The Role of Plant Hormones in the Interaction of Colletotrichum Species with Their Host Plants
Abstract
:1. Introduction
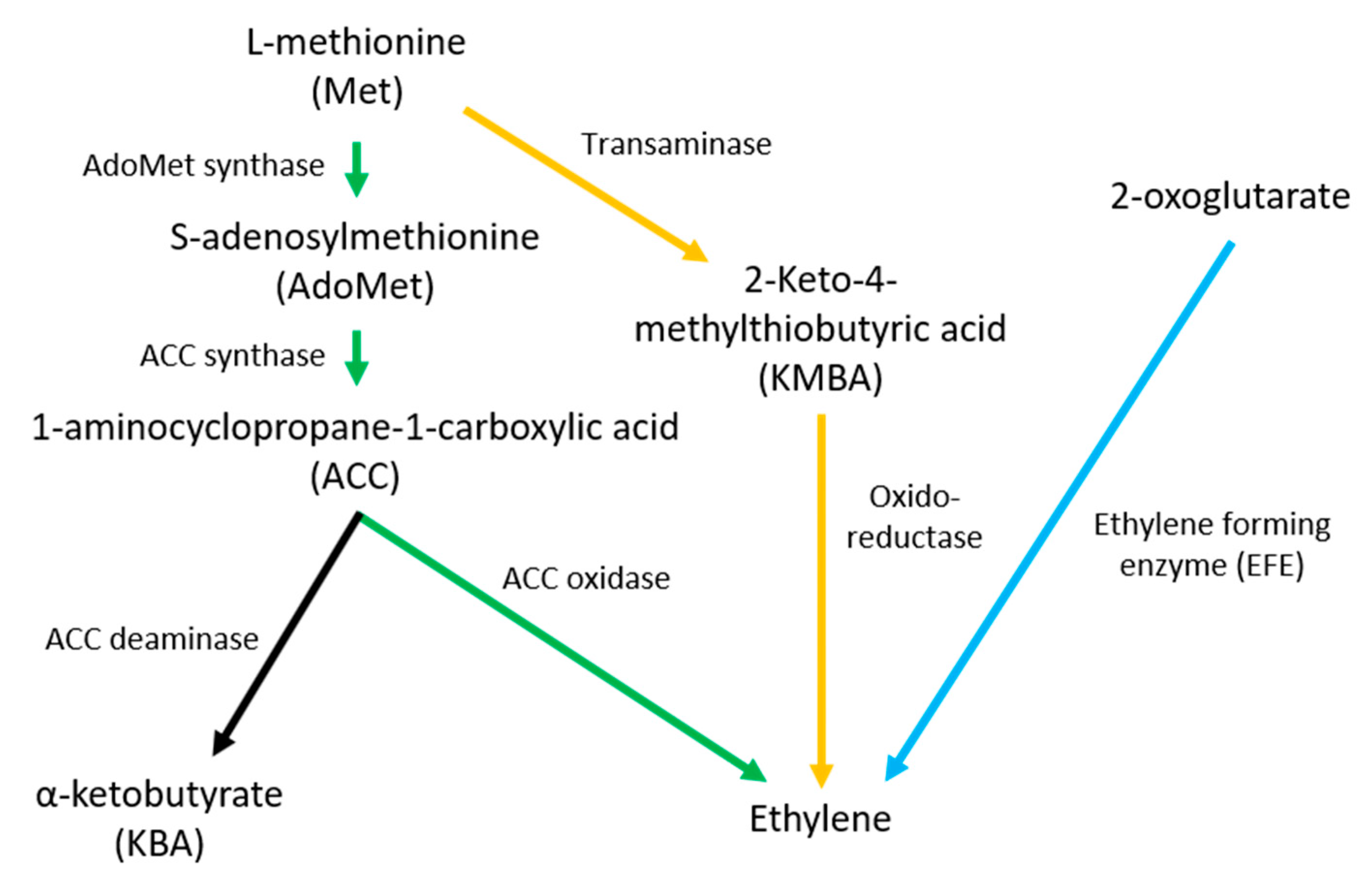
2. Ethylene
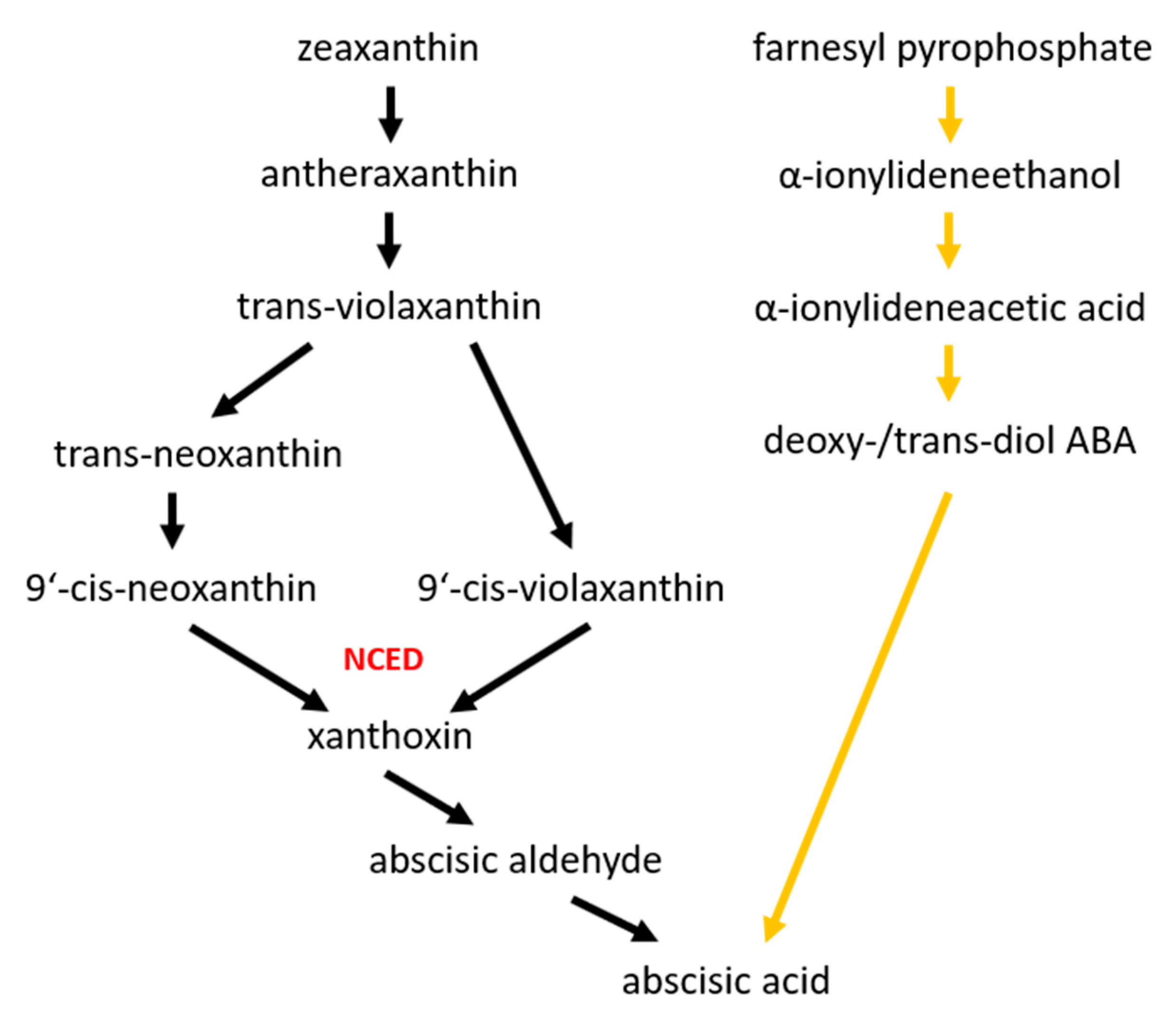
3. Abscisic Acid
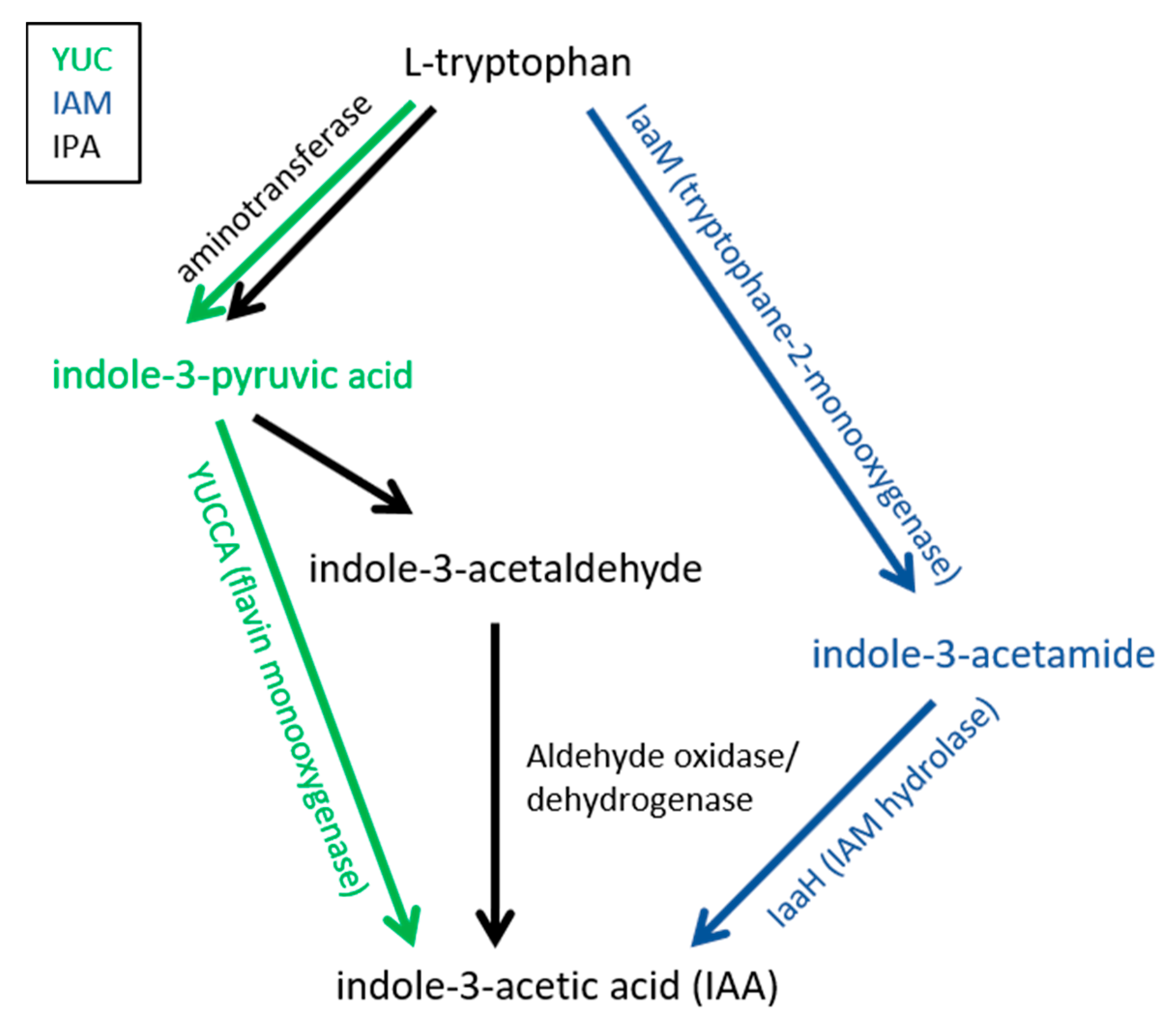
4. Auxin
5. Salicylic Acid
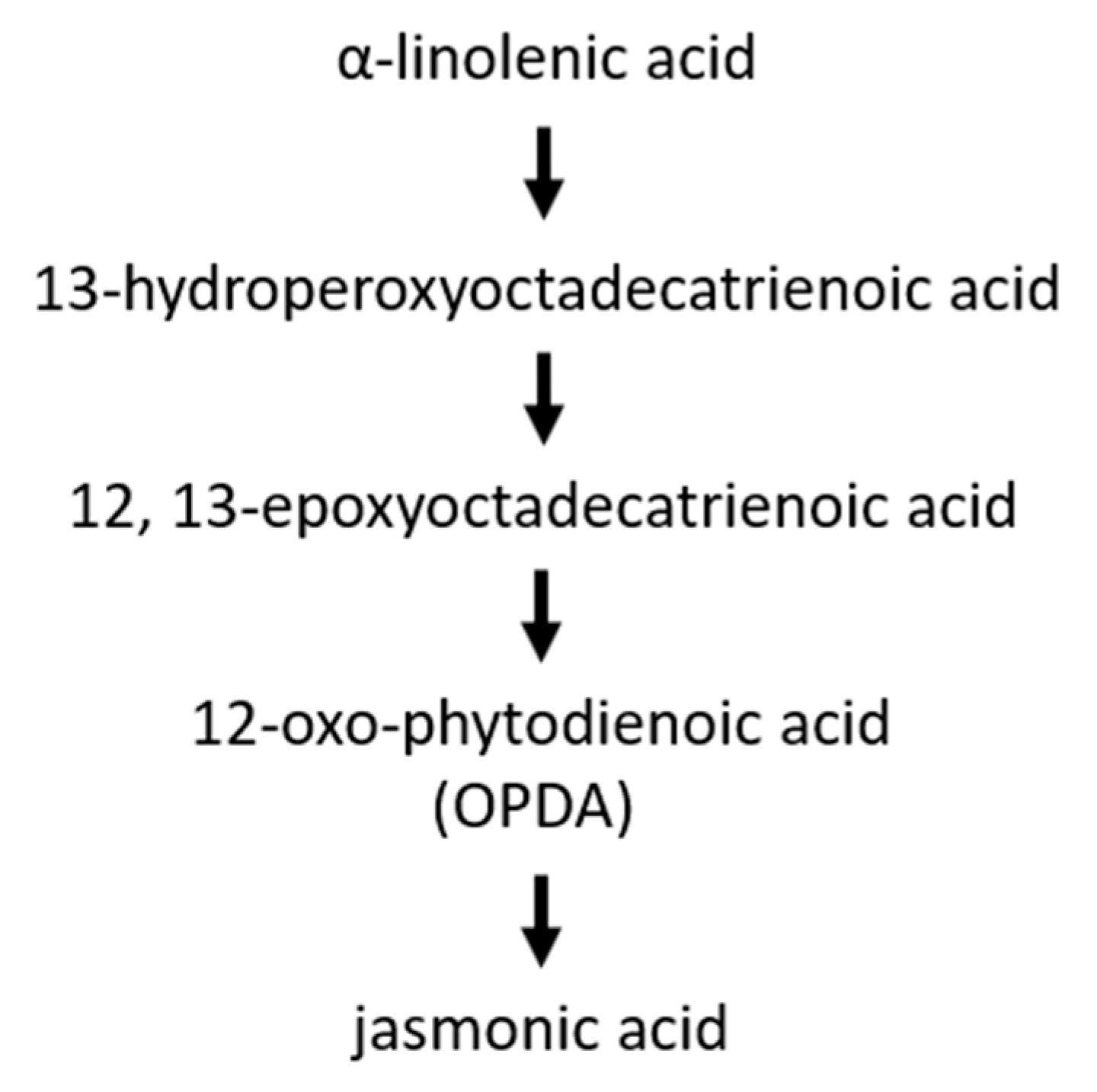
6. Jasmonic Acid
7. Brassinosteroids
8. Synopsis
Author Contributions
Funding
Conflicts of Interest
References
- Talhinhas, P.; Sreenivasaprasad, S.; Neves-Martins, J.; Oliveira, H. Molecular and Phenotypic Analyses Reveal Association of Diverse Colletotrichum acutatum Groups and a Low Level of C. gloeosporioides with Olive Anthracnose. Appl. Environ. Microbiol. 2005, 71, 2987–2998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sukno, S.A.; García, V.M.; Shaw, B.D.; Thon, M.R. Root Infection and Systemic Colonization of Maize by Colletotrichum graminicola. Appl. Environ. Microbiol. 2008, 74, 823–832. [Google Scholar] [CrossRef] [Green Version]
- Rogério, F.; Ciampi-Guillardi, M.; Barbieri, M.C.G.; Bragança, C.A.D.; Seixas, C.D.S.; Almeida, A.M.R.; Massola, N.S. Phylogeny and Variability of Colletotrichum truncatum Associated with Soybean Anthracnose in Brazil. J. Appl. Microbiol. 2017, 122, 402–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xavier, K.V.; Pfeiffer, T.; Parreira, D.F.; Chopra, S.; Vaillancourt, L. Aggressiveness of Colletotrichum sublineola Strains from Sorghum bicolor and S. halepense to Sweet Sorghum Variety Sugar Drip, and Their Impact on Yield. Plant Dis. 2017, 101, 1578–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.-Y.; Forcelini, B.B.; Peres, N.A. Anthracnose Fruit and Root Necrosis of Strawberry are Caused by a Dominant Species within the Colletotrichum acutatum Species Complex in the United States. Phytopathology 2019, 109, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Perfect, S.E.; Hughes, H.B.; O’Connell, R.J.; Green, J.R. Colletotrichum: A Model Genus for Studies on Pathology and Fungal-Plant Interactions. Fungal Genet. Biol. 1999, 27, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Münch, S.; Lingner, U.; Floss, D.S.; Ludwig, N.; Sauer, N.; Deising, H.B. The Hemibiotrophic Lifestyle of Colletotrichum Species. J. Plant Physiol. 2008, 165, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Nair, D.N.; Padmavathy, S. Impact of Endophytic Microorganisms on Plants, Environment and Humans. Sci. World J. 2014, 2014, 250693. [Google Scholar] [CrossRef] [Green Version]
- Baroncelli, R.; Talhinhas, P.; Pensec, F.; Sukno, S.A.; Le Floch, G.; Thon, M.R. The Colletotrichum Acutatum Species Complex as a Model System to Study Evolution and Host Specialization in Plant Pathogens. Front. Microbiol. 2017, 8, 2001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Zhou, Z.; Wu, J.; Ji, Z.; Zhang, J. Comparative Transcriptome Analysis Reveals Significant Differences in Gene Expression between Appressoria and Hyphae in Colletotrichum gloeosporioides. Gene 2018, 670, 63–69. [Google Scholar] [CrossRef]
- Liang, X.; Shang, S.; Dong, Q.; Wang, B.; Zhang, R.; Gleason, M.L.; Sun, G. Transcriptomic Analysis Reveals Candidate Genes Regulating Development and Host Interactions of Colletotrichum fructicola. BMC Genom. 2018, 19, 557. [Google Scholar] [CrossRef]
- O’Connell, R.J.; Thon, M.R.; Hacquard, S.; Amyotte, S.G.; Kleemann, J.; Torres, M.F.; Damm, U.; Buiate, E.A.; Epstein, L.; Alkan, N.; et al. Lifestyle Transitions in Plant Pathogenic Colletotrichum Fungi Deciphered by Genome and Transcriptome Analyses. Nat. Genet. 2012, 44, 1060–1065. [Google Scholar] [CrossRef]
- Wang, X.; Xu, X.; Liang, Y.; Wang, Y.; Tian, C. A Cdc42 Homolog in Colletotrichum gloeosporioides Regulates Morphological Development and is Required for ROS-Mediated Plant Infection. Curr. Genet. 2018, 64, 1153–1169. [Google Scholar] [CrossRef] [PubMed]
