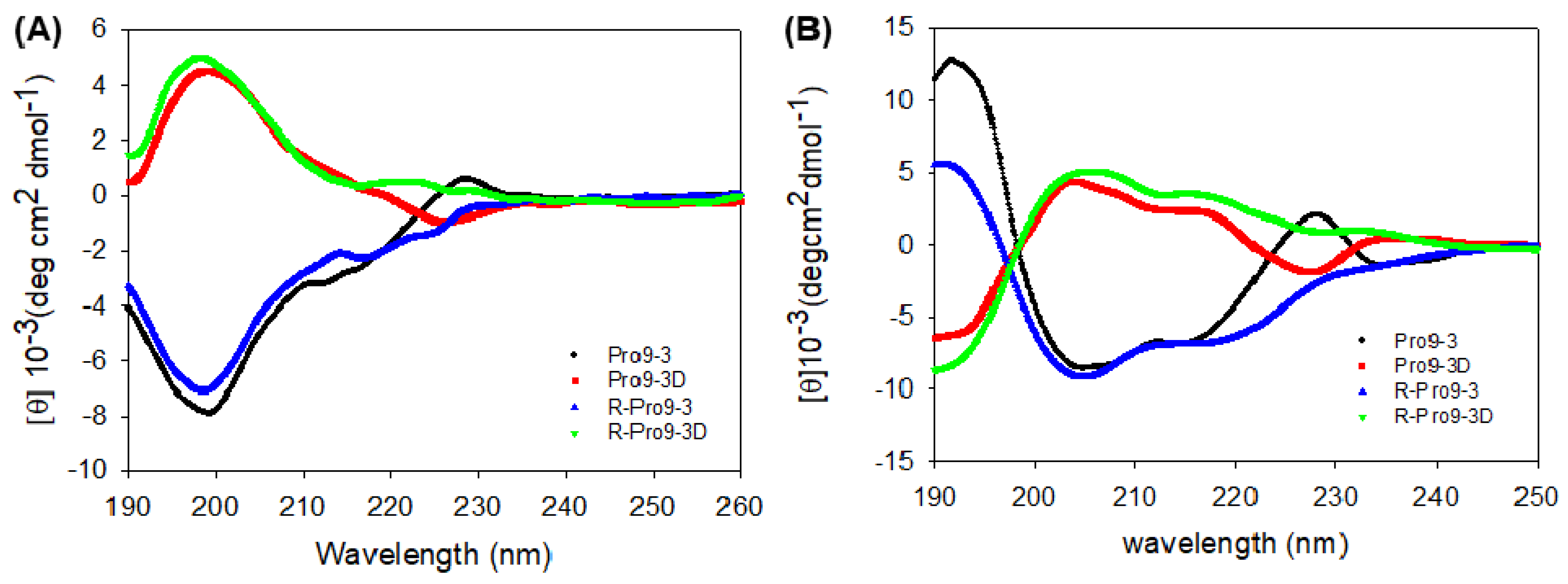
2.6. Circular Dichroism (CD) Spectroscopy of Peptides
2.7. Cytotoxicity of Peptides against Mammalian Cells
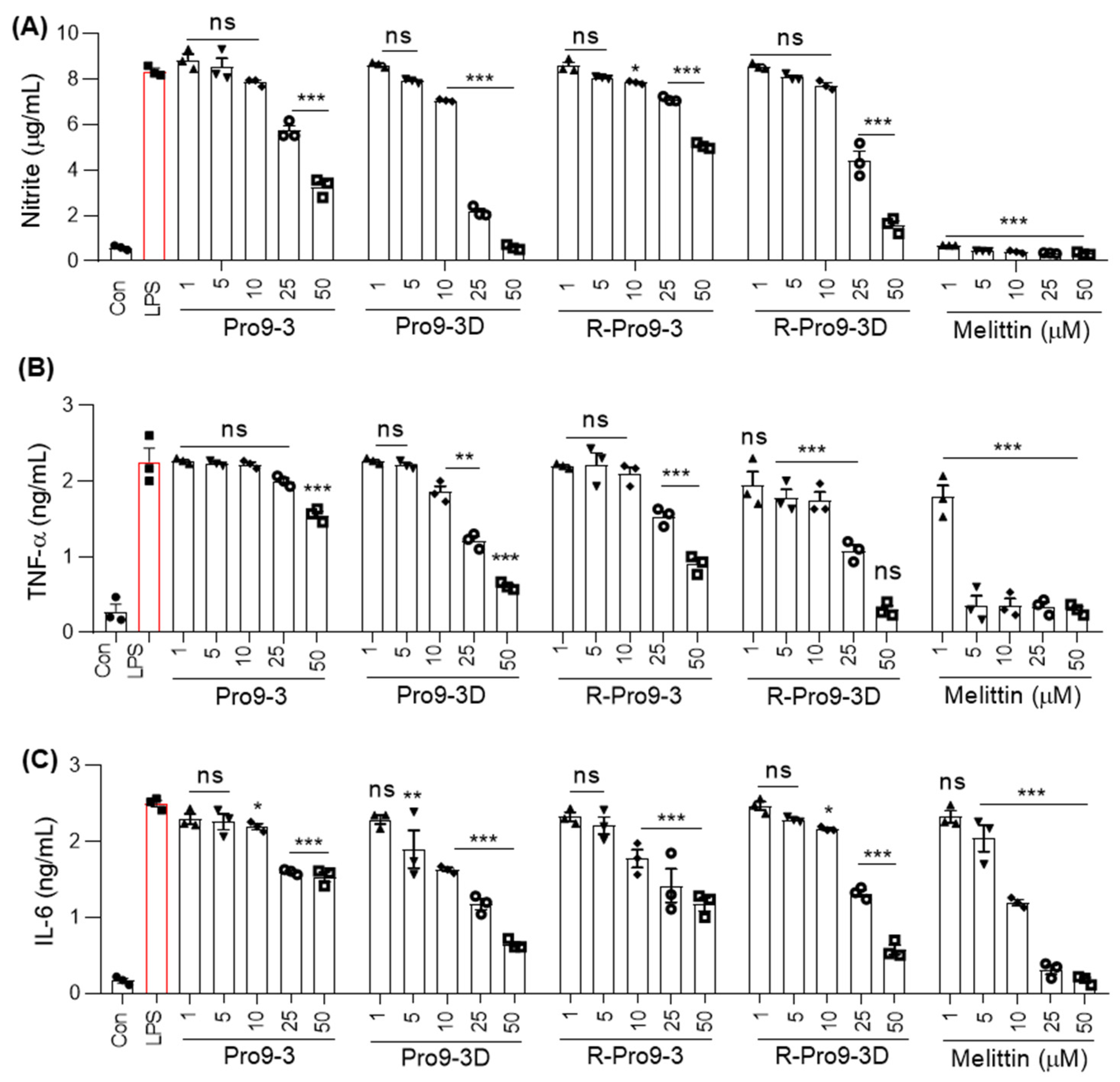
2.8. R-Pro9-3D Suppresses Inflammatory Cytokine Production in Lipopolysaccharide (LPS)-Stimulated RAW 264.7 Cells
2.9. R-Pro9-3D Effectively Damaged the Outer Membrane of CRAB C0
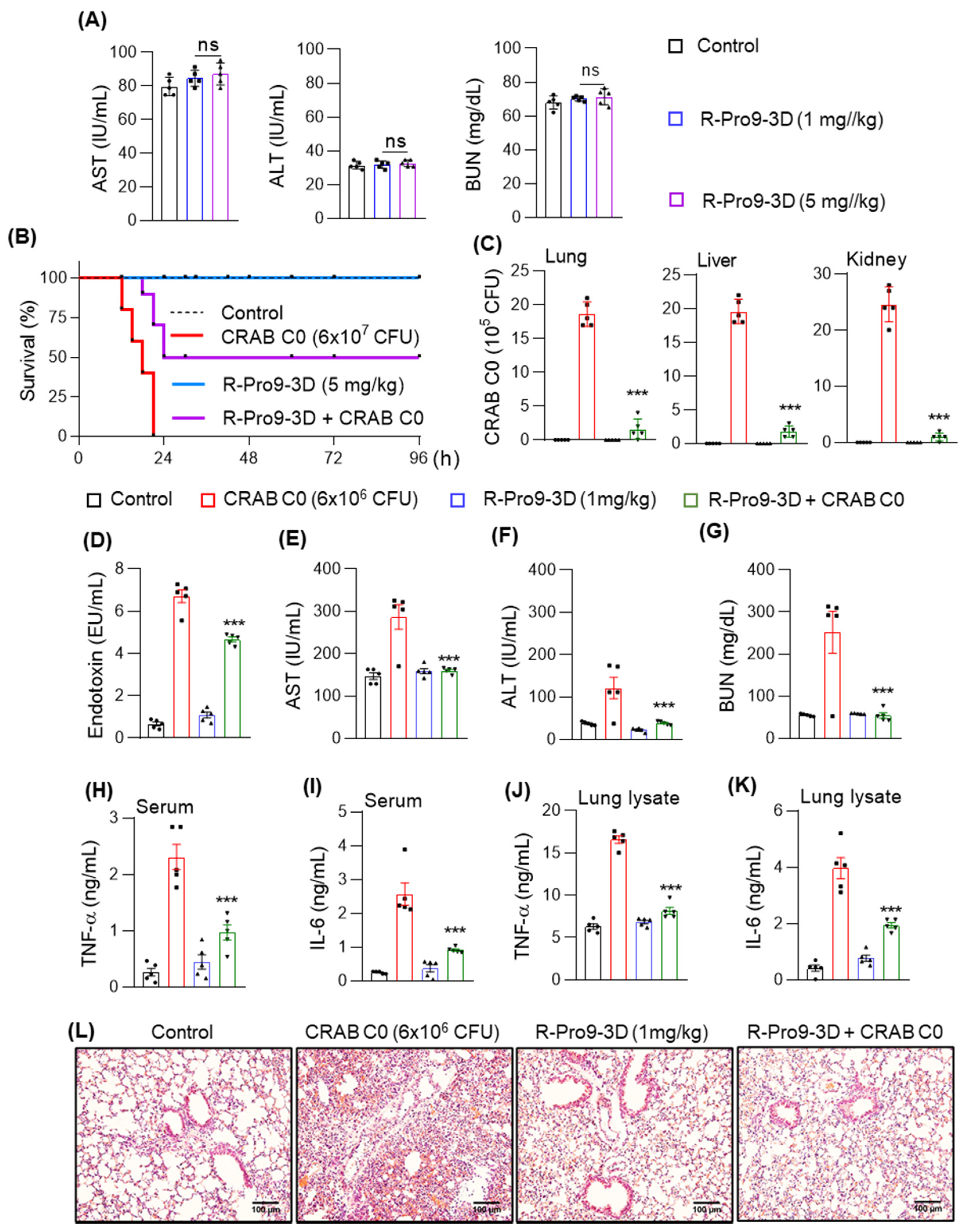
2.10. R-Pro9-3D Protects Mice against CRAB-Induced Septic Shock
3. Discussion
4. Materials and Methods
4.1. Peptide Synthesis
4.2. Bacterial Strains
4.3. Measurement of Antibacterial Activity
4.4. Protease Stability Assay
4.5. Time-Killing Assay
4.6. Peptide–LPS Binding Assay
4.7. Limulus Amebocyte Lysate (LAL) Assay
4.8. CRAB Depolarization Assay
4.9. Bacterial Outer Membrane Permeability Assay
4.10. Biofilm Assay
4.11. Circular Dichroism (CD) Analysis
4.12. Hemolytic Aassay
4.13. Cytotoxicity Assessment In Vitro
4.14. Scanning Electron Microscope Analysis
4.15. Quantification of Nitrite and Inflammatory Cytokine Production in LPS-Stimulated RAW264.7 Cells
4.16. Animals
4.17. In Vivo Toxicity Measurements
4.18. Survival Analysis
4.19. CRABC0 Sepsis Mouse Model
4.20. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, J.; Wei, H. Immune Intervention in Sepsis. Front. Pharmacol. 2021, 12, 718089. [Google Scholar] [CrossRef]
- Maldonado, R.F.; Sa-Correia, I.; Valvano, M.A. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol. Rev. 2016, 40, 480–493. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Howard, A.; O’Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Spellberg, B.; Rex, J.H. The value of single-pathogen antibacterial agents. Nat. Rev. Drug Discov. 2013, 12, 963. [Google Scholar] [CrossRef] [Green Version]
- Giammanco, A.; Cala, C.; Fasciana, T.; Dowzicky, M.J. Global Assessment of the Activity of Tigecycline against Multidrug-Resistant Gram-Negative Pathogens between 2004 and 2014 as Part of the Tigecycline Evaluation and Surveillance Trial. mSphere 2017, 2, e00310-16. [Google Scholar] [CrossRef] [Green Version]
- Rangel, K.; Chagas, T.P.G.; De-Simone, S.G. Acinetobacter baumannii Infections in Times of COVID-19 Pandemic. Pathogens 2021, 10, 1006. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019, 69 (Suppl. 7), S521–S528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piperaki, E.T.; Tzouvelekis, L.S.; Miriagou, V.; Daikos, G.L. Carbapenem-resistant Acinetobacter baumannii: In pursuit of an effective treatment. Clin. Microbiol. Infect. 2019, 25, 951–957. [Google Scholar] [CrossRef]
