Age-Related Changes in Lipidome of Rat Frontal Cortex and Cerebellum Are Partially Reversed by Methionine Restriction Applied in Old Age
Abstract
:1. Introduction
2. Results
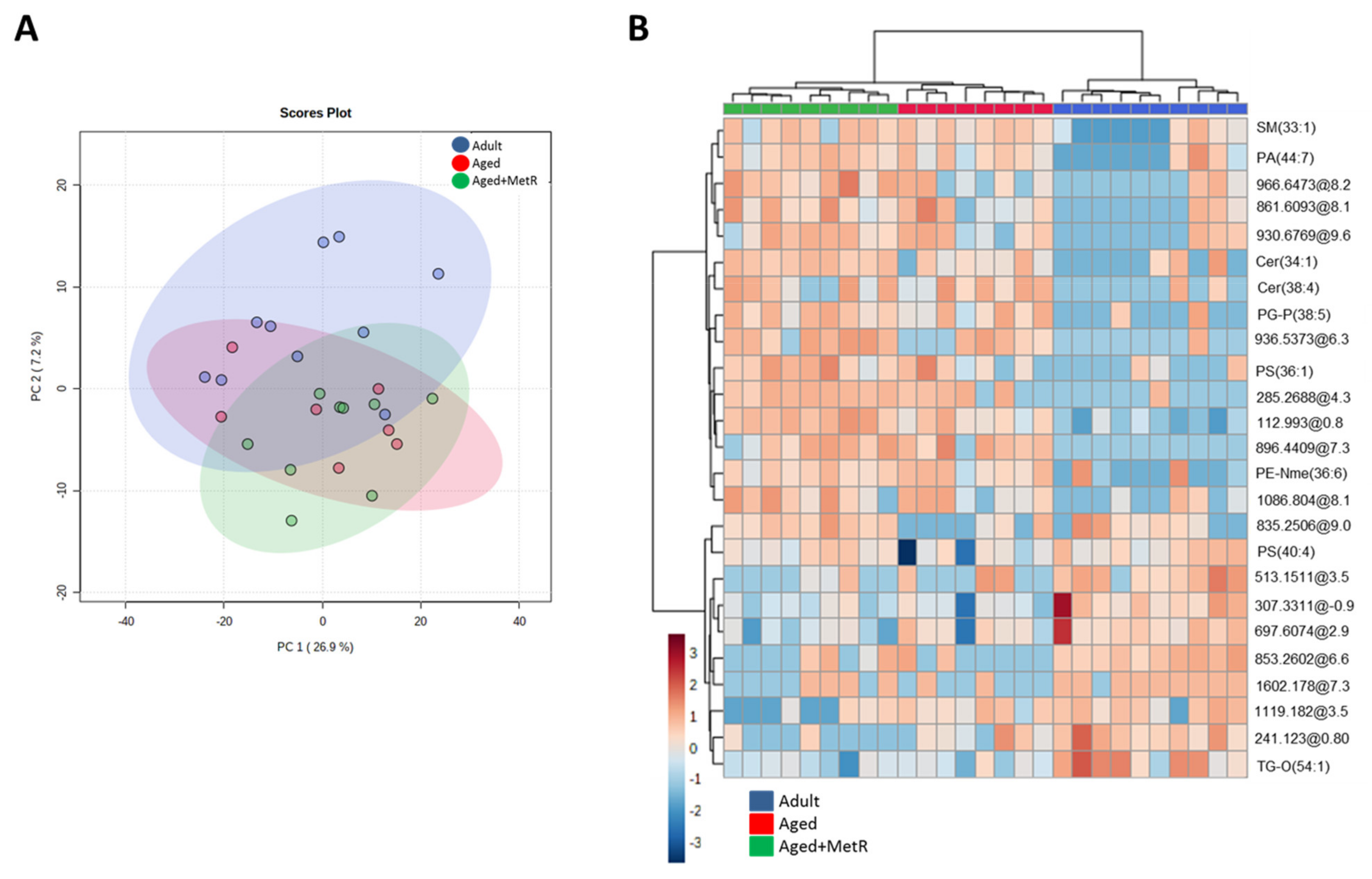
2.1. Effect of Aging and Methionine Restriction in the Cerebellum Lipidome
2.2. Effect of Aging and Methionine Restriction in the Frontal Cortex Lipidome
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Animals and Diets
4.3. Sample Homogenization
4.4. Untargeted Lipidomic Analysis
4.5. Fatty Acid Profile
4.6. Markers of Mitochondrial Stress and Tissue Protein Damage
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sastry, P.S. Lipids of nervous tissue: Composition and metabolism. Prog. Lipid Res. 1985, 24, 69–176. [Google Scholar] [CrossRef]
- Piomelli, D.; Astarita, G.; Rapaka, R.A. Neuroscientist’s guide to lipidomics. Nat. Rev. Neurosci. 2007, 8, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Naudí, A.; Cabré, R.; Jové, M.; Ayala, V.; Gonzalo, H.; Portero-Otín, M.; Ferrer, I.; Pamplona, R. Lipidomics of human brain aging and Alzheimer’s disease pathology. Int. Rev. Neurobiol. 2015, 122, 133–189. [Google Scholar] [PubMed]
- Purdon, A.D.; Rosenberger, T.A.; Shetty, H.U.; Rapoport, S.I. Energy consumption by phospholipid metabolism in mammalian brain. Neurochem. Res. 2002, 27, 1641–1647. [Google Scholar] [CrossRef]
- Hagen, R.M.; Rodriguez-Cuenca, S.; Vidal-Puig, A. An allostatic control of membrane lipid composition by SREBP1. FEBS Lett. 2010, 584, 2689–2698. [Google Scholar] [CrossRef] [Green Version]
- Aviram, R.; Manella, G.; Kopelman, N.; Neufeld-Cohen, A.; Zwighaft, Z.; Elimelech, M.; Adamovich, Y.; Golik, M.; Wang, C.; Han, X.; et al. Lipidomics analyses reveal temporal and spatial lipid organization and uncover daily oscillations in intracellular organelles. Mol. Cell 2016, 62, 636–648. [Google Scholar] [CrossRef]
- Khrameeva, E.; Kurochkin, I.; Bozek, K.; Giavalisco, P.; Khaitovich, P. Lipidome evolution in mammalian tissues. Mol. Biol. Evol. 2018, 35, 1947–1957. [Google Scholar] [CrossRef] [Green Version]
- Jové, M.; Mota-Martorell, N.; Pradas, I.; Galo-Licona, J.D.; Martín-Gari, M.; Obis, È.; Sol, J.; Pamplona, R. The lipidome fingerprint of longevity. Molecules 2020, 25, 4343. [Google Scholar] [CrossRef]
- Almeida, I.; Magalhães, S.; Nunes, A. Lipids: Biomarkers of healthy aging. Biogerontology 2021, 22, 273–295. [Google Scholar] [CrossRef]
- De Diego, I.; Peleg, S.; Fuchs, B. The role of lipids in aging-related metabolic changes. Chem. Phys. Lipids 2019, 222, 59–69. [Google Scholar] [CrossRef]
- Johnson, A.A.; Stolzing, A. The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell 2019, 18, e13048. [Google Scholar] [CrossRef] [Green Version]
- Jové, M.; Pradas, I.; Dominguez-Gonzalez, M.; Ferrer, I.; Pamplona, R. Lipids and lipoxidation in human brain aging. Mitochondrial ATP-synthase as a key lipoxidation target. Redox Biol. 2019, 23, 101082. [Google Scholar] [CrossRef]
