CNGB3 Missense Variant Causes Recessive Achromatopsia in Original Braunvieh Cattle
Abstract
:1. Introduction
2. Results
2.1. Clinical Description
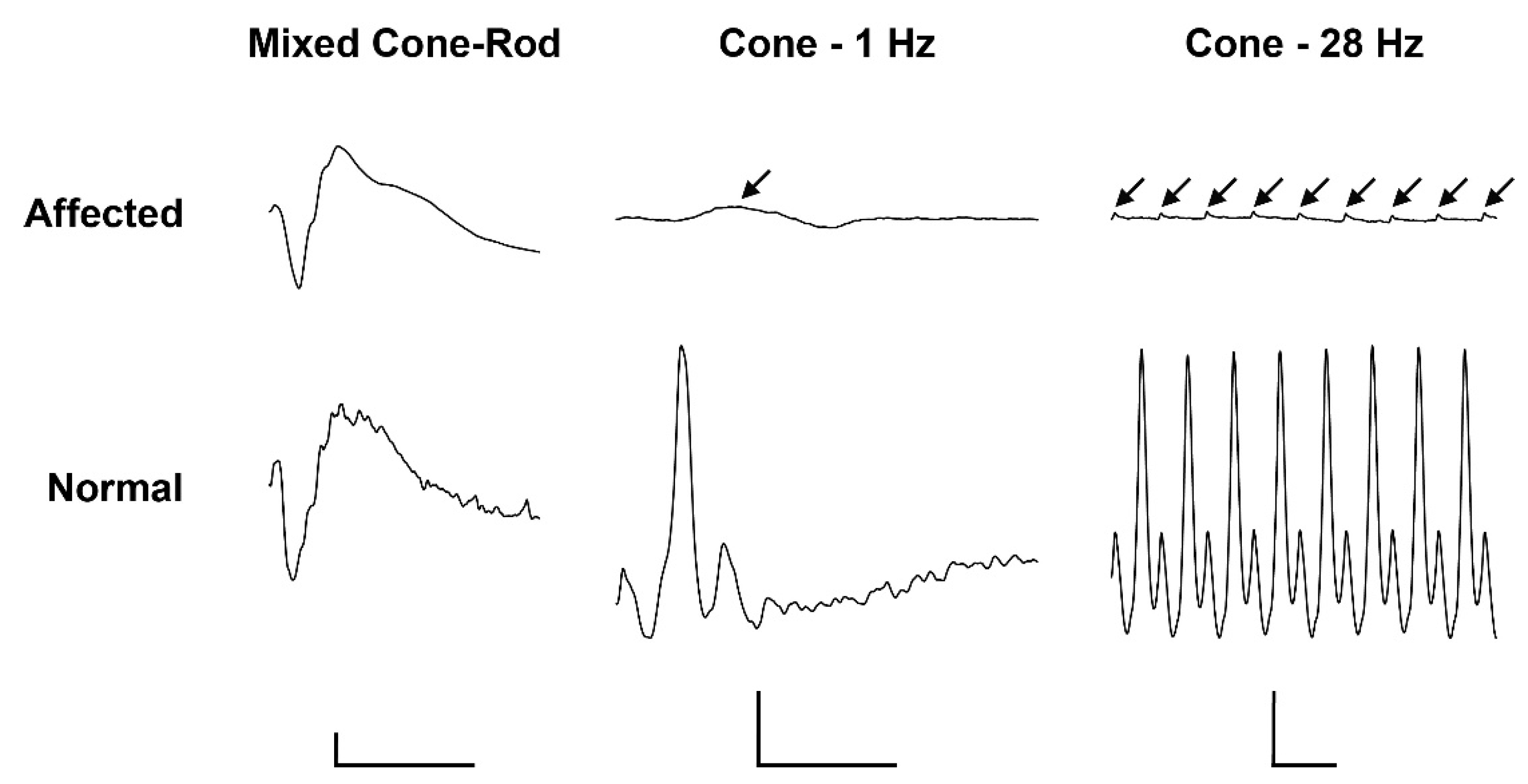
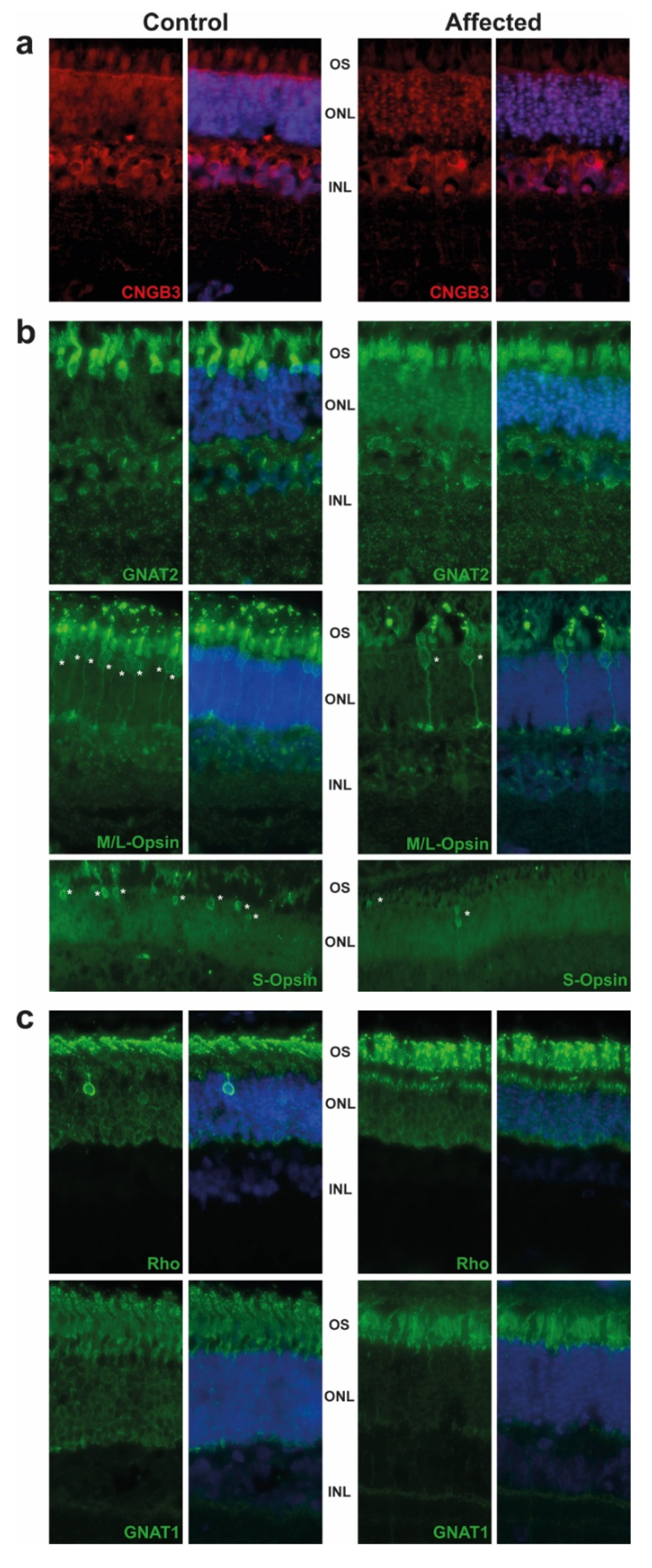
2.2. Pathological Phenotype
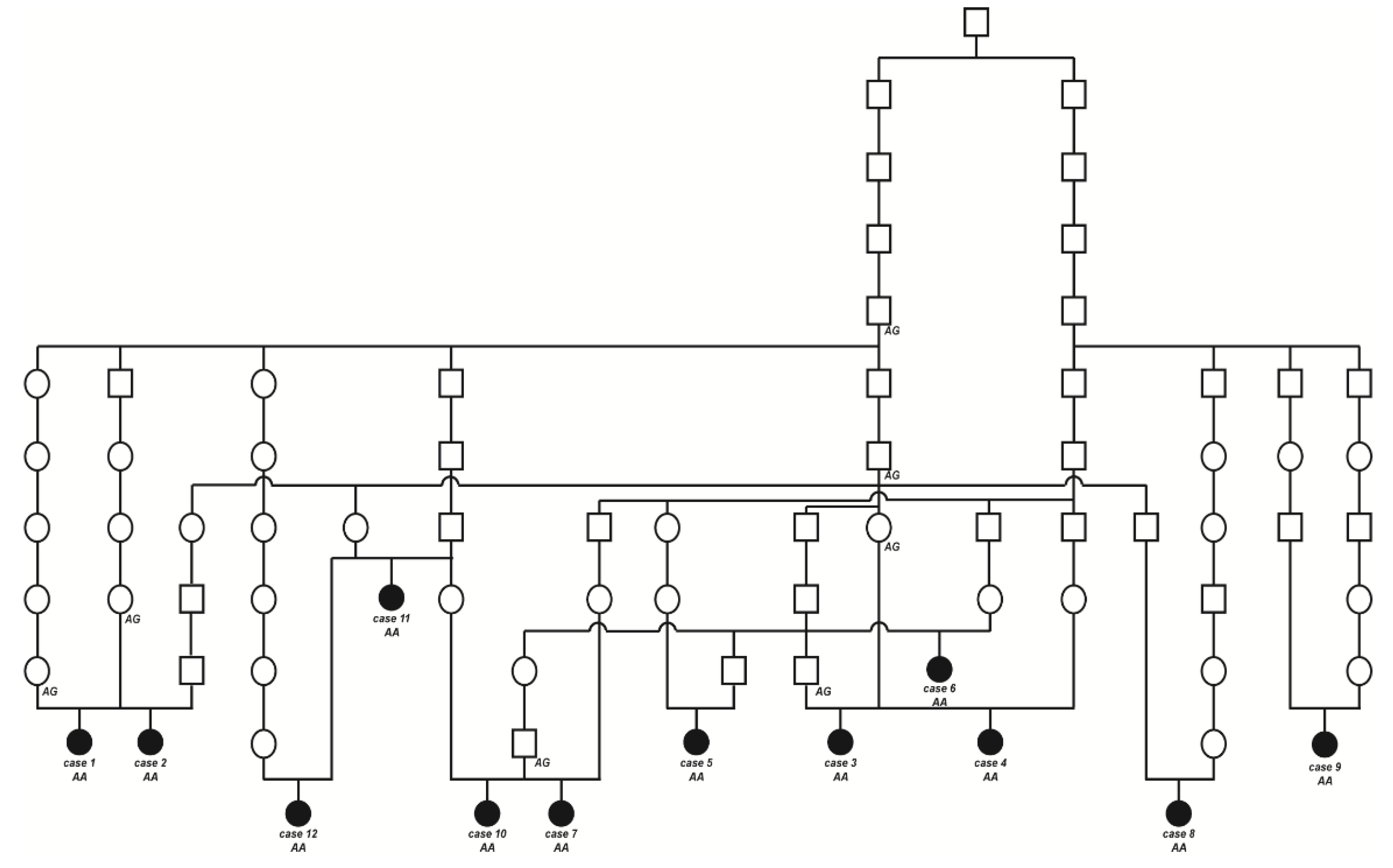
2.3. Pedigree Analysis
2.4. Genetic Analysis
3. Discussion
4. Materials and Methods
4.1. Animal Selection for Genetic Analysis
4.2. Ophthalmological Examination including Electroretinography
4.3. Targeting Cones by Immunofluorescence
4.4. Morphology and Histopathology of the Visual Pathway
4.5. SNP Genotyping and Subsequent Homozygosity Mapping with 12 Affected Cattle
4.6. Whole-Genome Resequencing and Variant Calling
4.7. Genotyping Assays
4.7.1. PCR and Sanger Sequencing
4.7.2. Axiom® Genotyping Array
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lavach, J.D. Large Animal Ophthalmology; Mosby: St. Louis, MO, USA, 1990. [Google Scholar]
- Williams, D.L. Production animal ophthalmology. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, xi–xii. [Google Scholar] [CrossRef]
- Williams, D.L. Welfare issues in farm animal ophthalmology. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Leipold, H.W. Congenital ocular defects in food-producing animals. Vet. Clin. N. Am. Large Anim. Pract. 1984, 6, 577–595. [Google Scholar] [CrossRef]
- Gelatt, K.N.; Ben-Shlomo, G.; Gilger, B.C.; Hendrix, D.V.; Kern, T.J.; Plummer, C.E. Veterinary Ophthalmology, 6th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2021; pp. 1983–2054. [Google Scholar]
- Williams, D.L. Congenital abnormalities in production animals. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 477–486. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Li, Y.; Li, M.; Jia, G.; Dong, H.; Zhang, Y.; He, C.; Wang, C.; Deng, L.; Yang, Y. Hypovitaminosis A coupled to secondary bacterial infection in beef cattle. BMC Vet. Res. 2012, 8, 222. [Google Scholar] [CrossRef] [Green Version]