- Asakura, M.; Yoshino, K.; Hill, A.M.; Kubo, Y.; Sakai, Y.; Takano, Y. Primary and Secondary Metabolism Regulates Lipolysis in Appressoria of Colletotrichum orbiculare. Fungal Genet. Biol. 2012, 49, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Jones, J.D.G. Role of Plant Hormones in Plant Defence Responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA Signaling in Stress-Response and Seed Development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in Integrating Plant Responses to Drought and Salt Stresses. Field Crop. Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Lata, C.; Prasad, M. Role of DREBs in Regulation of Abiotic Stress Responses in Plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef] [Green Version]
- Navarro, L.; Bari, R.; Achard, P.; Lisón, P.; Nemri, A.; Harberd, N.P.; Jones, J.D.G. DELLAs Control Plant Immune Responses by Modulating the Balance of Jasmonic Acid and Salicylic Acid Signaling. Curr. Biol. 2008, 18, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Watanabe, Y.; Leyva-Gonzalez, M.A.; Ha, C.V.; Fujita, Y.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Herrera-Estrella, L.; et al. Arabidopsis AHP2, AHP3 and AHP5 Histidine Phosphotransfer Proteins Function as Redundant Negative Regulators of Drought Stress Response. Proc. Natl. Acad. Sci. USA 2013, 110, 4840–4845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Jesus Miranda, V.; Porto, W.F.; da Rocha Fernandes, G.; Pogue, R.; Nolasco, D.O.; Araujo, A.C.G.; Cota, L.V.; de Freitas, C.G.; Dias, S.C.; Franco, O.L. Comparative Transcriptomic Analysis Indicates Genes Associated with Local and Systemic Resistance to Colletotrichum graminicola in Maize. Sci. Rep. 2017, 7, 2483. [Google Scholar] [CrossRef] [Green Version]
- Padder, B.A.; Kamfwa, K.; Awale, H.E.; Kelly, J.D. Transcriptome Profiling of the Phaseolus vulgaris—Colletotrichum lindemuthianum Pathosystem. PLoS ONE 2016, 11, e0165823. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, F.; Chen, M.; Liu, Z.; Zhang, Z.; Fu, J.; Ma, Y. Comparative Transcriptomics Reveals Differential Gene Expression Related to Colletotrichum gloeosporioides Resistance in the Octoploid Strawberry. Front. Plant Sci. 2017, 8, 779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, K.; Gong, D.; Zhang, L.; Hu, H.; Jia, Z.; Gu, H.; Song, K. Transcriptome Characterization and Expression Profiles of the Related Defense Genes in Postharvest Mango Fruit against Colletotrichum gloeosporioides. Gene 2016, 576, 275–283. [Google Scholar] [CrossRef]
- Johnson, P.R.; Ecker, J.R. The Ethylene Gas Signal Transduction Pathway: A Molecular Perspective. Annu. Rev. Genet. 1998, 32, 227–254. [Google Scholar] [CrossRef]
- Adams, D.O.; Yang, S.F. Ethylene Biosynthesis: Identification of 1-Aminocyclopropane-1-Carboxylic Acid as an Intermediate in the Conversion of Methionine to Ethylene. Proc. Natl. Acad. Sci. USA 1979, 76, 170–174. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, H.; Takahashi, M.; Fujii, T.; Tazaki, M.; Ogawa, T. An NADH:Fe(III)EDTA Oxidoreductase from Cryptococcus Albidus: An Enzyme Involved in Ethylene Production in vivo? FEMS Microbiol. Lett. 1989, 51, 107–111. [Google Scholar] [CrossRef] [Green Version]
- Young, R.E.; Pratt, H.K.; Biale, J.B. Identification of Ethylene as a Volatile Product of the Fungus Penicillium digitatum. Plant Physiol. 1951, 26, 304–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobsen, D.W.; Wang, C.H. The Biogenesis of Ethylene in Penicillium digitatum. Plant Physiol. 1968, 43, 1959–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagahama, K.; Ogawa, T.; Fujii, T.; Tazaki, M.; Tanase, S.; Morino, Y.; Fukuda, H. Purification and Properties of an Ethylene-Forming Enzyme from Pseudomonas syringae Pv. Phaseolicola PK2. J. Gen. Microbiol. 1991, 137, 2281–2286. [Google Scholar] [CrossRef] [Green Version]