- Surguchov, A. α-Synuclein interacts with lipoproteins in plasma. Front. Mol. Neurosci. 2017, 10, 362. [Google Scholar] [CrossRef] [Green Version]
- Smiljanic, K.; Vanmierlo, T.; Djordjevic, A.M.; Perovic, M.; Loncarevic-Vasiljkovic, N.; Tesic, V.; Rakic, L.; Ruzdijic, S.; Lutjohann, D.; Kanazir, S. Aging induces tissue-specific changes in cholesterol metabolism in rat brain and liver. Lipids 2013, 48, 1069–1077. [Google Scholar] [CrossRef]
- Giusto, N.M.; Salvador, G.A.; Castagnet, P.I.; Pasquaré, S.J.; Ilincheta de Boschero, M.G. Age-associated changes in central nervous system glycerolipid composition and metabolism. Neurochem. Res. 2002, 27, 1513–1523. [Google Scholar] [CrossRef]
- Favrelère, S.; Stadelmann-Ingrand, S.; Huguet, F.; De Javel, D.; Piriou, A.; Tallineau, C.; Durand, G. Age-related changes in ethanolamine glycerophospholipid fatty acid levels in rat frontal cortex and hippocampus. Neurobiol. Aging 2000, 21, 653–660. [Google Scholar] [CrossRef]
- Wackerlig, J.; Köfeler, H.C.; Korz, V.; Hussein, A.M.; Feyissa, D.D.; Höger, H.; Urban, E.; Langer, T.; Lubec, G.; Lubec, J. Differences in hypothalamic lipid profiles of young and aged male rats with impaired and unimpaired spatial cognitive abilities and memory. Front. Aging Neurosci. 2020, 12, 204. [Google Scholar] [CrossRef]
- Palestini, P.; Masserini, M.; Sonnino, S.; Giuliani, A.; Tettamanti, G. Changes in the ceramide composition of rat forebrain gangliosides with age. J. Neurochem. 1990, 54, 230–235. [Google Scholar] [CrossRef]
- Šmidák, R.; Köfeler, H.C.; Hoeger, H.; Lubec, G. Comprehensive identification of age-related lipidome changes in rat amygdala during normal aging. PLoS ONE 2017, 12, e0180675. [Google Scholar] [CrossRef] [Green Version]
- Pamplona, R.; Barja, G. Mitochondrial oxidative stress, aging and caloric restriction: The protein and methionine connection. Biochim. Biophys. Acta 2006, 1757, 496–508. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Roman, I.; Barja, G. Regulation of longevity and oxidative stress by nutritional interventions: Role of methionine restriction. Exp. Gerontol. 2013, 48, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Naudí, A.; Caro, P.; Jové, M.; Gómez, J.; Boada, J.; Ayala, V.; Portero-Otín, M.; Barja, G.; Pamplona, R. Methionine restriction decreases endogenous oxidative molecular damage and increases mitochondrial biogenesis and uncoupling protein 4 in rat brain. Rej. Res. 2007, 10, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Caro, P.; Gomez, J.; Sanchez, I.; Naudi, A.; Ayala, V.; López-Torres, M.; Pamplona, R.; Barja, G. Forty percent methionine restriction decreases mitochondrial oxygen radical production and leak at complex I during forward electron flow and lowers oxidative damage to proteins and mitochondrial DNA in rat kidney and brain mitochondria. Rej. Res. 2009, 12, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Jové, M.; Ayala, V.; Ramírez-Núñez, O.; Naudí, A.; Cabré, R.; Spickett, C.M.; Portero-Otín, M.; Pamplona, R. Specific lipidome signatures in central nervous system from methionine-restricted mice. J. Proteome Res. 2013, 12, 2679–2689. [Google Scholar] [CrossRef] [PubMed]