- Adams, M.D.; Nickel, G.C.; Bajaksouzian, S.; Lavender, H.; Murthy, A.R.; Jacobs, M.R.; Bonomo, R.A. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 2009, 53, 3628–3634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moffatt, J.H.; Harper, M.; Adler, B.; Nation, R.L.; Li, J.; Boyce, J.D. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3022–3024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinberg, S.E.; Villedieu, A.; Bagdasarian, N.; Karah, N.; Teare, L.; Elamin, W.F. Control and management of multidrug resistant Acinetobacter baumannii: A review of the evidence and proposal of novel approaches. Infect. Prev. Pract. 2020, 2, 100077. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Scribano, D.; Sarshar, M.; Di Bonaventura, G.; Palamara, A.T.; Ambrosi, C. Gram-Negative Bacteria Holding Together in a Biofilm: The Acinetobacter baumannii Way. Microorganisms 2021, 9, 1353. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.H.; Chan, B.K.; Chan, E.W.; Chen, S. Over-Expression of ISAba1-Linked Intrinsic and Exogenously Acquired OXA Type Carbapenem-Hydrolyzing-Class D-ss-Lactamase-Encoding Genes Is Key Mechanism Underlying Carbapenem Resistance in Acinetobacter baumannii. Front. Microbiol. 2019, 10, 2809. [Google Scholar] [CrossRef] [Green Version]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Cell Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Hancock, R.E.W.; Alford, M.A.; Haney, E.F. Antibiofilm activity of host defence peptides: Complexity provides opportunities. Nat. Rev. Microbiol. 2021, 19, 786–797. [Google Scholar] [CrossRef] [PubMed]
- Fernandez de Ullivarri, M.; Arbulu, S.; Garcia-Gutierrez, E.; Cotter, P.D. Antifungal Peptides as Therapeutic Agents. Front. Cell Infect. Microbiol. 2020, 10, 105. [Google Scholar] [CrossRef] [PubMed]
- van der Does, A.M.; Bogaards, S.J.; Ravensbergen, B.; Beekhuizen, H.; van Dissel, J.T.; Nibbering, P.H. Antimicrobial peptide hLF1-11 directs granulocyte-macrophage colony-stimulating factor-driven monocyte differentiation toward macrophages with enhanced recognition and clearance of pathogens. Antimicrob. Agents Chemother. 2010, 54, 811–816. [Google Scholar] [CrossRef] [Green Version]
- Lin, G.Y.; Chang, C.F.; Lan, C.Y. The interaction Between Carbohydrates and the Antimicrobial Peptide P-113Tri is Involved in the Killing of Candida albicans. Microorganisms 2020, 8, 299. [Google Scholar] [CrossRef] [Green Version]