- Etesami, H.; Maheshwari, D.K. Use of Plant Growth Promoting Rhizobacteria (PGPRs) with Multiple Plant Growth Promoting Traits in Stress Agriculture: Action Mechanisms and Future Prospects. Ecotoxicol. Environ. Saf. 2018, 156, 225–246. [Google Scholar] [CrossRef]
- Svoboda, T.; Parich, A.; Güldener, U.; Schöfbeck, D.; Twaruschek, K.; Václavíková, M.; Hellinger, R.; Wiesenberger, G.; Schuhmacher, R.; Adam, G. Biochemical Characterization of the Fusarium graminearum Candidate ACC-Deaminases and Virulence Testing of Knockout Mutant Strains. Front. Plant Sci. 2019, 10, 1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.; Hameed, S.; Shahid, M.; Iqbal, M.; Lazarovits, G.; Imran, A. Functional Characterization of Potential PGPR Exhibiting Broad-Spectrum Antifungal Activity. Microbiol. Res. 2020, 232, 126389. [Google Scholar] [CrossRef]
- Mishra, R.; Mohanty, J.N.; Chand, S.K.; Joshi, R.K. Can-MiRn37a Mediated Suppression of Ethylene Response Factors Enhances the Resistance of Chilli against Anthracnose Pathogen Colletotrichum truncatum L. Plant Sci. 2018, 267, 135–147. [Google Scholar] [CrossRef]
- Moyano-Cañete, E.; Bellido, M.L.; García-Caparrós, N.; Medina-Puche, L.; Amil-Ruiz, F.; González-Reyes, J.A.; Caballero, J.L.; Muñoz-Blanco, J.; Blanco-Portales, R. FaGAST2, a Strawberry Ripening-Related Gene, Acts Together with FaGAST1 to Determine Cell Size of the Fruit Receptacle. Plant Cell Physiol. 2013, 54, 218–236. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Steed, A.; Travella, S.; Keller, B.; Nicholson, P. Fusarium graminearum Exploits Ethylene Signalling to Colonize Dicotyledonous and Monocotyledonous Plants. New Phytol. 2009, 182, 975–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, N.; Goodwin, P.H.; Hsiang, T. The Role of Ethylene during the Infection of Nicotiana Tabacum by Colletotrichum destructivum. J. Exp. Bot. 2003, 54, 2449–2456. [Google Scholar] [CrossRef]
- Daundasekera, M.; Joyce, D.C.; Aked, J.; Adikaram, N.K.B. Ethylene Production by Colletotrichum musae in vitro. Physiol. Mol. Plant Pathol. 2003, 62, 21–28. [Google Scholar] [CrossRef]
- Liu, X.; Li, B.; Yang, Y.; Cai, J.; Shi, T.; Zheng, X.; Huang, G. Pathogenic Adaptations Revealed by Comparative Genome Analyses of Two Colletotrichum spp., the Causal Agent of Anthracnose in Rubber Tree. Front. Microbiol. 2020, 11, 1484. [Google Scholar] [CrossRef] [PubMed]
- Kleemann, J.; Rincon-Rivera, L.J.; Takahara, H.; Neumann, U.; van Themaat, E.V.L.; van der Does, H.C.; Hacquard, S.; Stüber, K.; Will, I.; Schmalenbach, W.; et al. Sequential Delivery of Host-Induced Virulence Effectors by Appressoria and Intracellular Hyphae of the Phytopathogen Colletotrichum higginsianum. PLoS Pathog. 2012, 8, e1002643. [Google Scholar] [CrossRef]
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of Abscisic Acid Synthesis and Signaling Mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef] [Green Version]
- Lievens, L.; Pollier, J.; Goossens, A.; Beyaert, R.; Staal, J. Abscisic Acid as Pathogen Effector and Immune Regulator. Front. Plant Sci. 2017, 8, 587. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.-J.; Bressan, R.A.; Song, C.-P.; Zhu, J.-K.; Zhao, Y. Abscisic Acid Dynamics, Signaling, and Functions in Plants. J. Integr. Plant. Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [Green Version]
