Dynamics of Fusarium Mycotoxins and Lytic Enzymes during Pea Plants’ Infection
Abstract
:1. Introduction
2. Results
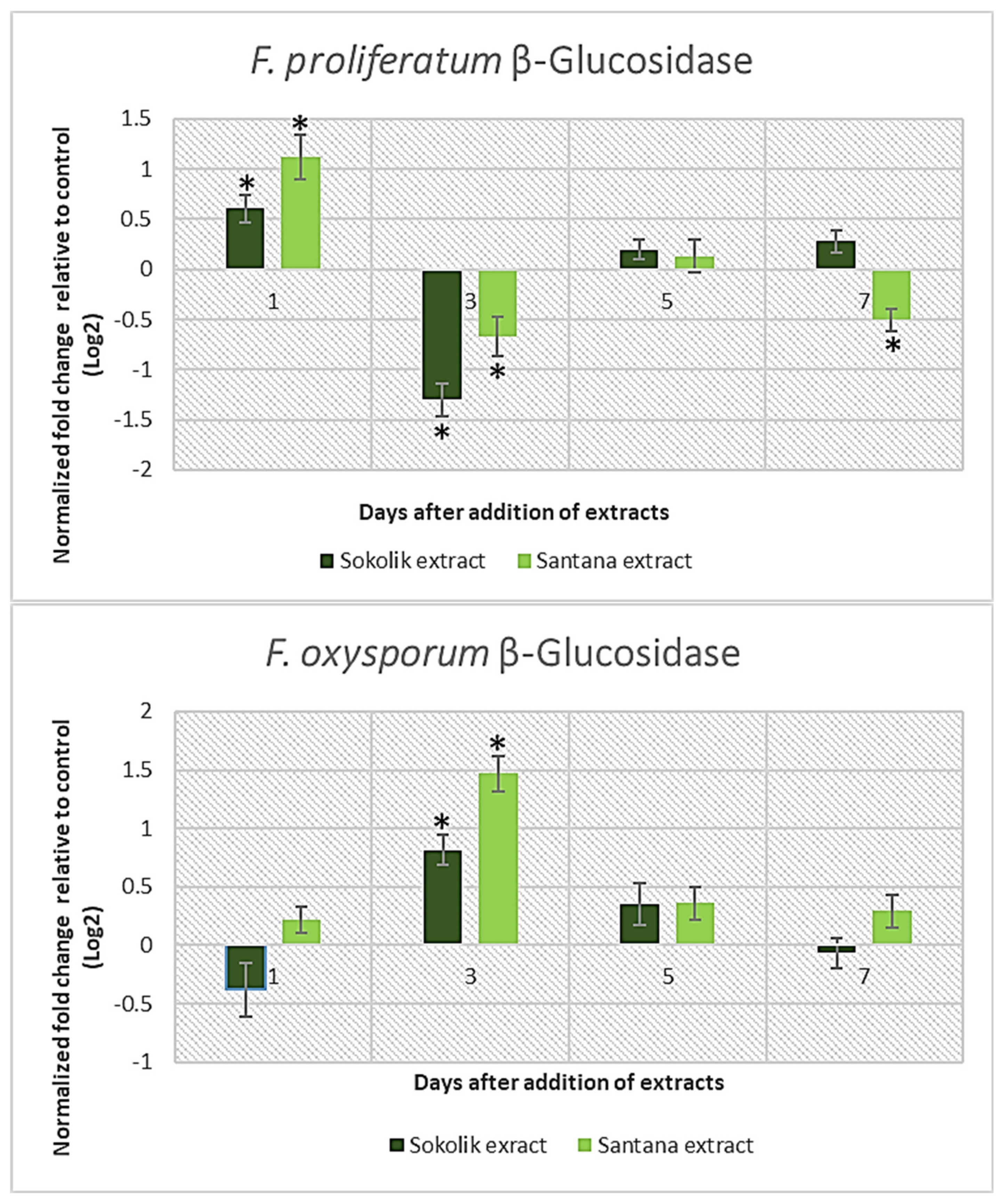
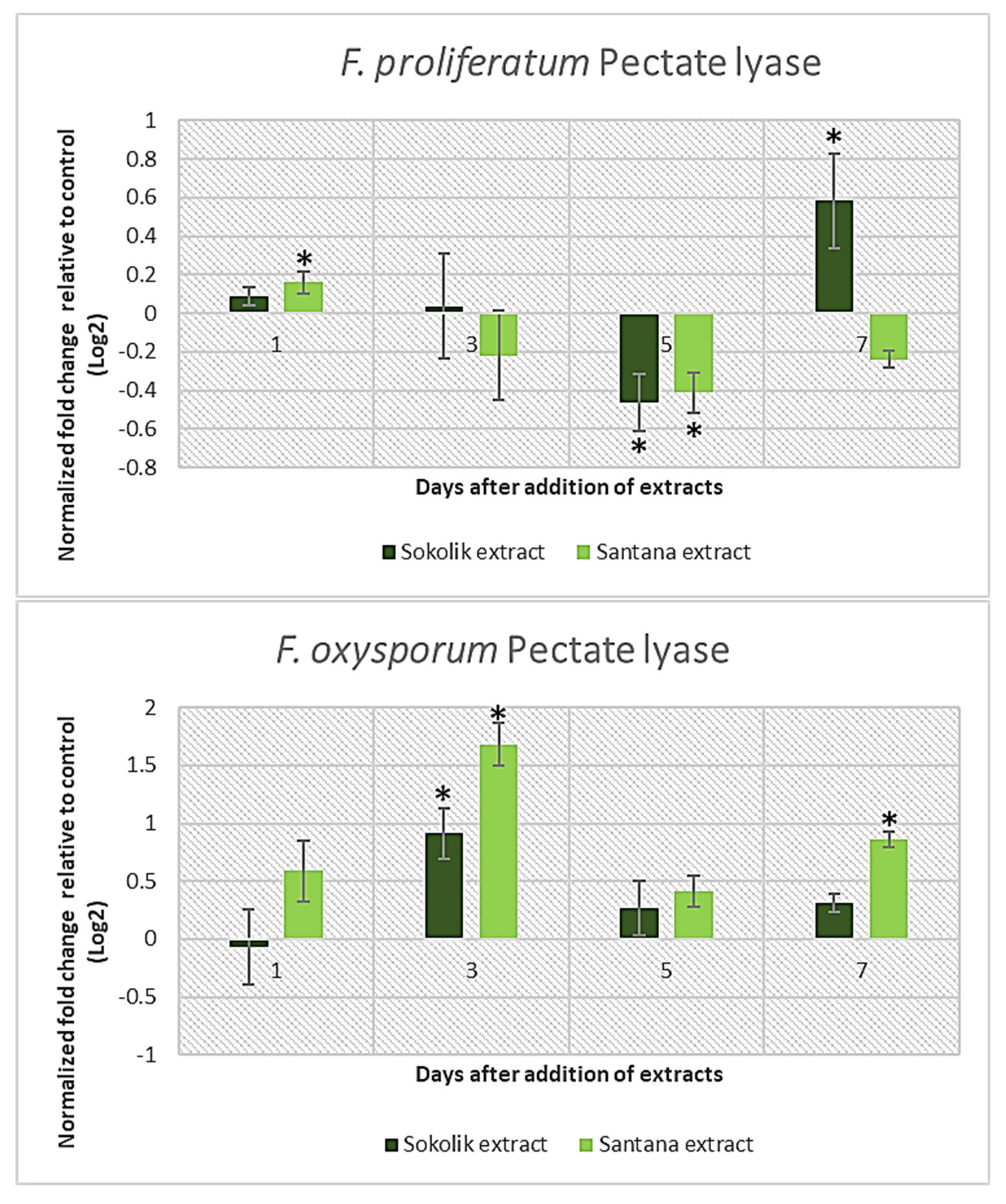
2.1. Fungal Gene Expression Studies
2.2. Plant Infection Studies