- Won, H.S.; Kang, S.J.; Choi, W.S.; Lee, B.J. Activity optimization of an undecapeptide analogue derived from a frog-skin antimicrobial peptide. Mol. Cells 2011, 31, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.; Siman-Tov, G.; Hall, G.; Bhalla, N.; Narayanan, A. Human Antimicrobial Peptides as Therapeutics for Viral Infections. Viruses 2019, 11, 704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carballar-Lejarazu, R.; Rodriguez, M.H.; de la Cruz Hernandez-Hernandez, F.; Ramos-Castaneda, J.; Possani, L.D.; Zurita-Ortega, M.; Reynaud-Garza, E.; Hernandez-Rivas, R.; Loukeris, T.; Lycett, G.; et al. Recombinant scorpine: A multifunctional antimicrobial peptide with activity against different pathogens. Cell Mol. Life Sci. 2008, 65, 3081–3092. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, S.; Li, D.; Wei, D.Q.; Zhao, J.; Wang, J. Human Intestinal Defensin 5 Inhibits SARS-CoV-2 Invasion by Cloaking ACE2. Gastroenterology 2020, 159, 1145–1147.e4. [Google Scholar] [CrossRef]
- Mwangi, J.; Yin, Y.; Wang, G.; Yang, M.; Li, Y.; Zhang, Z.; Lai, R. The antimicrobial peptide ZY4 combats multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii infection. Proc. Natl. Acad. Sci. USA 2019, 116, 26516–26522. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wu, Z.; Mao, C.; Guo, G.; Zeng, Z.; Fei, Y.; Wan, S.; Peng, J.; Wu, J. Antimicrobial Peptide Cec4 Eradicates the Bacteria of Clinical Carbapenem-Resistant Acinetobacter baumannii Biofilm. Front. Microbiol. 2020, 11, 1532. [Google Scholar] [CrossRef]
- Asif, M.; Alvi, I.A.; Rehman, S.U. Insight into Acinetobacter baumannii: Pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect. Drug Resist. 2018, 11, 1249–1260. [Google Scholar] [CrossRef] [Green Version]
- Kapil, S.; Sharma, V. d-Amino acids in antimicrobial peptides: A potential approach to treat and combat antimicrobial resistance. Can. J. Microbiol. 2021, 67, 119–137. [Google Scholar] [CrossRef] [PubMed]
- Hicks, R.P.; Abercrombie, J.J.; Wong, R.K.; Leung, K.P. Antimicrobial peptides containing unnatural amino acid exhibit potent bactericidal activity against ESKAPE pathogens. Bioorg. Med. Chem. 2013, 21, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.M.; Boyle, N.A.; MacDonald, M.T.; Janda, K.D. Peptidomimetics and peptide backbone modifications. Mini Rev. Med. Chem. 2002, 2, 463–473. [Google Scholar] [CrossRef]