- Patkar, R.N.; Naqvi, N.I. Fungal Manipulation of Hormone-Regulated Plant Defense. PLoS Pathog. 2017, 13, e1006334. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Yang, S.; Meng, L.; Wang, B.-G. The Plant Hormone Abscisic Acid Regulates the Growth and Metabolism of Endophytic Fungus Aspergillus nidulans. Sci. Rep. 2018, 8, 6504. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.K.; Kim, J.H.; Kim, Y.H.; Kim, H.T. ABA Increases Susceptibility of Pepper Fruits to Infection of Anthracnose by Collectotrichum acutatum. Plant Pathol. J. 2008, 24, 400–406. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, Y.; Cao, H.; Hao, X.; Zeng, J.; Yang, Y.; Wang, X. Transcriptome Analysis of an Anthracnose-Resistant Tea Plant Cultivar Reveals Genes Associated with Resistance to Colletotrichum camelliae. PLoS ONE 2016, 11, e0148535. [Google Scholar] [CrossRef] [PubMed]
- Mayek-PÉrez, N.; GarcÍa-Espinosa, R.; LÓpez-CastaÑeda, C.; Acosta-Gallegos, J.A.; Simpson, J. Water Relations, Histopathology and Growth of Common Bean (Phaseolus vulgaris L.) during Pathogenesis of Macrophomina phaseolina under Drought Stress. Physiol. Mol. Plant Pathol. 2002, 60, 185–195. [Google Scholar] [CrossRef]
- Ghassemian, M.; Nambara, E.; Cutler, S.; Kawaide, H.; Kamiya, Y.; McCourt, P. Regulation of Abscisic Acid Signaling by the Ethylene Response Pathway in Arabidopsis. Plant Cell 2000, 12, 1117–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhang, L.; Cai, X.; Li, X.; Bian, L.; Luo, Z.; Li, Z.; Chen, Z.; Xin, Z. (E)-Nerolidol is a Volatile Signal that Induces Defenses against Insects and Pathogens in Tea Plants. Hortic. Res. 2020, 7, 52. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Puebla, A.; De Santiago, C.; Trombotto, S.; David, L.; Larralde-Corona, C.P.; Shirai, K. Addition of Abscisic Acid Increases the Production of Chitin Deacetylase by Colletotrichum gloeosporioides in Submerged Culture. Process Biochem. 2016, 51, 959–966. [Google Scholar] [CrossRef]
- Pacheco, N.; Trombotto, S.; David, L.; Shirai, K. Activity of Chitin Deacetylase from Colletotrichum gloeosporioides on Chitinous Substrates. Carbohydr. Polym. 2013, 96, 227–232. [Google Scholar] [CrossRef]
- Shrestha, B.; Blondeau, K.; Stevens, W.F.; Hegarat, F.L. Expression of Chitin Deacetylase from Colletotrichum lindemuthianum in Pichia Pastoris: Purification and Characterization. Protein Expr. Purif. 2004, 38, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Tsigos, I.; Bouriotis, V. Purification and Characterization of Chitin Deacetylase from Colletotrichum lindemuthianum. J. Biol. Chem. 1995, 270, 26286–26291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, F.; Zhang, B.-S.; Zhao, J.-H.; Huang, J.-F.; Jia, P.-S.; Wang, S.; Zhang, J.; Zhou, J.-M.; Guo, H.-S. Deacetylation of Chitin Oligomers Increases Virulence in Soil-Borne Fungal Pathogens. Nat. Plants 2019, 5, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Langner, T.; Göhre, V. Fungal Chitinases: Function, Regulation and Potential Roles in Plant/Pathogen Interactions. Curr. Genet. 2016, 62, 243–254. [Google Scholar] [CrossRef]
- Ali, M.; Li, Q.-H.; Zou, T.; Wei, A.-M.; Gombojav, G.; Lu, G.; Gong, Z.-H. Chitinase Gene Positively Regulates Hypersensitive and Defense Responses of Pepper to Colletotrichum acutatum Infection. Int. J. Mol. Sci. 2020, 21, 6624. [Google Scholar] [CrossRef] [PubMed]
- Limon, T.; Birke, A.; Monribot-Villanueva, J.L.; Guerrero-Analco, J.A.; Altúzar-Molina, A.; Carrión, G.; Goycoolea, F.M.; Moerschbacher, B.M.; Aluja, M. Chitosan Coatings Reduce Fruit Fly (Anastrepha obliqua) Infestation and Development of the Fungus Colletotrichum gloeosporioides in Manila Mangoes. J. Sci. Food Agric. 2021, 101, 2756–2766. [Google Scholar] [CrossRef] [PubMed]
- Morffy, N.; Strader, L.C. Old Town Roads: Routes of Auxin Biosynthesis across Kingdoms. Curr. Opin. Plant Biol. 2020, 55, 21–27. [Google Scholar] [CrossRef]