- Murgiano, L.; Jagannathan, V.; Calderoni, V.; Joechler, M.; Gentile, A.; Drögemüller, C. Looking the cow in the eye: Deletion in the NID1 gene is associated with recessive inherited cataract in Romagnola cattle. PLoS ONE 2014, 9, e110628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollmann, A.K.; Dammann, I.; Wemheuer, W.M.; Wemheuer, W.E.; Chilla, A.; Tipold, A.; Schulz-Schaeffer, W.J.; Beck, J.; Schütz, E.; Brenig, B. Morgagnian cataract resulting from a naturally occurring nonsense mutation elucidates a role of CPAMD8 in mammalian lens development. PLoS ONE 2017, 12, e0180665. [Google Scholar]
- Sun, W.; Li, S.; Xiao, X.; Wang, P.; Zhang, Q. Genotypes and phenotypes of genes associated with achromatopsia: A reference for clinical genetic testing. Mol. Vis. 2020, 26, 588–602. [Google Scholar] [PubMed]
- Reicher, S.; Seroussi, E.; Gootwine, E. A mutation in gene CNGA3 is associated with day blindness in sheep. Genomics 2010, 95, 101–104. [Google Scholar] [CrossRef] [Green Version]
- Ross, M.; Ofri, R.; Aizenberg, I.; Abu-Siam, M.; Pe’er, O.; Arad, D.; Rosov, A.; Gootwine, E.; Dvir, H.; Honig, H.; et al. Naturally-occurring myopia and loss of cone function in a sheep model of achromatopsia. Sci. Rep. 2020, 10, 19314. [Google Scholar] [CrossRef]
- Komáromy, A.M. Day blind sheep and the importance of large animal disease models. Vet. J. 2010, 185, 241–242. [Google Scholar] [CrossRef] [PubMed]