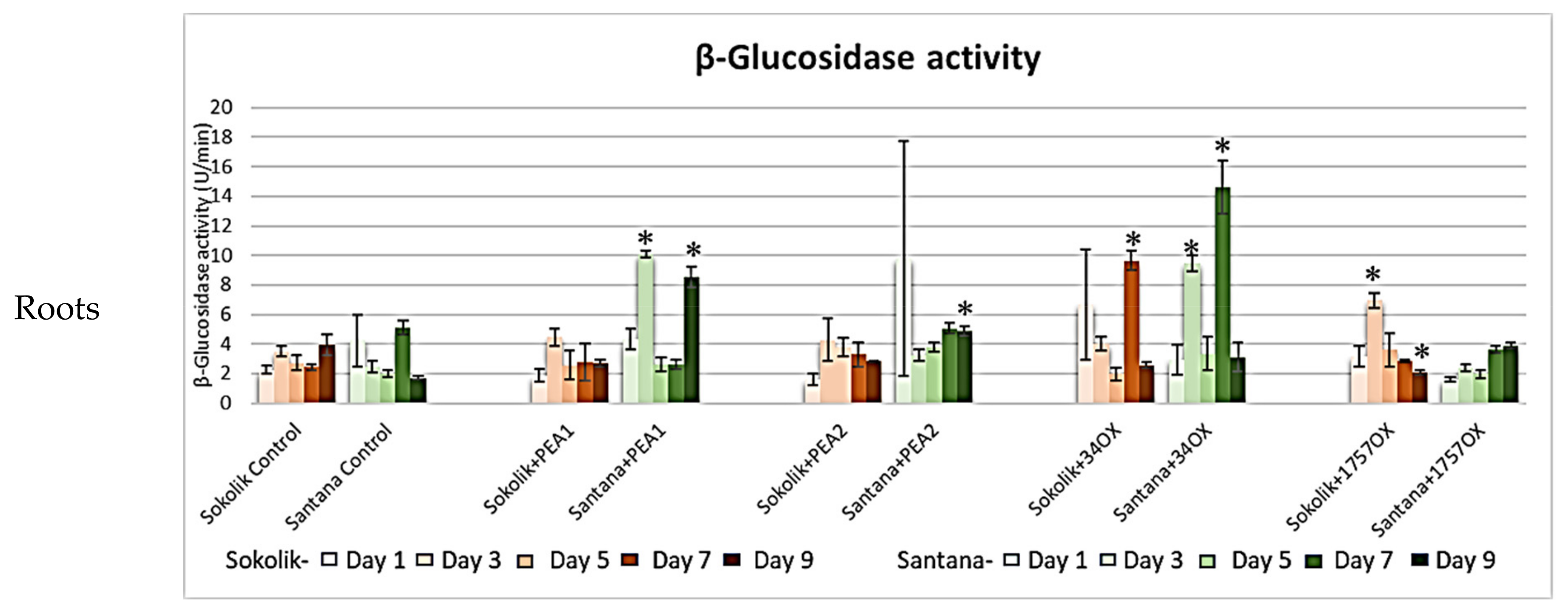
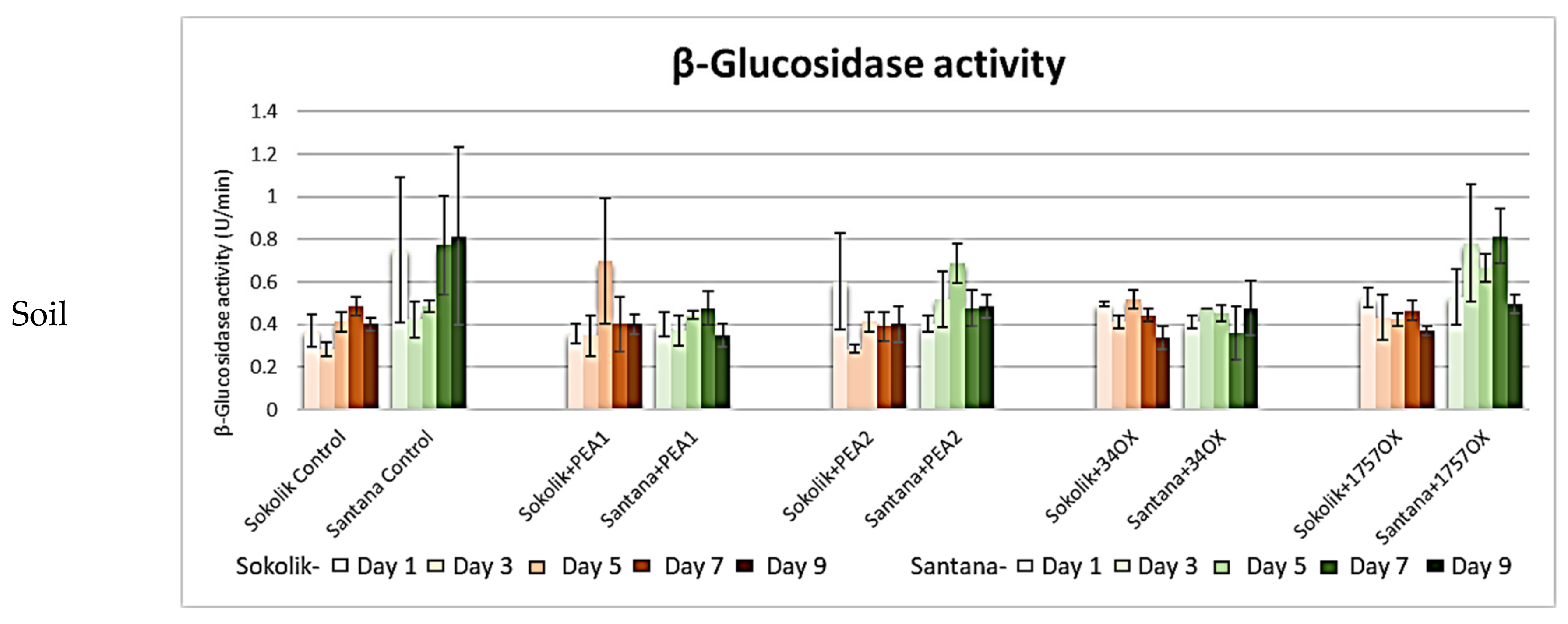
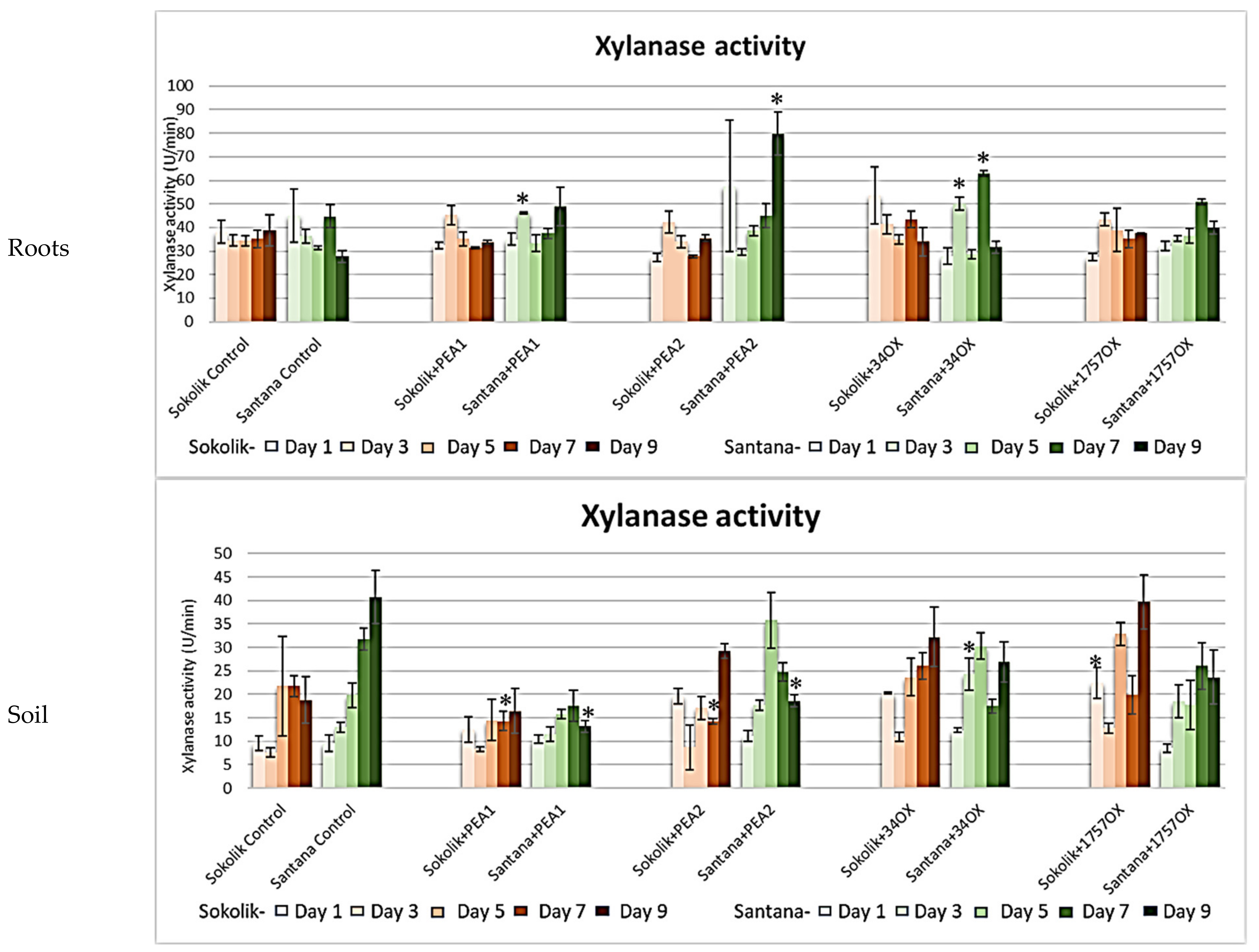
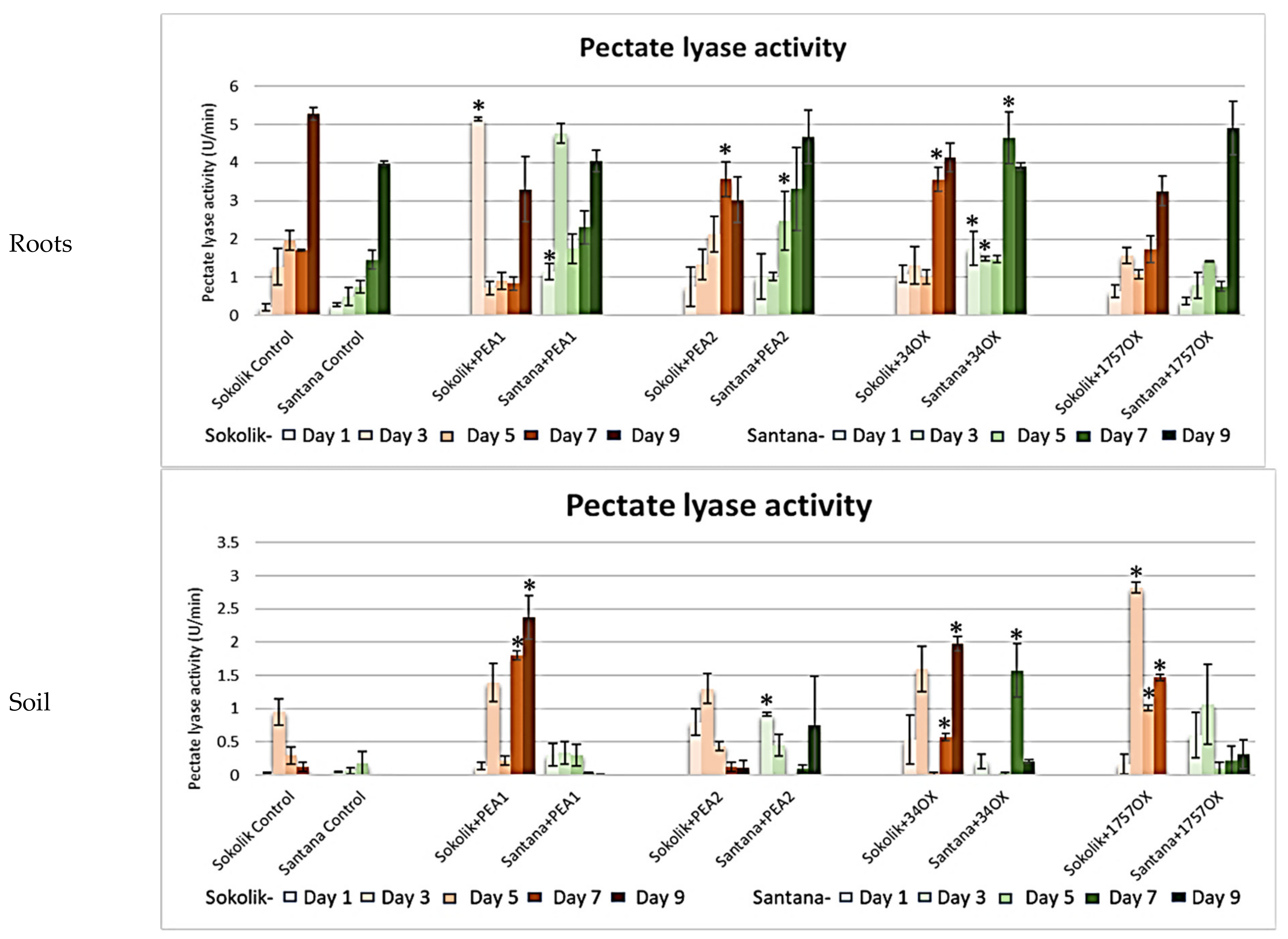
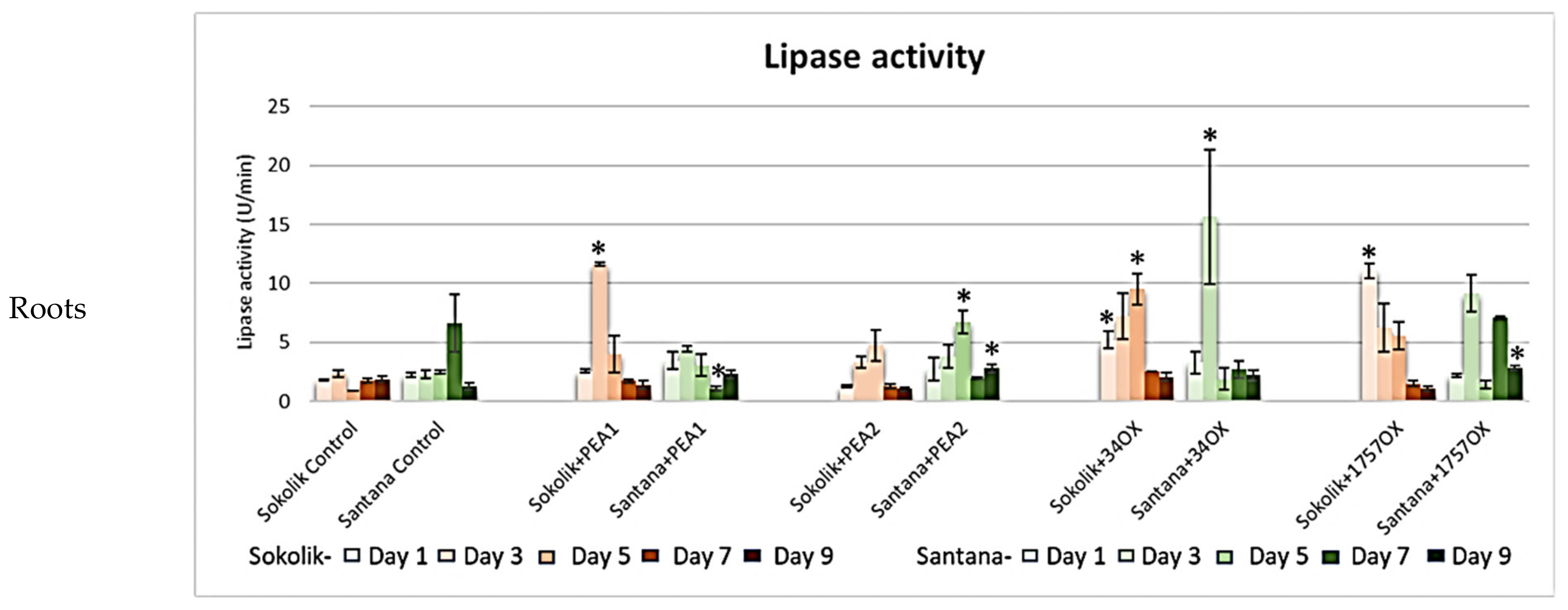
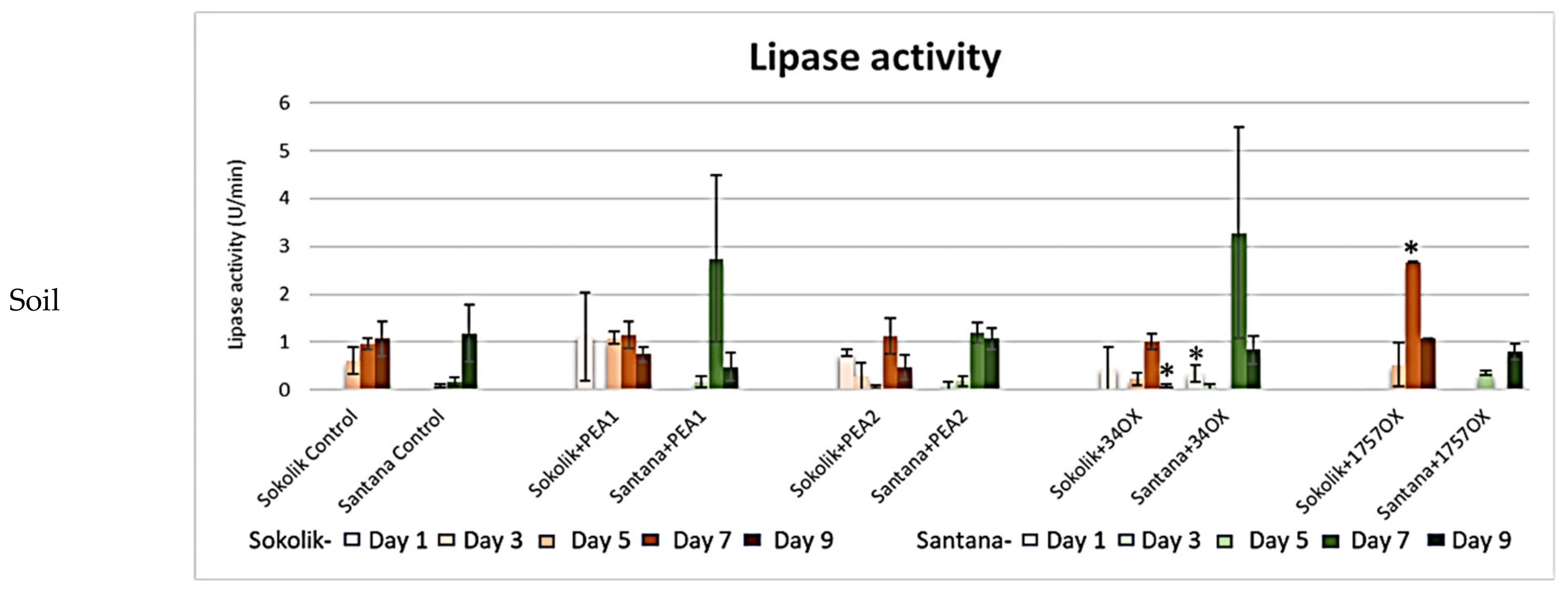
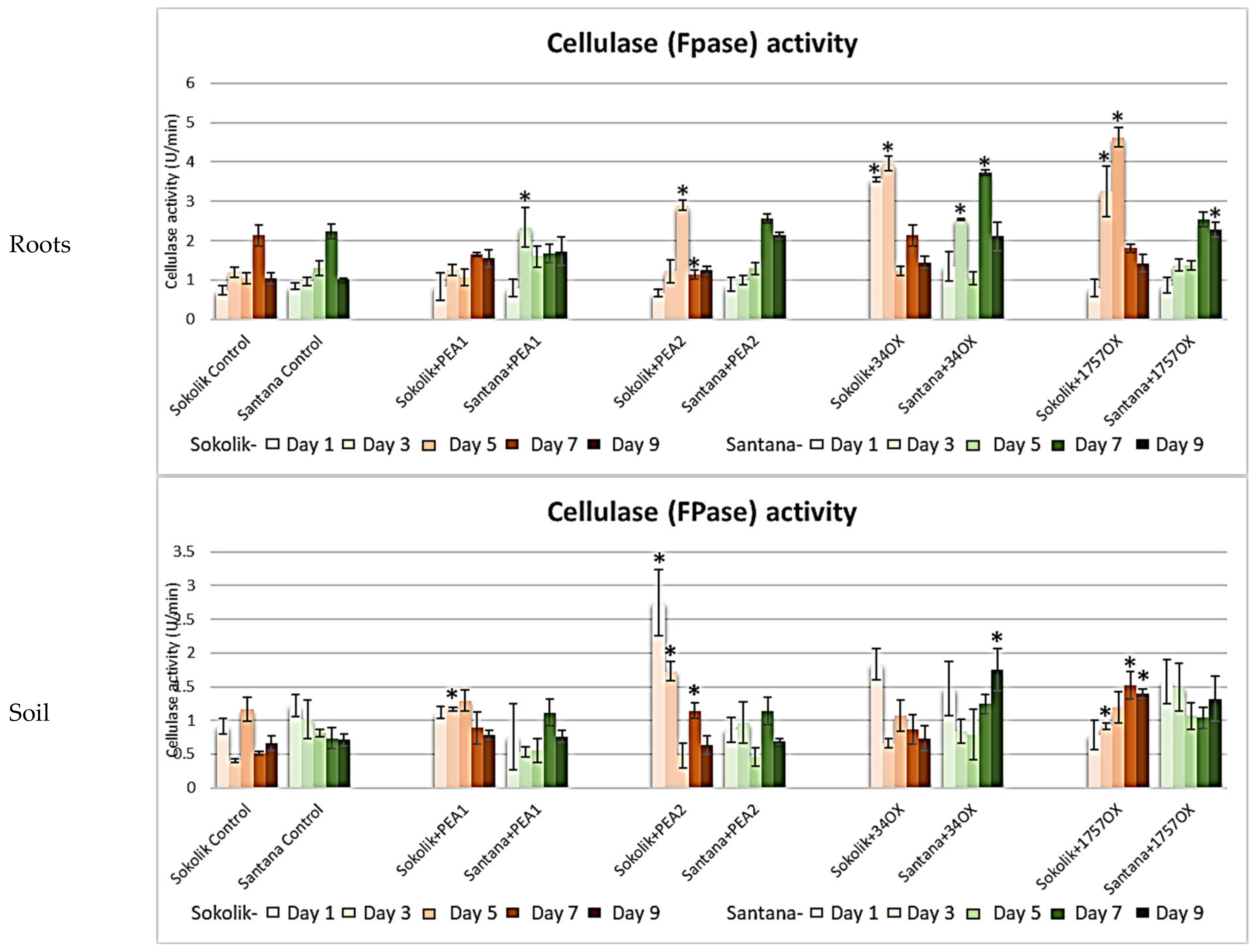
2.2.1. Cell Wall-Degrading Enzyme Assays
2.2.2. Mycotoxins Quantification
3. Discussion
4. Materials and Methods
4.1. Fungal Gene Expression Studies
4.1.1. Fungal Strains and Growth Conditions
4.1.2. Plant Extract Preparation
4.1.3. Gene Expression Studies
4.2. Plant Cultivation
4.3. Plant Infection Studies
4.3.1. Enzyme Activity Assays
β-Glucosidase
Chitinase
Xylanase
Pectate Lyase
Polygalacturonase
Lipase
Cellulase (Filter Paper Assay)
Endo beta-1, 4 glucanase (CMCase)
Exo-β-1,4-glucanase (avicelase)
Protease
4.4. Mycotoxins Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The Editors of Encyclopaedia Britannica. Fusarium Wilt. In Encyclopedia Britannica; Encyclopedia Britannica, Inc.: Chicago, IL, USA, 2017; pp. 1–2. Available online: https://www.britannica.com/science/fusarium-wilt (accessed on 1 August 2021).