- Mladenović, D.; Radosavljević, T.; Hrnčić, D.; Rasic-Markovic, A.; Stanojlović, O. The effects of dietary methionine restriction on the function and metabolic reprogramming in the liver and brain—Implications for longevity. Rev. Neurosci. 2019, 30, 581–593. [Google Scholar] [CrossRef]
- Filip, P.; Bares, M. Cerebellum—Aging of the neuronal machine. In Factors Affecting Neurological Aging. Genetics, Neurology, Behavior, and Diet; Martin, C.R., Preedy, V.R., Rajendram, R., Eds.; Elsevier, Academic Press: London, UK, 2021; pp. 281–288. [Google Scholar]
- Liang, K.J.; Carlson, E.S. Resistance, vulnerability and resilience: A review of the cognitive cerebellum in aging and neurodegenerative diseases. Neurobiol. Learn. Mem. 2020, 170, 106981. [Google Scholar] [CrossRef]
- Kolb, B. Functions of the frontal cortex of the rat: A comparative review. Brain Res. 1984, 320, 65–98. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Cabré Cucó, R. Vulnerabilitat Neuronal Selectiva Associada a L'envelliment Fisiològic del Sistema Nerviós Central Humà. Universitat de Lleida: Lleida, Spain, 15. PhD Thesis, Universitat de Lleida, Lleida, Spain, December 2015. [Google Scholar]
- Chan, R.B.; Oliveira, T.G.; Cortes, E.P.; Honig, L.S.; Duff, K.E.; Small, S.A.; Wenk, M.R.; Shui, G.; Di Paolo, G. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J. Biol. Chem. 2012, 287, 2678–2688. [Google Scholar] [CrossRef] [Green Version]
- Prasain, J.K.; Wilson, L.; Hoang, H.D.; Moore, R.; Miller, M.A. Comparative lipidomics of Caenorhabditis elegans metabolic disease models by SWATH non-targeted tandem mass spectrometry. Metabolites 2015, 5, 677–696. [Google Scholar] [CrossRef] [Green Version]
- Bozek, K.; Khrameeva, E.E.; Reznick, J.; Omerbašić, D.; Bennett, N.C.; Lewin, G.R.; Azpurua, J.; Gorbunova, V.; Seluanov, A.; Regnard, P.; et al. Lipidome determinants of maximal lifespan in mammals. Sci. Rep. 2017, 7, 5. [Google Scholar] [CrossRef]
- Ferrer, I. Proteomics and lipidomics in the human brain. Handb. Clin. Neurol. 2018, 150, 285–302. [Google Scholar]
- Naudí, A.; Cabré, R.; Dominguez-Gonzalez, M.; Ayala, V.; Jové, M.; Mota-Martorell, N.; Piñol-Ripoll, G.; Gil-Villar, M.P.; Rué, M.; Portero-Otín, M.; et al. Region-specific vulnerability to lipid peroxidation and evidence of neuronal mechanisms for polyunsaturated fatty acid biosynthesis in the healthy adult human central nervous system. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids. 2017, 1862, 485–495. [Google Scholar] [CrossRef]
- Díaz, M.; Mesa-Herrera, F.; Marín, R. DHA and its elaborated modulation of antioxidant defenses of the brain: Implications in aging and AD neurodegeneration. Antioxidants 2021, 10, 907. [Google Scholar] [CrossRef]
- Jové, M.; Mota-Martorell, N.; Torres, P.; Portero-Otin, M.; Ferrer, I.; Pamplona, R. New insights into human prefrontal cortex aging with a lipidomics approach. Expert Rev. Proteom. 2021, 18, 333–344. [Google Scholar] [CrossRef]