- Winkler, P.A.; Occelli, L.M.; Petersen-Jones, S.M. Large animal models of inherited retinal degenerations: A review. Cells 2020, 9, 882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothammer, S.; Seichter, D.; Förster, M.; Medugorac, I. A genome-wide scan for signatures of differential artificial selection in ten cattle breeds. BMC Genom. 2013, 14, 908. [Google Scholar] [CrossRef] [Green Version]
- Signer-Hasler, H.; Burren, A.; Neuditschko, M.; Frischknecht, M.; Garrick, D.; Stricker, C.; Gredler, B.; Bapst, B.; Flury, C. Population structure and genomic inbreeding in nine Swiss dairy cattle populations. Genet. Sel. Evol. 2017, 49, 83. [Google Scholar] [CrossRef] [Green Version]
- Bhati, M.; Kadri, N.K.; Crysnanto, D.; Pausch, H. Assessing genomic diversity and signatures of selection in Original Braunvieh cattle using whole-genome sequencing data. BMC Genom. 2020, 21, 27. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [Green Version]
- Miyadera, K.; Acland, G.M.; Aguirre, G.D. Genetic and phenotypic variations of inherited retinal diseases in dogs: The power of within- and across-breed studies. Mamm. Genome 2012, 23, 40–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, C.Y.; Goldstein, O.; Kukekova, A.V.; Holley, D.; Knollinger, A.M.; Huson, H.J.; Pearce-Kelling, S.E.; Acland, G.M.; Komáromy, A.M. Genomic deletion of CNGB3 is identical by descent in multiple canine breeds and causes achromatopsia. BMC Genet. 2013, 14, 27. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, N.; Delemotte, L.; Klein, M.L.; Komáromy, A.M.; Tanaka, J.C. A cyclic nucleotide-gated channel mutation associated with canine daylight blindness provides insight into a role for the S2 segment tri-Asp motif in channel biogenesis. PLoS ONE 2014, 9, e88768. [Google Scholar] [CrossRef]