- Robinson, M.; Riov, J.; Sharon, A. Indole-3-Acetic Acid Biosynthesis in Colletotrichum gloeosporioides f. sp. Aeschynomene. Appl. Environ. Microbiol. 1998, 64, 5030–5032. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Zou, W.X.; Meng, J.C.; Hu, J.; Tan, R.X. New Bioactive Metabolites Produced by Colletotrichum Sp., an Endophytic Fungus in Artemisia annua. Plant Sci. 2000, 151, 67–73. [Google Scholar] [CrossRef]
- Chung, K.R.; Shilts, T.; Ertürk, U.; Timmer, L.W.; Ueng, P.P. Indole Derivatives Produced by the Fungus Colletotrichum acutatum Causing Lime Anthracnose and Postbloom Fruit Drop of Citrus. FEMS Microbiol. Lett. 2003, 226, 23–30. [Google Scholar] [CrossRef] [Green Version]
- Numponsak, T.; Kumla, J.; Suwannarach, N.; Matsui, K.; Lumyong, S. Biosynthetic Pathway and Optimal Conditions for the Production of Indole-3-Acetic Acid by an Endophytic Fungus, Colletotrichum fructicola CMU-A109. PLoS ONE 2018, 13, e0205070. [Google Scholar] [CrossRef]
- Chanclud, E.; Morel, J.-B. Plant Hormones: A Fungal Point of View. Mol. Plant Pathol. 2016, 17, 1289–1297. [Google Scholar] [CrossRef]
- Benjamins, R.; Scheres, B. Auxin: The Looping Star in Plant Development. Annu. Rev. Plant Biol. 2008, 59, 443–465. [Google Scholar] [CrossRef] [PubMed]
- Maor, R.; Haskin, S.; Levi-Kedmi, H.; Sharon, A. In Planta Production of Indole-3-Acetic Acid by Colletotrichum Gloeosporioides f. sp. Aeschynomene. Appl. Environ. Microbiol. 2004, 70, 1852–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, B.A.; Amsellem, Z.; Maor, R.; Sharon, A.; Gressel, J. Transgenically Enhanced Expression of Indole-3-Acetic Acid Confers Hypervirulence to Plant Pathogens. Phytopathology 2002, 92, 590–596. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Huang, X.; He, C.; Zhang, Q.-Y.; Zou, X.; Duan, K.; Gao, Q. Novel Fungal Pathogenicity and Leaf Defense Strategies are Revealed by Simultaneous Transcriptome Analysis of Colletotrichum fructicola and Strawberry Infected by this Fungus. Front. Plant Sci. 2018, 9, 434. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Wang, Y.; Xiong, F.; Hao, X.; Zhang, X.; Li, N.; Wang, L.; Zeng, J.; Yang, Y.; Wang, X. Integrated Transcriptomic and Metabolomic Analyses Reveal the Effects of Callose Deposition and Multihormone Signal Transduction Pathways on the Tea Plant—Colletotrichum camelliae Interaction. Sci. Rep. 2020, 10, 12858. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraj, A.; Wang, X.; Wang, S.; Liu, S.; Zhang, R.; Wu, A.; Wei, C. Identification of Regulatory Networks of MicroRNAs and Their Targets in Response to Colletotrichum gloeosporioides in Tea Plant (Camellia sinensis L.). Front. Plant Sci. 2019, 10, 1096. [Google Scholar] [CrossRef] [Green Version]
- Kurepa, J.; Shull, T.E.; Karunadasa, S.S.; Smalle, J.A. Modulation of Auxin and Cytokinin Responses by Early Steps of the Phenylpropanoid Pathway. BMC Plant Biol. 2018, 18, 278. [Google Scholar] [CrossRef]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate Synthase is Required to Synthesize Salicylic Acid for Plant Defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.-L.; Sheng, Y.-Y.; Cai, Z.-Y.; Yang, R.; Li, Q.-S.; Li, X.-M.; Li, D.; Guo, X.-Y.; Lu, J.-L.; Ye, J.-H.; et al. Involvement of Salicylic Acid in Anthracnose Infection in Tea Plants Revealed by Transcriptome Profiling. Int. J. Mol. Sci. 2019, 20, 2439. [Google Scholar] [CrossRef] [Green Version]
- Shine, M.B.; Xiao, X.; Kachroo, P.; Kachroo, A. Signaling Mechanisms Underlying Systemic Acquired Resistance to Microbial Pathogens. Plant Sci. 2019, 279, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Liu, P.-P.; Cameron, R.K.; Park, S.-W.; Yang, Y.; Kumar, D.; Zhou, F.; Padukkavidana, T.; Gustafsson, C.; Pichersky, E.; et al. Identification of Likely Orthologs of Tobacco Salicylic Acid-Binding Protein 2 and Their Role in Systemic Acquired Resistance in Arabidopsis Thaliana. Plant J. 2008, 56, 445–456. [Google Scholar] [CrossRef]