- Buxton, E.W. Fusarium Diseases of Peas. Trans. Br. Mycol. Soc. 1955, 38. [Google Scholar] [CrossRef]
- Marcinkowska, J. Fungi Occurrence on Seeds of Field Pea. Acta Mycol. 2013, 43, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Sideman, E. Fusarium Wilt of Peas; Maine Organic Farmers and Gardeners: Unity, ME, USA, 2010; Available online: https://www.mofga.org/resources/plant-diseases/fusarium-wilt-of-peas/ (accessed on 1 August 2021).
- Waśkiewicz, A.; Stepień, Ł.; Wilman, K.; Kachlicki, P. Diversity of Pea-Associated, F. Proliferatum and F. Verticillioides Populations Revealed by FUM1 Sequence Analysis and Fumonisin Biosynthesis. Toxins 2013, 5, 488–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas, H.K.; Duke, S.O.; Tatsumi, T. Phytotoxicity of Fumonisins and Rfzated Compounds. J. Toxicol. Toxin Rev. 1993, 12, 225–251. [Google Scholar] [CrossRef]
- Doehlert, D.C.; Knutson, C.A.; Vesonder, R.F. Phytotoxic Effects of Fumonisin B1 on Maize Seedling Growth. Mycopathologia 1994, 127, 117–121. [Google Scholar] [CrossRef]
- Górna, K.; Pawłowicz, I.; Waśkiewicz, A.; Stepień, Ł. Fusarium proliferatum Strains Change Fumonisin Biosynthesis and Accumulation When Exposed to Host Plant Extracts. Fungal Biol. 2016, 120, 884–893. [Google Scholar] [CrossRef]
- Perincherry, L.; Ajmi, C.; Oueslati, S.; Waśkiewicz, A.; Stępień, Ł. Induction of Fusarium Lytic Enzymes by Extracts from Resistant and Susceptible Cultivars of Pea (Pisum sativum L.). Pathogens 2020, 9, 976. [Google Scholar] [CrossRef]
- Lalaoui, F.; Halama, P.; Dumortier, V.; Paul, B. Cell Wall-Degrading Enzymes, Produced in Vitro by Isolates of Phaeosphaeria Nodorum Differing in Aggressiveness. Plant Pathol. 2000, 49, 727–733. [Google Scholar] [CrossRef]
- Tonukari, N.J.; Scott-Craig, J.S.; Walton, J.D. The Cochliobolus carbonum SNF1 Gene Is Required for Cell Wall-Degrading Enzyme Expression and Virulence on Maize. Plant Cell 2000, 12, 237–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maia, M.D.M.D.; Camargo De Morais, M.M.; De Morais, M.A.; Melo, E.H.M.; De Lima Filho, J.L. Production of Extracellular Lipase by the Phytopathogenic Fungus Fusarium Solani Fs1. Rev. Microbiol. 1999, 30, 304–309. [Google Scholar] [CrossRef]
- Kikot, G.E.; Hours, R.A.; Alconada, T.M. Contribution of Cell Wall Degrading Enzymes to Pathogenesis of Fusarium Graminearum: A Review. J. Basic Microbiol. 2009, 49, 231–241. [Google Scholar] [CrossRef]
- Cooper, R.M.; Wood, R.K.S. Regulation of Synthesis of Cell Wall Degrading Enzymes by Verticillium Albo-Atrum and Fusarium oxysporum f. sp. lycopersici. Physiol. Plant Pathol. 1975, 5, 135–156. [Google Scholar] [CrossRef]
- Pekkarinen, A.; Mannonen, L.; Jones, B.L.; Niku-Paavola, M.L. Production of Proteases by Fusarium Species Grown on Barley Grains and in Media Containing Cereal Proteins. J. Cereal Sci. 2000, 31, 253–261. [Google Scholar] [CrossRef]
- Panagiotou, G.; Kekos, D.; Macris, B.J.; Christakopoulos, P. Production of Cellulolytic and Xylanolytic Enzymes by Fusarium Oxysporum Grown on Corn Stover in Solid State Fermentation. Ind. Crops Prod. 2003, 18, 37–45. [Google Scholar] [CrossRef]
- Jorge, I.; de la Rosa, O.; Navas-Cortés, J.A.; Jiménez-Díaz, R.M.; Tena, M. Extracellular Xylanases from Two Pathogenic Races of Fusarium oxysporum f. sp. ciceris: Enzyme Production in Culture and Purification and Characterization of a Major Isoform as an Alkaline Endo-β-(1,4)-Xylanase of Low Molecular Weight. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2005, 88, 48–59. [Google Scholar] [CrossRef]
- Naraian, R.; Gautam, R.L. Penicillium Enzymes for the Saccharification of Lignocellulosic Feedstocks. In New and Future Developments in Microbial Biotechnology and Bioengineering: Penicillium System Properties and Applications; Elsevier: Amsterdam, The Netherlands, 2018; pp. 121–136. [Google Scholar] [CrossRef]
- Brunner, G. Processing of Biomass with Hydrothermal and Supercritical Water. In Supercritical Fluid Science and Technology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 5, pp. 395–509. [Google Scholar] [CrossRef]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef]
- Peretto, R.; Favaron, F.; Bettini, V.; De Lorenzo, G.; Marinp, S.; Alghisi, P.; Cervone, F.; Bonfante, P. Expression and Localization of Polygalacturonase during the Outgrowth of Lateral Roots in Allium porrum L. Planta 1992, 188, 164–172. [Google Scholar] [CrossRef]
- Hahn, M.G.; Bucheli, P.; Cervone, F.; Doares, S.H.; O’Neill, R.A.; Darvill, A.; Albersheim, P. The Role of the Cell Wall Plant-Microbe, Constituents in Plant-Pathogen Interactions. In Interactions; Nester, E.K., Ed.; McGraw-Hill: New York, NY, USA, 1989; pp. 1–38. [Google Scholar]
- Annamalai, N.; Rajeswari, M.V.; Balasubramanian, T. Endo-1,4-β-Glucanases: Role, Applications and Recent Developments. Springer: Cham, Germany, 2016; pp. 37–45. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant Cell Wall-Degrading Enzymes and Their Secretion in Plant-Pathogenic Fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial Diversity and Soil Functions. Eur. J. Soil Sci. 2003, 54, 655–670. [Google Scholar] [CrossRef]