- Tanaka, N.; Dutrow, E.V.; Miyadera, K.; Delemotte, L.; MacDermaid, C.M.; Reinstein, S.L.; Crumley, W.R.; Dixon, C.J.; Casal, M.L.; Klein, M.L.; et al. Canine CNGA3 Gene Mutations Provide Novel Insights into Human Achromatopsia-Associated Channelopathies and Treatment. PLoS ONE 2015, 10, e0138943. [Google Scholar] [CrossRef]
- Sidjanin, D.J.; Lowe, J.K.; McElwee, J.L.; Milne, B.S.; Phippen, T.M.; Sargan, D.R.; Aguirre, G.D.; Acland, G.M.; Ostrander, E.A. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum. Mol. Genet. 2002, 11, 1823–1833. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.T.; Huang, H.; Chen, Y.H.; Dong, L.J.; Li, X.R.; Zhang, X.M. Achromatopsia caused by novel missense mutations in the CNGA3 gene. Int. J. Ophthalmol. 2015, 8, 910–915. [Google Scholar]
- Hagger, C. Estimates of genetic diversity in the brown cattle population of Switzerland obtained from pedigree information. J. Anim. Breed. Genet. 2005, 122, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sargolzaei, M.; Chesnais, J.P.; Schenkel, F.S. FImpute—An efficient imputa- tion algorithm for dairy cattle populations. J. Dairy Sci. 2011, 94, 421. [Google Scholar]
- Häfliger, I.M.; Wiedemar, N.; Švara, T.; Starič, J.; Cociancich, V.; Šest, K.; Gombač, M.; Paller, T.; Agerholm, J.S.; Drögemüller, C. Identification of small and large genomic candidate variants in bovine pulmonary hypoplasia and anasarca syndrome. Anim. Genet. 2020, 51, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Daetwyler, H.D. 1000 Bull Genomes Project to Map Simple and Complex Genetic Traits in Cattle: Applications and Outcomes. Annu. Rev. Anim. Biosci. 2019, 7, 89–102. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| GG | AG | AA | |
|---|---|---|---|
| Achromatopsia-affected calves | 12 | ||
| Obligate carriers a | 5 | ||
| Other Original Braunvieh cattle b,c | 2477 | 463 | 12 |
| Brown Swiss cattle c | 14,976 | 52 | |
| Holstein cattle c | 14,825 | ||