- Cabré, R.; Naudí, A.; Dominguez-Gonzalez, M.; Jové, M.; Ayala, V.; Mota-Martorell, N.; Pradas, I.; Nogueras, L.; Rué, M.; Portero-Otín, M.; et al. Lipid profile in human frontal cortex is sustained throughout healthy adult life span to decay at advanced ages. J. Gerontol. A Biol. Sci. Med. Sci. 2018, 73, 703–710. [Google Scholar]
- Cabré, R.; Naudí, A.; Dominguez-Gonzalez, M.; Ayala, V.; Jové, M.; Mota-Martorell, N.; Piñol-Ripoll, G.; Gil-Villar, M.P.; Rué, M.; Portero-Otín, M.; et al. Sixty years old is the breakpoint of human frontal cortex aging. Free Radic. Biol. Med. 2017, 103, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Norris, S.E.; Friedrich, M.G.; Mitchell, T.W.; Truscott, R.J.W.; Else, P.L. Human prefrontal cortex phospholipids containing docosahexaenoic acid increase during normal adult aging, whereas those containing arachidonic acid decrease. Neurobiol. Aging 2015, 36, 1659–1669. [Google Scholar] [CrossRef]
- Pamplona, R.; Dalfó, E.; Ayala, V.; Bellmunt, M.J.; Prat, J.; Ferrer, I.; Portero-Otín, M. Proteins in human brain cortex are modified by oxidation, glycoxidation, and lipoxidation. Effects of Alzheimer disease and identification of lipoxidation targets. J. Biol. Chem. 2005, 280, 21522–21530. [Google Scholar] [CrossRef] [Green Version]
- Almena, M.; Mérida, I. Shaping up the membrane: Diacylglycerol coordinates spatial orientation of signaling. Trends Biochem. Sci. 2011, 36, 593–603. [Google Scholar] [CrossRef]
- Posse de Chaves, E.; Sipione, S. Sphingolipids and gangliosides of the nervous system in membrane function and dysfunction. FEBS Lett. 2010, 584, 1748–1759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meikle, P.J.; Summers, S.A. Sphingolipids and phospholipids in insulin resistance and related metabolic disorders. Nat. Rev. Endocrinol. 2017, 13, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Trayssac, M.; Hannun, Y.A.; Obeid, L.M. Role of sphingolipids in senescence: Implication in aging and age-related diseases. J. Clin. Invest. 2018, 128, 2702–2712. [Google Scholar] [CrossRef] [PubMed]
- Pradas, I.; Jové, M.; Huynh, K.; Puig, J.; Ingles, M.; Borras, C.; Viña, J.; Meikle, P.J.; Pamplona, R. Exceptional human longevity is associated with a specific plasma phenotype of ether lipids. Redox Biol. 2019, 21, 101127. [Google Scholar] [CrossRef]
- Dean, J.M.; Lodhi, I.J. Structural and functional roles of ether lipids. Protein Cell 2018, 9, 196–206. [Google Scholar] [CrossRef]
- Teixeira, V.; Maciel, P.; Costa, V. Leading the way in the nervous system: Lipid droplets as new players in health and disease. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158820. [Google Scholar] [CrossRef]
- Vance, J.E.; Vance, D.E. Phospholipid biosynthesis in mammalian cells. Biochem. Cell Biol. 2004, 82, 113–128. [Google Scholar] [CrossRef]
- Domingo-Almenara, X.; Montenegro-Burke, J.R.; Paul Benton, H.; Siuzdak, G. Annotation: A computational solution for streamlining metabolomics analysis. Anal. Chem. 2018, 90, 480–489. [Google Scholar] [CrossRef] [Green Version]