- Falanga, A.; Nigro, E.; De Biasi, M.G.; Daniele, A.; Morelli, G.; Galdiero, S.; Scudiero, O. Cyclic Peptides as Novel Therapeutic Microbicides: Engineering of Human Defensin Mimetics. Molecules 2017, 22, 1217. [Google Scholar] [CrossRef] [Green Version]
- Staskiewicz, A.; Ledwon, P.; Rovero, P.; Papini, A.M.; Latajka, R. Triazole-Modified Peptidomimetics: An Opportunity for Drug Discovery and Development. Front. Chem. 2021, 9, 674705. [Google Scholar] [CrossRef]
- Vagner, J.; Qu, H.; Hruby, V.J. Peptidomimetics, a synthetic tool of drug discovery. Curr. Opin. Chem. Biol. 2008, 12, 292–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruvo, M.; Fassina, G. End-group modified retro-inverso isomers of tripeptide oxytocin analogues: Binding to neurophysin II and enhancement of its self-association properties. Int. J. Pept. Protein Res. 1995, 45, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Doti, N.; Mardirossian, M.; Sandomenico, A.; Ruvo, M.; Caporale, A. Recent Applications of Retro-Inverso Peptides. Int. J. Mol. Sci. 2021, 22, 8677. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Kamysz, W.; Mickiewicz, B.; Rodziewicz-Motowidlo, S.; Greber, K.; Okroj, M. Temporin A and its retro-analogues: Synthesis, conformational analysis and antimicrobial activities. J. Pept. Sci. 2006, 12, 533–537. [Google Scholar] [CrossRef]
- Shin, S.; Kim, J.K.; Lee, J.Y.; Jung, K.W.; Hwang, J.S.; Lee, J.; Lee, D.G.; Kim, I.; Shin, S.Y.; Kim, Y. Design of potent 9-mer antimicrobial peptide analogs of protaetiamycine and investigation of mechanism of antimicrobial action. J. Pept. Sci. 2009, 15, 559–568. [Google Scholar] [CrossRef]
- Lee, E.; Kim, J.K.; Shin, S.; Jeong, K.W.; Lee, J.; Lee, D.G.; Hwang, J.S.; Kim, Y. Enantiomeric 9-mer peptide analogs of protaetiamycine with bacterial cell selectivities and anti-inflammatory activities. J. Pept. Sci. 2011, 17, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, M.; Choi, J.; Jang, A.; Kim, Y. A Novel Peptide Antibiotic, Pro10-1D, Designed from Insect Defensin Shows Antibacterial and Anti-Inflammatory Activities in Sepsis Models. Int. J. Mol. Sci. 2020, 21, 6216. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente-Nunez, C.; Reffuveille, F.; Fernandez, L.; Hancock, R.E. Bacterial biofilm development as a multicellular adaptation: Antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 2013, 16, 580–589. [Google Scholar] [CrossRef]