- Li, T.; Jian, Q.; Wang, Y.; Chen, F.; Yang, C.; Gong, L.; Duan, X.; Yang, B.; Jiang, Y. Inhibitory Mechanism of Butylated Hydroxyanisole against Infection of Fusarium Proliferatum Based on Comparative Proteomic Analysis. J. Proteom. 2016, 148, 1–11. [Google Scholar] [CrossRef]
- Huertas-González, M.D.; Ruiz-Roldán, M.C.; Maceira, F.I.G.; Roncero, M.I.G.; Di Pietro, A. Cloning and Characterization of Pl1 Encoding an in Planta-Secreted Pectate Lyase of Fusarium oxysporum. Curr. Genet. 1999, 35, 36–40. [Google Scholar] [CrossRef]
- Gómez-Gómez, E.; Ruíz-Roldán, M.C.; Di Pietro, A.; Roncero, M.I.G.; Hera, C. Role in Pathogenesis of Two Endo-β-1,4-Xylanase Genes from the Vascular Wilt Fungus Fusarium Oxysporum. Fungal Genet. Biol. 2002, 35, 213–222. [Google Scholar] [CrossRef]
- Juge, N. Plant Protein Inhibitors of Cell Wall Degrading Enzymes. Trends Plant Sci. 2006, 11, 359–367. [Google Scholar] [CrossRef]
- Breitenbach, L.C.; Coelho, B. Antimicrobial Activity of Secondary Metabolites and Lectins from Plants. Curr. Res. Technol. Educ. Top. Appl. Microbiol. Microb. Biotechnol. 2010, 1, 396–406. [Google Scholar]
- Woo, K.H.; Hwan, Y.J.; Hwan, K.S.; Beom, H.S.; Cheon, Y.; Ju, K.S. Detection of Extracellular Enzyme Activities in Various Fusarium spp. Mycobiology 2007, 35, 162–165. [Google Scholar] [CrossRef] [Green Version]
- Dharmawardhana, D.P.; Ellis, B.E.; Carlson, J.E. A Beta-Glucosidase from Lodgepole Pine Xylem Specific for the Lignin Precursor Coniferin. Plant Physiol. 1995, 107, 331–339. [Google Scholar] [CrossRef]
- Kristoffersen, P.; Brzobohaty, B.; Höhfeld, I.; Bako, L.; Melkonian, M.; Palme, K. Developmental Regulation of the Maize Zm-P60.1 Gene Encoding a β-Glucosidase Located to Plastids. Planta 2000, 210, 407–415. [Google Scholar] [CrossRef]
- Suzuki, H.; Takahashi, S.; Watanabe, R.; Fukushima, Y.; Fujita, N.; Noguchi, A.; Yokoyama, R.; Nishitani, K.; Nishino, T.; Nakayama, T. An Isoflavone Conjugate-Hydrolyzing β-Glucosidase from the Roots of Soybean (Glycine Max) Seedlings: Purification, Gene Cloning, Phylogenetics, and Cellular Localization. J. Biol. Chem. 2006, 281, 30251–30259. [Google Scholar] [CrossRef] [Green Version]
- Mann, B. Role of Pectic Enzymes in the Fusarium Wilt Syndrome of Tomato. Trans. Br. Mycol. Soc. 1962, 45, 169. [Google Scholar] [CrossRef]
- Palusa, S.G.; Golovkin, M.; Shin, S.B.; Richardson, D.N.; Reddy, A.S.N. Organ-Specific, Developmental, Hormonal and Stress Regulation of Expression of Putative Pectate Lyase Genes in Arabidopsis. New Phytol. 2007, 174, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, M.; Biller, S.; Stanley, K.; Kajstura, T.; Prusty, R. Expression Profiling of Auxin-Treated Arabidopsis Roots: Toward a Molecular Analysis of Lateral Root Emergence. Plant Cell Physiol. 2006, 47, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Palanisamy, A.; Thangavelu, V. Lipase Catalyzed Ester Synthesis for Food Processing Industries. Braz. Arch. Biol. Technol. 2009, 52, 207–219. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Ma, B.; Duan, K.X.; Li, X.K.; Lu, X.; Yin, C.C.; Tao, J.J.; Wei, W.; Zhang, W.K.; Xin, P.Y.; et al. The GDSL Lipase MHZ11 Modulates Ethylene Signaling in Rice Roots. Plant Cell 2020, 32, 1626–1643. [Google Scholar] [CrossRef] [Green Version]
- Tagele, S.B.; Kim, S.W.; Lee, H.G.; Lee, Y.S. Aggressiveness and Fumonisins Production of Fusarium subglutinans and Fusarium temperatum on Korean Maize Cultivars. Agronomy 2019, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Otani, H.; Kohmoto, K.; Nishimura, S.; Nakashima, T.; Ueno, T.; Fukami, H. Studies on Black Spot Disease of Japanese Pear. XXII. Biological Activities of AK-Toxins I and II, Host-Specific Toxins from Alternaria Alternata Japanese Pear Pathotype. Jpn. J. Phytopathol. 1985, 51, 285–293. [Google Scholar] [CrossRef]
- Baayen, R.P.; Van Eijk, C.; Elgersma, D.M. Histology of Roots of Resistant and Susceptible Carnation Cultivars from Soil Infested with Fusarium oxysporum f. sp. dianthi. Neth. J. Plant Pathol. 1989, 95, 3–13. [Google Scholar] [CrossRef]
- Bani, M.; Pérez-de-Luque, A.; Rubiales, D.; Rispail, N. Physical and Chemical Barriers in Root Tissues Contribute to Quantitative Resistance to Fusarium oxysporum f. sp. pisi in Pea. Front. Plant Sci. 2018, 9, 199. [Google Scholar] [CrossRef]
- Wilman, K.; Stępień, Ł.; Fabiańska, I.; Kachlicki, P. Plant-Pathogenic Fungi in Seeds of Different Pea Cultivars in Poland. Arh. Hig. Rada Toksikol. 2014, 65, 329–338. [Google Scholar] [CrossRef] [Green Version]
- López-Errasquín, E.; Vázquez, C.; Jiménez, M.; González-Jaén, M.T. Real-Time RT-PCR Assay to Quantify the Expression of Fum1 and Fum19 Genes from the Fumonisin-Producing Fusarium verticillioides. J. Microbiol. Methods 2007, 68, 312–317. [Google Scholar] [CrossRef]