- Pradas, I.; Jové, M.; Cabré, R.; Ayala, V.; Mota-Martorell, N.; Pamplona, R. Effects of aging and methionine restriction on rat kidney metabolome. Metabolites 2019, 9, 280. [Google Scholar] [CrossRef] [Green Version]
- Castro-Perez, J.M.; Kamphorst, J.; DeGroot, J.; Lafeber, F.; Goshawk, J.; Yu, K.; Shockcor, J.P.; Vreeken, R.J.; Hankemeier, T. Comprehensive LC-MS E lipidomic analysis using a shotgun approach and its application to biomarker detection and identification in osteoarthritis patients. J. Proteome Res. 2010, 9, 2377–2389. [Google Scholar] [CrossRef]
- Pizarro, C.; Arenzana-Rámila, I.; Pérez-del-Notario, N.; Pérez-Matute, P.; González-Sáiz, J.-M. Plasma lipidomic profiling method based on ultrasound extraction and liquid chromatography mass spectrometry. Anal. Chem. 2013, 85, 12085–12092. [Google Scholar] [CrossRef]
- Andrés-Benito, P.; Gelpi, E.; Jové, M.; Mota-Martorell, N.; Obis, È.; Portero-Otin, M.; Povedano, M.; Pujol, A.; Pamplona, R.; Ferrer, I. Lipid alterations in human frontal cortex in ALS-FTLD-TDP43 proteinopathy spectrum are partly related to peroxisome impairment. Neuropathol. Appl. Neurobiol. 2021, 47, 544–563. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Human Serum Metabolome (HUSERMET) consortium. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef] [Green Version]

| Class | ID | Post Hoc | Biological Meaning (Biomarker) |
|---|---|---|---|
| GL | TG (52:0) a | A < R | |
| TG (47:0) b | A < R | ||
| TG-O (54:1) b | Ag, R < A | aging | |
| TG-O (60:1) b | Ag, R < A | aging | |
| TG-O (47:0) b | A, Ag < R | MetR | |
| TG-O (56:1) b | Ag < R, A | healthy aging | |
| GP | PS (36:1) a | A < R | |
| PE-P (38:4) a | A < R | ||
| PG-P (38:5) b | A < Ag, R | aging | |
| PE-P (40:4)/PE-O (40:5) a | Ag, R < A | aging | |
| PA (44:7) a | A < Ag, R | aging | |
| PE-Nme (36:6) b | A < Ag, R | aging | |
| PC-O (38:4)/PC-P (38:3) b | A < Ag, R | aging | |
| PE-Nme (34:3)/PE-NMe2 (33:3)/PC (32:3)/PE (35:3) b | A, Ag < R | MetR | |
| PC (36:4)/PC-O (35:4) a | A, Ag < R | MetR | |
| PS (40:4) a | Ag < R, A | healthy aging | |
| SP | Cer (34:1) a | Ag < R, A | healthy aging |
| Cer (38:4) a | A < Ag, R | aging | |
| SM (33:1) b | A < Ag, R | aging | |
| Ganglioside GA2 (44:1) b | Ag, R < A | aging | |
| N-(2R-Hydroxyhexadecanoyl)-2S-amino-9-methyl-4E,8E-octadecadiene-1,3R-diol b | A < Ag, R | aging | |
| SL | CE (22:4) b | Ag, R < A | aging |
| cholest-5-en-3b-yl (11Z,14Z-eicosadienoate) c | A, Ag < R | MetR | |
| CE (xx) | Ag < R, A | healthy aging |
| Fatty Acid | Adult | Aged | Aged+MetR |
|---|---|---|---|
| 14:0 | 0.26 ± 0.00 | 0.29 ± 0.01 a,* | 0.32 ± 0.01 a,*** |
| 16:0 | 17.93 ± 0.16 | 18.00 ± 0.04 | 17.83 ± 0.14 |
| 16:1n-7 | 0.38 ± 0.01 | 0.41 ± 0.02 | 0.40 ± 0.01 |
| 18:0 | 17.63 ± 0.06 | 17.70 ± 0.09 | 17.66 ± 0.08 |
| 18:1n-9 | 26.00 ± 0.14 | 26.53 ± 0.25 | 26.74 ± 0.21 a,* |
| 18:2n-6 | 1.14 ± 0.02 | 1.19 ± 0.04 | 1.37 ± 0.05 a,***; b,** |