- Longo, F.; Vuotto, C.; Donelli, G. Biofilm formation in Acinetobacter baumannii. New Microbiol. 2014, 37, 119–127. [Google Scholar] [PubMed]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Eze, E.C.; El Zowalaty, M.E.; Pillay, M. Antibiotic resistance and biofilm formation of Acinetobacter baumannii isolated from high-risk effluent water in tertiary hospitals in South Africa. J. Glob. Antimicrob. Resist. 2021, 27, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, M.; Novak-Frazer, L.; Bates, M.; Baguneid, M.; Alonso-Rasgado, T.; Xia, G.; Rautemaa-Richardson, R.; Bayat, A. Validation of biofilm formation on human skin wound models and demonstration of clinically translatable bacteria-specific volatile signatures. Sci. Rep. 2018, 8, 9431. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, M.; Benoliel, C.; Drider, D.; Dhulster, P.; Chihib, N.E. Biofilm formation and persistence on abiotic surfaces in the context of food and medical environments. Arch. Microbiol. 2014, 196, 453–472. [Google Scholar] [CrossRef]
- Lee, A.C.; Harris, J.L.; Khanna, K.K.; Hong, J.H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Karmakar, S.; Babu, S.P. TLR2 and TLR4 mediated host immune responses in major infectious diseases: A review. Braz. J. Infect. Dis. 2016, 20, 193–204. [Google Scholar] [CrossRef] [Green Version]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2011, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Pazgier, M.; Li, J.; Li, C.; Liu, M.; Zou, G.; Li, Z.; Chen, J.; Tarasov, S.G.; Lu, W.Y.; et al. Limitations of peptide retro-inverso isomerization in molecular mimicry. J. Biol. Chem. 2010, 285, 19572–19581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.; Kim, J.K.; Jeon, D.; Jeong, K.W.; Shin, A.; Kim, Y. Functional Roles of Aromatic Residues and Helices of Papiliocin in its Antimicrobial and Anti-inflammatory Activities. Sci. Rep. 2015, 5, 12048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chorev, M.; Goodman, M. Recent developments in retro peptides and proteins--an ongoing topochemical exploration. Trends Biotechnol. 1995, 13, 438–445. [Google Scholar] [CrossRef]
- Rai, J. Peptide and protein mimetics by retro and retroinverso analogs. Chem. Biol. Drug Des. 2019, 93, 724–736. [Google Scholar] [CrossRef]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef] [PubMed]
- Le, C.F.; Fang, C.M.; Sekaran, S.D. Intracellular Targeting Mechanisms by Antimicrobial Peptides. Antimicrob. Agents Chemother. 2017, 61, e02340-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, L.; Yang, L.; Weiss, T.M.; Waring, A.J.; Lehrer, R.I.; Huang, H.W. Interaction of antimicrobial peptides with lipopolysaccharides. Biochemistry 2003, 42, 12251–12259. [Google Scholar] [CrossRef] [Green Version]