- Stepień, Ł.; Waśkiewicz, A.; Wilman, K. Host Extract Modulates Metabolism and Fumonisin Biosynthesis by the Plant-Pathogenic Fungus fusarium proliferatum. Int. J. Food Microbiol. 2015, 193, 74–81. [Google Scholar] [CrossRef]
- Jurado, M.; Marín, P.; Callejas, C.; Moretti, A.; Vázquez, C.; González-Jaén, M.T. Genetic Variability and Fumonisin Production by Fusarium proliferatum. Food Microbiol. 2010, 27, 50–57. [Google Scholar] [CrossRef]
- Taylor, A.; Vágány, V.; Jackson, A.C.; Harrison, R.J.; Rainoni, A.; Clarkson, J.P. Identification of Pathogenicity-Related Genes in Fusarium oxysporum f. sp. cepae. Mol. Plant. Pathol. 2016, 17, 1032–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panke-Buisse, K.; Poole, A.C.; Goodrich, J.K.; Ley, R.E.; Kao-Kniffin, J. Selection on Soil Microbiomes Reveals Reproducible Impacts on Plant Function. ISME J. 2015, 9, 980–989. [Google Scholar] [CrossRef]
- Parry, N.J.; Beever, D.E.; Owen, E.; Vandenberghe, I.; Van Beeumen, J.; Bhat, M.K. Biochemical Characterization and Mechanism of Action of a Thermostable β-Glucosidase Purified from Thermoascus aurantiacus. Biochem. J. 2001, 353, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.-M.; Tseng, C.-S.; Cheng, C.-Y.; Li, Y.-K. Purification, Characterization and Cloning of a Chitinase from Bacillus sp. NCTU2. Biotechnol. Appl. Biochem. 2002, 35, 213. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Abramić, M.; Leščić, I.; Korica, T.; Vitale, L.; Saenger, W.; Pigac, J. Purification and Properties of Extracellular Lipase from Streptomyces rimosus. Enzym. Microb. Technol. 1999, 25, 522–529. [Google Scholar] [CrossRef]
- Yu, X.; Liu, Y.; Cui, Y.; Cheng, Q.; Zhang, Z.; Lu, J.H.; Meng, Q.; Teng, L.; Ren, X. Measurement of Filter Paper Activities of Cellulase with Microplate-Based Assay. Saudi J. Biol. Sci. 2016, 23, 93–98. [Google Scholar] [CrossRef] [Green Version]
- Kole, M.M.; Draper, I.; Gerson, D.F. Production of Protease by Bacillus Subtilis Using Simultaneous Control of Glucose and Ammonium Concentrations. J. Chem. Technol. Biotechnol. 2007, 41, 197–206. [Google Scholar] [CrossRef]
- Urbaniak, M.; Waśkiewicz, A.; Trzebny, A.; Koczyk, G.; Stępień, Ł. Cyclodepsipeptide Biosynthesis in Hypocreales Fungi and Sequence Divergence of The Non-Ribosomal Peptide Synthase Genes. Pathogens 2020, 9, 552. [Google Scholar] [CrossRef] [PubMed]
- Witaszak, N.; Lalak-Kańczugowska, J.; Waśkiewicz, A.; Stępień, L. The Impacts of Asparagus Extract Fractions on Growth and Fumonisins Biosynthesis in Fusarium Proliferatum. Toxins 2020, 12, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]



|
Cell Wall-Degrading Enzymes | F. proliferatum PEA1 | F. proliferatum PEA2 | F. oxysporum 34OX | F. oxysporum 1757OX | ||||
|---|---|---|---|---|---|---|---|---|
| Sokolik | Santana | Sokolik | Santana | Sokolik | Santana | Sokolik | Santana | |
| Xylanase | √ | √ | √ | √ | ||||
| Protease | √ | √ | ||||||
| Lipase | √ | √ | √ | √ | √ | √ | ||
| Polygalacturonase | √ | √ | √ | |||||
| Pectin Lyase | √ | √ | √ | |||||
| Chitinase | √ | |||||||
| Endo β-1, 4 glucanase | √ | √ | √ | |||||
| Exo-β-1,4-glucanase | ||||||||
| β-glucosidase | √ | √ | √ | √ | ||||
| Cellulase | √ | √ | √ | √ | √ | √ | ||
| PEA1 | 1757 OX | |
|---|---|---|
| β-Glucosidase (bgl1 gene) | BG1f-CTATCCCTCGCTGCAAGAAC BG1r-GTGGGCAACAAGAAGGTTGT | BGf-CCCAAGCAACTCCGAGGTTT BGr-TGCTGGAGCCGACGAAAATG |
| Pectate lyase (pl1 gene) | PL1f-CTAGCCTCTGTTTGCCAAGG PL1r-TCAGCATGAGAAACGGTGAG | PL4f-GGTGAGCAAGTTTCTCTCGACT PL4r-CACTGGTCTGCTTGAGGGTG |
| Xylanase (xyl4 (F. proliferatum) and xyl2 gene (F. oxysporum)) | XYL4f-CCATCAACTATGGCGGTTCT XYL4fr-GTAGACGGTGCCCTTGTGTT | XYL2f-CAGGTCGTCAACTTTGCTCA XYL2r-TTAACCCACTGAGGGAGCTG |
| β-tubulin | Fpbtf-ACATCCAGACAGCCCTTTGTG Fpbtr-AGTTTCCGATGAAGGTCGAAGA [47] | Fobtf-TTCTGCTGTCATGTCCGGTGT Fobtr-TCAGAGGAGCAAAGCCAACCA [48] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perincherry, L.; Urbaniak, M.; Pawłowicz, I.; Kotowska, K.; Waśkiewicz, A.; Stępień, Ł. Dynamics of Fusarium Mycotoxins and Lytic Enzymes during Pea Plants’ Infection. Int. J. Mol. Sci. 2021, 22, 9888. https://doi.org/10.3390/ijms22189888
Perincherry L, Urbaniak M, Pawłowicz I, Kotowska K, Waśkiewicz A, Stępień Ł. Dynamics of Fusarium Mycotoxins and Lytic Enzymes during Pea Plants’ Infection. International Journal of Molecular Sciences. 2021; 22(18):9888. https://doi.org/10.3390/ijms22189888
Chicago/Turabian StylePerincherry, Lakshmipriya, Monika Urbaniak, Izabela Pawłowicz, Karolina Kotowska, Agnieszka Waśkiewicz, and Łukasz Stępień. 2021. "Dynamics of Fusarium Mycotoxins and Lytic Enzymes during Pea Plants’ Infection" International Journal of Molecular Sciences 22, no. 18: 9888. https://doi.org/10.3390/ijms22189888
APA StylePerincherry, L., Urbaniak, M., Pawłowicz, I., Kotowska, K., Waśkiewicz, A., & Stępień, Ł. (2021). Dynamics of Fusarium Mycotoxins and Lytic Enzymes during Pea Plants’ Infection. International Journal of Molecular Sciences, 22(18), 9888. https://doi.org/10.3390/ijms22189888