| 18:3n-6 | 0.02 ± 0.00 | 0.02 ± 0.00 | 0.01 ± 0.00 |
| 18:3n-3 | 0.02 ± 0.00 | 0.02 ± 0.00 | 0.01 ± 0.00 a,* |
| 18:4n-3 | 0.03 ± 0.00 | 0.03 ± 0.00 | 0.03 ± 0.00 |
| 20:0 | 0.70 ± 0.04 | 0.83 ± 0.01 a,* | 0.79 ± 0.04 |
| 20:1n-9 | 4.06 ± 0.14 | 4.48 ± 0.07 a,* | 4.49 ± 0.10 a,* |
| 20:2n-6 | 0.33 ± 0.01 | 0.29 ± 0.01 a,** | 0.30 ± 0.01 |
| 20:3n-6 | 0.31 ± 0.01 | 0.30 ± 0.01 | 0.32 ± 0.01 |
| 20:4n-6 | 7.69 ± 0.08 | 7.27 ± 0.09 a,** | 7.36 ± 0.09 a,* |
| 20:5n-3 | 0.04 ± 0.00 | 0.04 ± 0.00 a,* | 0.04 ± 0.00 |
| 22:0 | 0.74 ± 0.02 | 0.75 ± 0.01 | 0.76 ± 0.02 |
| 22:1n-9 | 1.08 ± 0.09 | 1.01 ± 0.02 | 1.03 ± 0.03 |
| 22:4n-6 | 2.60 ± 0.03 | 2.22 ± 0.05 a,*** | 2.22 ± 0.05 a,*** |
| 22:5n-6 | 0.43 ± 0.03 | 0.45 ± 0.03 | 0.41 ± 0.03 |
| 22:5n-3 | 0.14 ± 0.00 | 0.14 ± 0.01 | 0.12 ± 0.00 |
| 22:6n-3 | 13.43 ± 0.12 | 12.96 ± 0.14 | 12.89 ± 0.17 a,* |
| 24:0 | 1.11 ± 0.05 | 1.00 ± 0.02 | 1.03 ± 0.02 |
| 24:1 | 2.26 ± 0.05 | 2.54 ± 0.10 | 2.41 ± 0.08 |
| 24:5n-3 | 0.04 ± 0.00 | 0.04 ± 0.00 | 0.04 ± 0.00 |
| 24:6n-3 | 1.31 ± 0.04 | 1.38 ± 0.03 | 1.45 ± 0.03 a,* |
| Fatty Acid Index | Adult | Aged | Aged+MetR |
|---|---|---|---|
| ACL | 18.91 ± 0.01 | 18.75 ± 0.08 | 18.89 ± 0.00 |
| SFA | 38.37 ± 0.17 | 38.35 ± 0.16 | 38.24 ± 0.14 |
| UFA | 61.52 ± 0.15 | 61.41 ± 0.12 | 61.58 ± 0.20 |
| MUFA | 33.64 ± 0.35 | 35.27 ± 0.16 a,** | 35.01 ± 0.33 a,** |
| PUFA | 27.53 ± 0.19 | 26.05 ± 0.19 a,*** | 26.57 ± 0.16 a,** |
| PUFAn-3 | 15.00 ± 0.10 | 14.59 ± 0.16 | 14.58 ± 0.15 |
| PUFAn-6 | 12.53 ± 0.12 | 11.65 ± 0.12 a,*** | 11.99 ± 0.15 a,* |
| DBI | 170.56 ± 0.96 | 165.22 ± 0.86 a,*** | 166.91 ± 0.64 a,* |
| PI | 166.05 ± 1.14 | 158.66 ± 1.08 a,*** | 160.06 ± 1.27 a,** |
| D9D (n-7) | 0.02 ± 0.00 | 0.02 ± 0.00 | 0.02 ± 0.00 |
| D9D (n-9) | 1.47 ± 0.01 | 1.50 ± 0.02 | 1.51 ± 0.02 |
| D5D (n-6) | 24.65 ± 0.42 | 24.45 ± 0.73 | 23.36 ± 0.63 |
| D6D (n-3) (a) | 1.84 ± 0.05 | 1.83 ± 0.08 | 1.99 ± 0.09 |
| D6D (n-3) (b) | 31.22 ± 1.76 | 34.49 ± 1.58 | 36.78 ± 1.87 |
| Elovl3 (n-9) | 0.16 ± 0.01 | 0.17 ± 0.00 | 0.17 ± 0.00 |
| Elovl6 | 0.98 ± 0.01 | 0.98 ± 0.00 | 0.99 ± 0.01 |
| Elovl1-3-7 (a) | 0.04 ± 0.00 | 0.05 ± 0.00 a,* | 0.04 ± 0.00 |
| Elovl1-3-7 (b) | 1.09 ± 0.05 | 0.91 ± 0.01 a,** | 0.97 ± 0.04 |
| Elovl1-3-7 (c) | 1.49 ± 0.06 | 1.34 ± 0.03 a,* | 1.33 ± 0.01 a,* |
| Elovl5 (n-6) | 0.29 ± 0.01 | 0.23 ± 0.00 a,*** | 0.22 ± 0.00 a,*** |
| Elovl2-5 (n-6) | 0.34 ± 0.00 | 0.31 ± 0.01 a,*** | 0.30 ± 0.01 a,*** |
| Elovl2-5 (n-3) | 3.76 ± 0.07 | 3.30 ± 0.18 a,* | 3.11 ± 0.12 a,** |
| Elovl2 (n-3) | 0.31 ± 0.01 | 0.30 ± 0.01 | 0.34 ± 0.01 |
| Peroxisomal β-oxidation | 10.39 ± 0.44 | 9.42 ± 0.15 | 8.91 ± 0.28 a,** |
| Damage Marker | Adult | Aged | Aged+MetR |
|---|---|---|---|
| GSA | 5013.14 ± 144.38 | 4376.78 ± 116.83 a,* | 4410.87 ± 77.14 a,* |
| CEL | 579.82 ± 39.05 | 725.42 ± 93.56 | 660.52 ± 37.93 |
| CML | 1408.30 ± 59.83 | 1387.38 ± 105.08 | 1302.08 ± 43.07 |
| MDAL | 196.20 ± 8.66 | 179.29 ± 5.01 | 195.32 ± 8.66 |
| 2-SC | 39.79 ± 4.34 | 26.85 ± 1.80 a,* | 26.79 ± 2.27 a,* |