- Loffredo, M.R.; Ghosh, A.; Harmouche, N.; Casciaro, B.; Luca, V.; Bortolotti, A.; Cappiello, F.; Stella, L.; Bhunia, A.; Bechinger, B.; et al. Membrane perturbing activities and structural properties of the frog-skin derived peptide Esculentin-1a(1-21)-NH2 and its Diastereomer Esc(1-21)-1c: Correlation with their antipseudomonal and cytotoxic activity. Biochim. Biophys. Acta Biomembr. 2017, 1859, 2327–2339. [Google Scholar] [CrossRef]
- Huang, J.; Hao, D.; Chen, Y.; Xu, Y.; Tan, J.; Huang, Y.; Li, F.; Chen, Y. Inhibitory effects and mechanisms of physiological conditions on the activity of enantiomeric forms of an alpha-helical antibacterial peptide against bacteria. Peptides 2011, 32, 1488–1495. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.; Prodromou, R.; Day, K.N.; Schneible, J.D.; Bacon, K.B.; Bowen, J.D.; Kilgore, R.E.; Catella, C.M.; Moore, B.D.; Mabe, M.D.; et al. Peptides and pseudopeptide ligands: A powerful toolbox for the affinity purification of current and next-generation biotherapeutics. J. Chromatogr. A 2021, 1635, 461632. [Google Scholar] [CrossRef]
- Rajasekaran, G.; Kim, E.Y.; Shin, S.Y. LL-37-derived membrane-active FK-13 analogs possessing cell selectivity, anti-biofilm activity and synergy with chloramphenicol and anti-inflammatory activity. Biochim. Biophys. Acta Biomembr. 2017, 1859, 722–733. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Dietz, M.J.; Li, B. Antimicrobial peptide LL-37 is bactericidal against Staphylococcus aureus biofilms. PLoS ONE 2019, 14, e0216676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Pletzer, D.; Haney, E.F.; Rahanjam, N.; Cheng, J.T.J.; Yue, M.; Aljehani, W.; Hancock, R.E.W.; Kizhakkedathu, J.N.; Straus, S.K. Aurein-Derived Antimicrobial Peptides Formulated with Pegylated Phospholipid Micelles to Target Methicillin-Resistant Staphylococcus aureus Skin Infections. ACS Infect. Dis. 2019, 5, 443–453. [Google Scholar] [CrossRef]
- Hamidian, M.; Nigro, S.J. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genom. 2019, 5, e000306. [Google Scholar] [CrossRef]
- Lenhard, J.R.; Nation, R.L.; Tsuji, B.T. Synergistic combinations of polymyxins. Int. J. Antimicrob. Agents 2016, 48, 607–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.; Jang, A.; Yoon, Y.K.; Kim, Y. Development of Novel Peptides for the Antimicrobial Combination Therapy against Carbapenem-Resistant Acinetobacter baumannii Infection. Pharmaceutics 2021, 13, 1800. [Google Scholar] [CrossRef]