- Amil-Ruiz, F.; Garrido-Gala, J.; Gadea, J.; Blanco-Portales, R.; Muñoz-Mérida, A.; Trelles, O.; de los Santos, B.; Arroyo, F.T.; Aguado-Puig, A.; Romero, F.; et al. Partial Activation of SA- and JA-Defensive Pathways in Strawberry upon Colletotrichum acutatum Interaction. Front. Plant Sci. 2016, 7, 1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoi, Y.; Tanaka, K.; Cook, S.D.; Hayashi, K.-I.; Kasahara, H. GH3 Auxin-Amido Synthetases Alter the Ratio of Indole-3-Acetic Acid and Phenylacetic Acid in Arabidopsis. Plant Cell Physiol. 2020, 61, 596–605. [Google Scholar] [CrossRef]
- Wang, D.; Pajerowska-Mukhtar, K.; Culler, A.H.; Dong, X. Salicylic Acid Inhibits Pathogen Growth in Plants through Repression of the Auxin Signaling Pathway. Curr. Biol. 2007, 17, 1784–1790. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, Q.; Li, Z.; Staswick, P.E.; Wang, M.; Zhu, Y.; He, Z. Dual Regulation Role of GH3.5 in Salicylic Acid and Auxin Signaling during Arabidopsis-Pseudomonas Syringae Interaction. Plant Physiol. 2007, 145, 450–464. [Google Scholar] [CrossRef] [Green Version]
- Lahey, K.A.; Yuan, R.; Burns, J.K.; Ueng, P.P.; Timmer, L.W.; Kuang-Ren, C. Induction of Phytohormones and Differential Gene Expression in Citrus Flowers Infected by the Fungus Colletotrichum acutatum. Mol. Plant Microbe Interact. 2004, 17, 1394–1401. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.-Y.; Zhang, L.-Q.; Song, L.-L.; Duan, K.; Li, N.; Wang, Y.-X.; Gao, Q.-H. The Different Interactions of Colletotrichum gloeosporioides with Two Strawberry Varieties and the Involvement of Salicylic Acid. Hortic. Res. 2016, 3, 16007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayat, Q.; Hayat, S.; Irfan, M.; Ahmad, A. Effect of Exogenous Salicylic Acid under Changing Environment: A Review. Environ. Exp. Bot. 2010, 68, 14–25. [Google Scholar] [CrossRef]
- Yoosomboon, P.; Sojikul, P.; Viboonjun, U.; Narangajavana, J. Salicylic Acid-Induced Syntaxin Gene Expression Coexists with Enhanced Resistance against Colletotrichum gloeosporioides Infection in Cassava. Trop. Plant Biol. 2021, 14, 50–62. [Google Scholar] [CrossRef]
- Sangpueak, R.; Phansak, P.; Thumanu, K.; Siriwong, S.; Wongkaew, S.; Buensanteai, N. Effect of Salicylic Acid Formulations on Induced Plant Defense against Cassava Anthracnose Disease. Plant Pathol. J. 2021, 37, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Gorman, Z.; Christensen, S.A.; Yan, Y.; He, Y.; Borrego, E.; Kolomiets, M.V. Green Leaf Volatiles and Jasmonic Acid Enhance Susceptibility to Anthracnose Diseases Caused by Colletotrichum graminicola in Maize. Mol. Plant Pathol. 2020, 21, 702–715. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Duan, K.; Zhang, L.; Zhang, L.; Song, L.; Yang, J.; Zou, X.; Wang, Y.; Gao, Q. Fast Quenching the Burst of Host Salicylic Acid is Common in Early Strawberry/Colletotrichum fructicola Interaction. Phytopathology 2019, 109, 531–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebauer, P.; Korn, M.; Engelsdorf, T.; Sonnewald, U.; Koch, C.; Voll, L.M. Sugar Accumulation in Leaves of Arabidopsis Sweet11/Sweet12 Double Mutants Enhances Priming of the Salicylic Acid-Mediated Defense Response. Front. Plant Sci. 2017, 8, 1378. [Google Scholar] [CrossRef] [Green Version]
- Kubota, M.; Nishi, K. Salicylic Acid Accumulates in the Roots and Hypocotyl after Inoculation of Cucumber Leaves with Colletotrichum lagenarium. J. Plant Physiol. 2006, 163, 1111–1117. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonates: News on Occurrence, Biosynthesis, Metabolism and Action of an Ancient Group of Signaling Compounds. Int. J. Mol. Sci. 2018, 19, 2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Li, X.; Sun, Z.; Shao, S.; Hu, L.; Ye, M.; Zhou, Y.; Xia, X.; Yu, J.; Shi, K. Antagonism between Phytohormone Signalling Underlies the Variation in Disease Susceptibility of Tomato Plants under Elevated CO2. J. Exp. Bot. 2015, 66, 1951–1963. [Google Scholar] [CrossRef] [Green Version]