| Class | ID | Post Hoc | Biological Meaning (Biomarker) |
|---|---|---|---|
| FA | 12-hydroxyheptadecanoic acid b | Ag, R < A | aging |
| GL | TG (50:0) b | R < A | |
| TG (48:1) b | Ag, R < A | aging | |
| TG (54:6) b | Ag, R < A | aging | |
| TG (50:1) b | Ag, R < A | aging | |
| TG (64:14) b | A < Ag, R | aging | |
| TG (47:0) b | Ag < R | ||
| TG-O (58:10) a | R < Ag | ||
| DG (40:3) b | R < Ag | ||
| TG (64:14) a | A, Ag < R | MetR | |
| TG-O (60:9) b | R, A < Ag | healthy aging | |
| TG (62:12) b | Ag < R, A | healthy aging | |
| GP | PE (36:1) a | R < A | |
| PE (39:5)/PE-P (40:4) a | R < A | ||
| PC (36:2) a | Ag, R < A | aging | |
| PE-P (40:6) a | Ag, R < A | aging | |
| PE-P (38:6) a | Ag, R < A | aging | |
| PE-NMe (34:3)/PE (35:4)/PC (32:3) b | A, Ag < R | MetR | |
| PE-NMe (34:3)/PC (32:3) b | A, Ag < R | MetR | |
| PA (44:7) a | A, Ag < R | MetR | |
| PE (40:4) a | R < A, Ag | MetR | |
| PE (38:1) a | R < A, Ag | MetR | |
| PE-P (38:1) a | R < A, Ag | MetR | |
| PI (38:4) a | Ag < R, A | healthy aging | |
| PI (38:5) a | Ag < A | ||
| PG (34:1) a | Ag < R, A | healthy aging | |
| PL | Dolichol-20 d | A < Ag, R | aging |
| SP | Cer (36:1) a | Ag < A | |
| LacCer (34:2)/GalCer (44:2) b | R < A, Ag | MetR | |
| Cer (36:1) b | R < A, Ag | MetR | |
| N-(2R-Hydroxyhexadecanoyl)-2S-amino-9-methyl-4E,8E-octadecadiene-1,3R-diol b | R < A, Ag | MetR | |
| Cer (34:1) c | R < A, Ag | MetR | |
| SL | CE (20:4) b | A < Ag, R | aging |
| CE (5:0) b | A, Ag < R | MetR |
| Fatty Acid | Adult | Aged | Aged+MetR |
|---|---|---|---|
| 14:0 | 0.92 ± 0.02 | 0.91 ± 0.01 | 0.92 ± 0.02 |
| 16:0 | 29.03 ± 0.014 | 29.31 ± 0.26 | 28.83 ± 0.12 |
| 16:1n-7 | 0.54 ± 0.02 | 0.56 ± 0.03 | 0.52 ± 0.01 |
| 18:0 | 24.17 ± 0.23 | 24.19 ± 0.11 | 24.20 ± 0.10 |
| 18:1n-9 | 19.71 ± 0.26 | 19.18 ± 0.19 | 19.47 ± 0.20 |
| 18:2n-6 | 0.94 ± 0.02 | 1.00 ± 0.03 | 1.06 ± 0.06 |
| 18:3n-6 | 0.03 ± 0.00 | 0.04 ± 0.00 | 0.04 ± 0.00 |
| 18:3n-3 | 0.03 ± 0.00 | 0.04 ± 0.00 | 0.04 ± 0.00 |
| 18:4n-3 | 0.04 ± 0.00 | 0.04 ± 0.00 | 0.05 ± 0.00 |
| 20:0 | 0.18 ± 0.01 | 0.18 ± 0.01 | 0.19 ± 0.01 |
| 20:1n-9 | 0.71 ± 0.04 | 0.68 ± 0.04 | 0.71 ± 0.04 |
| 20:2n-6 | 0.11 ± 0.00 | 0.11 ± 0.00 | 0.12 ± 0.01 |
| 20:3n-6 | 0.20 ± 0.00 | 0.22 ± 0.01 | 0.24 ± 0.01 a,*** |
| 20:4n-6 | 10.20 ± 0.26 | 10.49 ± 0.11 | 10.53 ± 0.18 |
| 20:5n-3 | 0.04 ± 0.00 | 0.04 ± 0.00 | 0.04 ± 0.00 |
| 22:0 | 0.18 ± 0.01 | 0.16 ± 0.01 | 0.16 ± 0.00 |
| 22:1n-9 | 0.57 ± 0.05 | 0.58 ± 0.03 | 0.62 ± 0.02 |
| 22:4n-6 | 2.38 ± 0.08 | 2.35 ± 0.05 | 2.35 ± 0.07 |
| 22:5n-6 | 0.80 ± 0.07 | 0.99 ± 0.09 | 0.91 ± 0.08 |
| 22:5n-3 | 0.09 ± 0.00 | 0.10 ± 0.00 | 0.10 ± 0.00 |
| 22:6n-3 | 7.95 ± 0.25 | 8.31 ± 0.19 | 8.36 ± 0.17 |
| 24:0 | 0.15 ± 0.01 | 0.17 ± 0.00 | 0.17 ± 0.01 |
| 24:1 | 0.17 ± 0.01 | 0.14 ± 0.01 | 0.14 ± 0.01 |
| 24:5n-3 | 0.04 ± 0.00 | 0.04 ± 0.00 | 0.04 ± 0.00 |
| 24:6n-3 | 0.19 ± 0.01 | 0.15 ± 0.00 a,* | 0.15 ± 0.01 a,** |
| Fatty Acid Index | Adult | Aged | Aged+MetR |
|---|---|---|---|