- Sheng, W.H.; Wang, J.T.; Li, S.Y.; Lin, Y.C.; Cheng, A.; Chen, Y.C.; Chang, S.C. Comparative in vitro antimicrobial susceptibilities and synergistic activities of antimicrobial combinations against carbapenem-resistant Acinetobacter species: Acinetobacter baumannii versus Acinetobacter genospecies 3 and 13TU. Diagn. Microbiol. Infect. Dis. 2011, 70, 380–386. [Google Scholar] [CrossRef]
- Maragakis, L.L.; Perl, T.M. Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 2008, 46, 1254–1263. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, M.; Choi, J.; Choi, S.; Kim, Y. Anti-Endotoxin 9-Meric Peptide with Therapeutic Potential for the Treatment of Endotoxemia. J. Microbiol. Biotechnol. 2021, 31, 25–32. [Google Scholar] [CrossRef]
- Jeon, D.; Jeong, M.C.; Jacob, B.; Bang, J.K.; Kim, E.H.; Cheong, C.; Jung, I.D.; Park, Y.; Kim, Y. Investigation of cationicity and structure of pseudin-2 analogues for enhanced bacterial selectivity and anti-inflammatory activity. Sci. Rep. 2017, 7, 1455. [Google Scholar] [CrossRef] [PubMed]
- Taheri, B.; Mohammadi, M.; Nabipour, I.; Momenzadeh, N.; Roozbehani, M. Identification of novel antimicrobial peptide from Asian sea bass (Lates calcarifer) by in silico and activity characterization. PLoS ONE 2018, 13, e0206578. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jacob, B.; Jang, M.; Kwak, C.; Lee, Y.; Son, K.; Lee, S.; Jung, I.D.; Jeong, M.S.; Kwon, S.H.; et al. Development of a novel short 12-meric papiliocin-derived peptide that is effective against Gram-negative sepsis. Sci. Rep. 2019, 9, 3817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
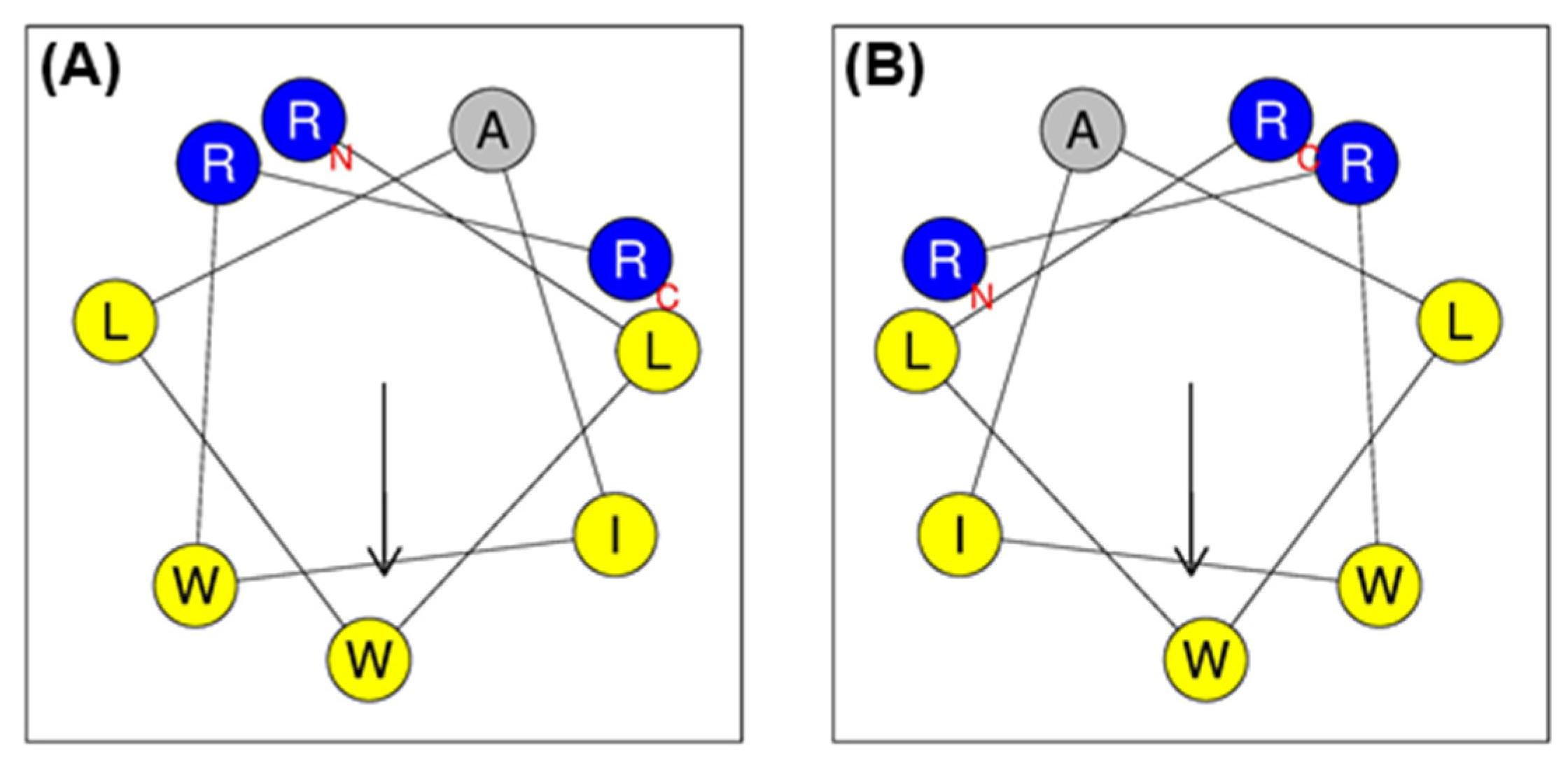


| Peptides | Sequence a | Length | Molecular Weight | Charge | Hydrophobic Moment <µH> b | Hydrophobicity <H> b |
|---|---|---|---|---|---|---|
| Pro9-3 | RLWLAIWRR-NH2 | 9 | 1269 | +3 | 0.692 | 0.776 |
| Pro9-3D | rlwlaiwrr-NH2 | 9 | 1269 | +3 | 0.692 | 0.776 |
| R-Pro9-3 | RRWIALWLR-NH2 | 9 | 1269 | +3 | 0.692 | 0.776 |
| R-Pro9-3D | rrwialwlr-NH2 | 9 | 1269 | +3 | 0.692 | 0.776 |
| Microorganisms | Minimal Inhibitory Concentration (MIC) in μM | ||||||
|---|---|---|---|---|---|---|---|
| Pro9-3 | Pro9-3D | R-Pro9-3 | R-Pro9-3D | Melittin | Imipenem | Meropenem | |
| Standard Gram-negative bacteria | |||||||
| E. coli | 16 | 8 | 16 | 8 | 8 | 0.5 | 0.25 |
| A. baumannii | 16 | 4 | 16 | 4 | 4 | 1 | 0.5 |
| P. aeruginosa | 64 | 8 | 64 | 8 | 16 | 1 | 1 |
| K. pneumoniae | 64 | 8 | 32 | 8 | 32 | <0.25 | <0.25 |