- Diniz, I.; Figueiredo, A.; Loureiro, A.; Batista, D.; Azinheira, H.; Várzea, V.; Pereira, A.P.; Gichuru, E.; Moncada, P.; Guerra-Guimarães, L.; et al. A First Insight into the Involvement of Phytohormones Pathways in Coffee Resistance and Susceptibility to Colletotrichum kahawae. PLoS ONE 2017, 12, e0178159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loureiro, A.; Nicole, M.R.; Várzea, V.; Moncada, P.; Bertrand, B.; Silva, M.C. Coffee Resistance to Colletotrichum Kahawae is Associated with Lignification, Accumulation of Phenols and Cell Death at Infection Sites. Physiol. Mol. Plant Pathol. 2012, 77, 23–32. [Google Scholar] [CrossRef]
- Silva, M.D.C.; Várzea, V.; Guerra-Guimarães, L.; Azinheira, H.G.; Fernandez, D.; Petitot, A.-S.; Bertrand, B.; Lashermes, P.; Nicole, M. Coffee resistance to the main diseases: Leaf rust and coffee berry disease. Braz. J. Plant Physiol. 2006, 18, 119–147. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Ahammed, G.J.; Li, Z.; Tang, M.; Yan, P.; Han, W. Decreased Biosynthesis of Jasmonic Acid via Lipoxygenase Pathway Compromised Caffeine-Induced Resistance to Colletotrichum gloeosporioides Under Elevated CO2 in Tea Seedlings. Phytopathology 2016, 106, 1270–1277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.J.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 Reveals the Versatile Functions of Jasmonic Acid in Maize Development and Defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Xie, D. Jasmonate in Plant Defence: Sentinel or Double Agent? Plant Biotechnol. J. 2015, 13, 1233–1240. [Google Scholar] [CrossRef] [Green Version]
- Bajguz, A.; Chmur, M.; Gruszka, D. Comprehensive Overview of the Brassinosteroid Biosynthesis Pathways: Substrates, Products, Inhibitors and Connections. Front. Plant Sci. 2020, 11, 1034. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses [OPEN]. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.-H.; Lee, M.; Park, C.-M. A Transcriptional Feedback Loop Modulating Signaling Crosstalks between Auxin and Brassinosteroid in Arabidopsis. Mol. Cells 2010, 29, 449–456. [Google Scholar] [CrossRef]
- Vert, G.; Walcher, C.L.; Chory, J.; Nemhauser, J.L. Integration of Auxin and Brassinosteroid Pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. USA 2008, 105, 9829–9834. [Google Scholar] [CrossRef] [Green Version]
- Davière, J.-M.; Achard, P. Gibberellin Signaling in Plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, M.-H.; Zhao, Z.-Z.; He, J.-X. Brassinosteroid Signaling in Plant-Microbe Interactions. Int. J. Mol. Sci. 2018, 19, 4091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nolan, T.; Chen, J.; Yin, Y. Cross-Talk of Brassinosteroid Signaling in Controlling Growth and Stress Responses. Biochem. J. 2017, 474, 2641–2661. [Google Scholar] [CrossRef]
- Furio, R.N.; Albornoz, P.L.; Coll, Y.; Zamora, G.M.M.; Salazar, S.M.; Martos, G.G.; Ricci, J.C.D. Effect of Natural and Synthetic Brassinosteroids on Strawberry Immune Response against Colletotrichum acutatum. Eur. J. Plant Pathol. 2019, 153, 167–181. [Google Scholar] [CrossRef]
- Sudheeran, P.K.; Sela, N.; Carmeli-Weissberg, M.; Ovadia, R.; Panda, S.; Feygenberg, O.; Maurer, D.; Oren-Shamir, M.; Aharoni, A.; Alkan, N. Induced Defense Response in Red Mango Fruit against Colletotrichum gloeosporioides. Hortic. Res. 2021, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Nakashita, H.; Yasuda, M.; Nitta, T.; Asami, T.; Fujioka, S.; Arai, Y.; Sekimata, K.; Takatsuto, S.; Yamaguchi, I.; Yoshida, S. Brassinosteroid Functions in a Broad Range of Disease Resistance in Tobacco and Rice. Plant J. 2003, 33, 887–898. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Hong, G.; Zhang, H.; Tan, X.; Li, L.; Kong, Y.; Sang, T.; Xie, K.; Wei, J.; Li, J.; et al. The OsGSK2 Kinase Integrates Brassinosteroid and Jasmonic Acid Signaling by Interacting with OsJAZ4. Plant Cell 2020, 32, 2806–2822. [Google Scholar] [CrossRef]