| ACL | 18.12 ± 0.01 | 18.13 ± 0.01 | 18.12 ± 0.02 |
| SFA | 54.63 ± 0.19 | 54.89 ± 0.35 | 54.45 ± 0.15 |
| UFA | 44.67 ± 0.58 | 45.09 ± 0.34 | 45.43 ± 0.16 |
| MUFA | 21.77 ± 0.33 | 21.15 ± 0.25 | 21.40 ± 0.27 |
| PUFA | 23.05 ± 0.58 | 23.94 ± 0.30 | 24.03 ± 0.33 |
| PUFAn-3 | 8.57 ± 0.17 | 8.74 ± 0.18 | 8.78 ± 0.17 |
| PUFAn-6 | 14.67 ± 0.36 | 15.20 ± 0.19 | 15.25 ± 0.27 |
| DBI | 128.67 ± 2.74 | 132.49 ± 1.52 | 132.94 ± 1.43 |
| PI | 123.50 ± 3.31 | 128.55 ± 1.91 | 128.69 ± 1.97 |
| D9D (n-7) | 0.02 ± 0.00 | 0.02 ± 0.00 | 0.02 ± 0.00 |
| D9D (n-9) | 0.82 ± 0.01 | 0.79 ± 0.01 | 0.80 ± 0.01 |
| D5D (n-6) | 49.68 ± 0.31 | 47.75 ± 2.05 | 43.81 ± 1.12 a,* |
| D6D (n-3) (a) | 1.03 ± 0.16 | 1.10 ± 0.07 | 1.18 ± 0.06 |
| D6D (n-3) (b) | 5.10 ± 0.55 | 3.88 ± 0.34 a,* | 3.86 ± 0.36 |
| Elovl3 (n-9) | 0.04 ± 0.00 | 0.04 ± 0.00 | 0.04 ± 0.00 |
| Elovl6 | 0.83 ± 0.01 | 0.83 ± 0.00 | 0.84 ± 0.01 |
| Elovl1-3-7 (a) | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.00 |
| Elovl1-3-7 (b) | 1.02 ± 0.04 | 0.86 ± 0.03 a,** | 0.86 ± 0.02 a,** |
| Elovl1-3-7 (c) | 0.85 ± 0.05 | 1.01 ± 0.15 | 1.07 ± 0.08 |
| Elovl5 (n-6) | 0.12 ± 0.00 | 0.11 ± 0.00 | 0.11 ± 0.00 |
| Elovl2-5 (n-6) | 0.23 ± 0.00 | 0.22 ± 0.00 | 0.22 ± 0.01 |
| Elovl2-5 (n-3) | 2.76 ± 0.22 | 2.65 ± 0.15 | 2.43 ± 0.14 |
| Elovl2 (n-3) | 0.41 ± 0.03 | 0.40 ± 0.02 | 0.42 ± 0.03 |
| Peroxisomal β-oxidation | 43.85 ± 3.05 | 54.04 ± 2.12 a,* | 56.40 ± 1.92 a,** |
| Damage Marker | Adults | Aged | Aged+MetR |
|---|---|---|---|
| GSA | 3615.40 ± 79.03 | 3700.19 ± 138.23 | 3623.83 ± 246.00 |
| CEL | 270.70 ± 15.58 | 259.79 ± 29.33 | 233.04 ± 11.36 |
| CML | 966.24 ± 40.79 | 1016.19 ± 63.22 | 949.69 ± 85.01 |
| MDAL | 372.59 ± 30.48 | 345.74 ± 6.80 | 376.28 ± 21.86 |
| 2-SC | 28.43 ± 4.18 | 20.78 ± 2.28 | 35.55 ± 4.99 b,* |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jové, M.; Cabré, R.; Mota-Martorell, N.; Martin-Garí, M.; Obis, È.; Ramos, P.; Canales, I.; Galo-Licona, J.D.; Sol, J.; Nogueras, L.; et al. Age-Related Changes in Lipidome of Rat Frontal Cortex and Cerebellum Are Partially Reversed by Methionine Restriction Applied in Old Age. Int. J. Mol. Sci. 2021, 22, 12517. https://doi.org/10.3390/ijms222212517
Jové M, Cabré R, Mota-Martorell N, Martin-Garí M, Obis È, Ramos P, Canales I, Galo-Licona JD, Sol J, Nogueras L, et al. Age-Related Changes in Lipidome of Rat Frontal Cortex and Cerebellum Are Partially Reversed by Methionine Restriction Applied in Old Age. International Journal of Molecular Sciences. 2021; 22(22):12517. https://doi.org/10.3390/ijms222212517
Chicago/Turabian StyleJové, Mariona, Rosanna Cabré, Natàlia Mota-Martorell, Meritxell Martin-Garí, Èlia Obis, Paula Ramos, Iván Canales, José Daniel Galo-Licona, Joaquim Sol, Lara Nogueras, and et al. 2021. "Age-Related Changes in Lipidome of Rat Frontal Cortex and Cerebellum Are Partially Reversed by Methionine Restriction Applied in Old Age" International Journal of Molecular Sciences 22, no. 22: 12517. https://doi.org/10.3390/ijms222212517