| Carbapenem-resistant Gram-negative bacteria | |||||||
| CREC E1 | 16 | 8 | 16 | 8 | 8 | >128 | >128 |
| CREC E2 | 16 | 8 | 16 | 8 | 8 | 16 | 16 |
| CRAB C0 | 8 | 4 | 8 | 4 | 4 | >128 | 128 |
| CRAB C1 | 16 | 8 | 32 | 8 | 4 | >128 | >128 |
| CRAB C2 | 16 | 8 | 32 | 4 | 4 | >128 | 128 |
| CRAB C3 | 16 | 8 | 32 | 8 | 4 | 128 | 128 |
| CRAB C4 | 16 | 8 | 32 | 4 | 4 | 128 | >128 |
| CRAB C5 | 16 | 8 | 32 | 4 | 4 | 128 | >128 |
| CRAB C6 | 16 | 8 | 32 | 4 | 4 | 128 | >128 |
| CRAB C7 | 16 | 8 | 32 | 4 | 4 | 128 | 128 |
| CRAB C8 | 16 | 8 | 32 | 4 | 4 | >128 | >128 |
| CRAB C9 | 16 | 8 | 16 | 4 | 4 | >128 | >128 |
| CRAB C10 | 16 | 8 | 16 | 4 | 4 | >128 | >128 |
| CRKP K1 | 64 | 16 | 32 | 16 | 64 | 32 | 64 |
| CRKP K2 | 64 | 8 | 32 | 8 | 64 | >128 | >128 |
| a GM | 25.6 | 8.0 | 27.4 | 6.3 | 13.1 | 90.3 | 91.9 |
| b HC10 | 200 | 200 | 200 | 200 | 3.1 | na | na |
| c Relative selective index | 7.8 | 25.0 | 7.3 | 31.7 | 0.2 | na | na |
| Microorganisms | Minimal Inhibitory Concentration (MIC) in μM | |||
|---|---|---|---|---|
| Pro9-3 | Pro9-3D | R-Pro9-3 | R-Pro9-3D | |
| E. coli + Trypsin | >64 | 8 | >64 | 8 |
| E. coli + α-Chymotrypsin | >64 | 8 | 64 | 8 |
| A. baumannii + Trypsin | >64 | 4 | >64 | 4 |
| A. baumannii + α-Chymotrypsin | >64 | 4 | >64 | 4 |
| CRAB C0 + Trypsin | >64 | 4 | 64 | 4 |
| CRAB C0 + α-Chymotrypsin | >64 | 4 | 64 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishnan, M.; Choi, J.; Jang, A.; Yoon, Y.K.; Kim, Y. Antiseptic 9-Meric Peptide with Potency against Carbapenem-Resistant Acinetobacter baumannii Infection. Int. J. Mol. Sci. 2021, 22, 12520. https://doi.org/10.3390/ijms222212520
Krishnan M, Choi J, Jang A, Yoon YK, Kim Y. Antiseptic 9-Meric Peptide with Potency against Carbapenem-Resistant Acinetobacter baumannii Infection. International Journal of Molecular Sciences. 2021; 22(22):12520. https://doi.org/10.3390/ijms222212520
Chicago/Turabian StyleKrishnan, Manigandan, Joonhyeok Choi, Ahjin Jang, Young Kyung Yoon, and Yangmee Kim. 2021. "Antiseptic 9-Meric Peptide with Potency against Carbapenem-Resistant Acinetobacter baumannii Infection" International Journal of Molecular Sciences 22, no. 22: 12520. https://doi.org/10.3390/ijms222212520
APA StyleKrishnan, M., Choi, J., Jang, A., Yoon, Y. K., & Kim, Y. (2021). Antiseptic 9-Meric Peptide with Potency against Carbapenem-Resistant Acinetobacter baumannii Infection. International Journal of Molecular Sciences, 22(22), 12520. https://doi.org/10.3390/ijms222212520