- Izquierdo-Bueno, I.; González-Rodríguez, V.E.; Simon, A.; Dalmais, B.; Pradier, J.-M.; Le Pêcheur, P.; Mercier, A.; Walker, A.-S.; Garrido, C.; Collado, I.G.; et al. Biosynthesis of Abscisic Acid in Fungi: Identification of a Sesquiterpene Cyclase as the Key Enzyme in Botrytis cinerea. Environ. Microbiol. 2018, 20, 2469–2482. [Google Scholar] [CrossRef]
- Luo, K.; Rocheleau, H.; Qi, P.-F.; Zheng, Y.-L.; Zhao, H.-Y.; Ouellet, T. Indole-3-Acetic Acid in Fusarium graminearum: Identification of Biosynthetic Pathways and Characterization of Physiological Effects. Fungal Biol. 2016, 120, 1135–1145. [Google Scholar] [CrossRef]
- Oliw, E.H.; Hamberg, M. An Allene Oxide and 12-Oxophytodienoic Acid Are Key Intermediates in Jasmonic Acid Biosynthesis by Fusarium oxysporum. J. Lipid Res. 2017, 58, 1670–1680. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.P.; Shelke, G.M.; Kumar, A.; Jha, P.N. Biochemistry and Genetics of ACC Deaminase: A Weapon to “Stress Ethylene” Produced in Plants. Front. Microbiol. 2015, 6, 937. [Google Scholar] [CrossRef] [PubMed]



| Host Plant | Cultivar/Strain | Tissue | Pathogen | SA-Levels | Time Point | Reference |
|---|---|---|---|---|---|---|
| Maize | W438 | Leaf | C. graminicola | ~100 pmol/g FW | 1 dpi | [85] |
| lox10-3 | ~100 pmol/g FW | |||||
| B73 | Leaf | <100 pmol/g FW | ||||
| opr7.5 opr8.2 | >200 pmol/g FW | |||||
| Strawberry | Jiuxiang | Leaf | C. fructicula | 0.36 ng/mg FW | 1 hpi | [86] |
| Benihoppe | 0.38 ng/mg FW | |||||
| Arabidopsis | Col-0 | Leaf | C. higginsianum | ~90 µg·m−2 | 2 dpi | [87] |
| sweet11 | ~110 µg·m−2 | |||||
| sweet12 | ~140 µg·m−2 | |||||
| sweet11/12 | ~190 µg·m−2 | |||||
| Cucumber | Cucumis sativus | Cotyledon inoculated: | C. lagenarium | [88] | ||
| Roots | 2 µg/g | 6 dpi | ||||
| Leaf | 0.6 µg/g | |||||
| Hypocotyl | 2 µg/g | |||||
| First leaf inoculated: | ||||||
| Roots | >2.5 µg/g | 7 dpi | ||||
| Leaf | >3.0 µg/g | |||||
| Hypocotyl | ~3.0 µg/g | |||||
| Tea plants | Longjing 43 and Zhenong 139 | Healthy leaves | Colletotrichum spp. | ~8 µg/g FW | Collected in July | [73] |
| Infected leaves | ~13 µg/g FW | |||||
| Strawberry | Camarosa | Control leaf | C. acutatum | 74.42 ng/g DW | 3 dpi | [76] |
| Infected leaf | 202.21 ng/g DW | |||||
| Control leaf | 52.33 ng/g DW | 5 dpi | ||||
| Infected leaf | 354.77 ng/g DW |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Svoboda, T.; Thon, M.R.; Strauss, J. The Role of Plant Hormones in the Interaction of Colletotrichum Species with Their Host Plants. Int. J. Mol. Sci. 2021, 22, 12454. https://doi.org/10.3390/ijms222212454
Svoboda T, Thon MR, Strauss J. The Role of Plant Hormones in the Interaction of Colletotrichum Species with Their Host Plants. International Journal of Molecular Sciences. 2021; 22(22):12454. https://doi.org/10.3390/ijms222212454
Chicago/Turabian StyleSvoboda, Thomas, Michael R. Thon, and Joseph Strauss. 2021. "The Role of Plant Hormones in the Interaction of Colletotrichum Species with Their Host Plants" International Journal of Molecular Sciences 22, no. 22: 12454. https://doi.org/10.3390/ijms222212454
APA StyleSvoboda, T., Thon, M. R., & Strauss, J. (2021). The Role of Plant Hormones in the Interaction of Colletotrichum Species with Their Host Plants. International Journal of Molecular Sciences, 22(22), 12454. https://doi.org/10.3390/ijms222212454