| Simmental cattle c | 2021 | ||
| Sequenced cattle genomes from various breeds (local Swiss cohort) d | 552 | 15 | |
| Control cattle from various breeds (1000 Bull Genomes project) e | 3298 | 7 f | 1 g |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Häfliger, I.M.; Marchionatti, E.; Stengård, M.; Wolf-Hofstetter, S.; Paris, J.M.; Jacinto, J.G.P.; Watté, C.; Voelter, K.; Occelli, L.M.; Komáromy, A.M.; et al. CNGB3 Missense Variant Causes Recessive Achromatopsia in Original Braunvieh Cattle. Int. J. Mol. Sci. 2021, 22, 12440. https://doi.org/10.3390/ijms222212440
Häfliger IM, Marchionatti E, Stengård M, Wolf-Hofstetter S, Paris JM, Jacinto JGP, Watté C, Voelter K, Occelli LM, Komáromy AM, et al. CNGB3 Missense Variant Causes Recessive Achromatopsia in Original Braunvieh Cattle. International Journal of Molecular Sciences. 2021; 22(22):12440. https://doi.org/10.3390/ijms222212440
Chicago/Turabian StyleHäfliger, Irene M., Emma Marchionatti, Michele Stengård, Sonja Wolf-Hofstetter, Julia M. Paris, Joana G. P. Jacinto, Christine Watté, Katrin Voelter, Laurence M. Occelli, András M. Komáromy, and et al. 2021. "CNGB3 Missense Variant Causes Recessive Achromatopsia in Original Braunvieh Cattle" International Journal of Molecular Sciences 22, no. 22: 12440. https://doi.org/10.3390/ijms222212440
APA StyleHäfliger, I. M., Marchionatti, E., Stengård, M., Wolf-Hofstetter, S., Paris, J. M., Jacinto, J. G. P., Watté, C., Voelter, K., Occelli, L. M., Komáromy, A. M., Oevermann, A., Goepfert, C., Borgo, A., Roduit, R., Spengeler, M., Seefried, F. R., & Drögemüller, C. (2021). CNGB3 Missense Variant Causes Recessive Achromatopsia in Original Braunvieh Cattle. International Journal of Molecular Sciences, 22(22), 12440. https://doi.org/10.3390/ijms222212440








