Integrated Multi-Omics Investigations of Metalloproteinases in Colon Cancer: Focus on MMP2 and MMP9
Abstract
1. Introduction
2. Results
- (1)
- Collagenases: MMP1, MMP8, and MMP13;
- (2)
- Gelatinases: MMP2 and MMP9;
- (3)
- Stromelysins: MMP3, MMP10, and MMP11;
- (4)
- Matrilysins: MMP7 and MMP26;
- (5)
- Membrane-bound/associated MMPs: MMP14 to MMP17, MMP24, and MMP25;
- (6)
- Others MMPs: MMP12, MMP19 to MMP23, MMP27, and MMP28.
2.1. Differential Expression of MMP Members between Normal and Cancer Tissues
2.2. Genetic and Epigenetic Alterations of MMP Members
2.3. Prognostic Value of MMP Expression in CRC
2.4. Correlation between MMPs and Immune Infiltration Levels in CRC
2.5. Correlation Analysis between MMP2 and MMP9 Expression and Immune Marker Sets
2.6. MMP2 and MMP9-Gene Co-Expression Networks
2.7. Activity Levels of Gelatinases MMP9 and MMP2 in Colon Cancer Tissues and Corresponding Sera Samples
3. Discussion
4. Materials and Methods
4.1. Expression Analysis of MMP Members Using Oncomine, UALCAN, and Colonomics
4.1.1. Oncomine Database
4.1.2. UALCAN Database
4.1.3. Colonomics Database
4.2. Genetic Alterations and Epigenetic Regulation of MMP Members Using cBioPortal and UALCAN
4.3. Evaluation of the Prognostic Value of MMPs Using GEPIA, UALCAN, Colonomics, and UCSC Xena
4.4. Association between MMP Expression and Immune Infiltration by TIMER
4.5. MMP2 and MMP9 Co-Expressed Genes and Pathways Enrichment Analysis
4.6. Tissue and Sera Samples from CRC Patients
4.7. Gelatin Zymography
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Piperigkou, Z.; Kyriakopoulou, K.; Koutsakis, C.; Mastronikolis, S.; Karamanos, N.K. Key Matrix Remodeling Enzymes: Functions and Targeting in Cancer. Cancers 2021, 13, 1441. [Google Scholar] [CrossRef]
- Carey, P.; Low, E.; Harper, E.; Stack, M. Metalloproteinases in Ovarian Cancer. Int. J. Mol. Sci. 2021, 22, 3403. [Google Scholar] [CrossRef]
- Raeeszadeh-Sarmazdeh, M.; Do, L.D.; Hritz, B.G. Metalloproteinases and Their Inhibitors: Potential for the Development of New Therapeutics. Cells 2020, 9, 1313. [Google Scholar] [CrossRef]
- Sohail, A.; Sun, Q.; Zhao, H.; Bernardo, M.M.; Cho, J.-A.; Fridman, R. MT4-(MMP17) and MT6-MMP (MMP25), A unique set of membrane-anchored matrix metalloproteinases: Properties and expression in cancer. Cancer Metastasis Rev. 2008, 27, 289–302. [Google Scholar] [CrossRef]
- Zucker, S.; Pei, D.; Cao, J.; Lopez-Otin, C. Membrane type-matrix metalloproteinases (MT-MMP). Curr. Top. Dev. Biol. 2003, 54, 1–74. [Google Scholar] [CrossRef]
- Asano, T.; Tada, M.; Cheng, S.; Takemoto, N.; Kuramae, T.; Abe, M.; Takahashi, O.; Miyamoto, M.; Hamada, J.-I.; Moriuchi, T.; et al. Prognostic Values of Matrix Metalloproteinase Family Expression in Human Colorectal Carcinoma. J. Surg. Res. 2008, 146, 32–42. [Google Scholar] [CrossRef]
- Benbow, U.; Brinckerhoff, C.E. The AP-1 site and MMP gene regulation: What is all the fuss about? Matrix Biol. 1997, 15, 519–526. [Google Scholar] [CrossRef]
- Nakamura, Y.; Tanaka, F.; Yoshikawa, Y.; Mimori, K.; Inoue, H.; Yanaga, K.; Mori, M. PDGF-BB is a Novel Prognostic Factor in Colorectal Cancer. Ann. Surg. Oncol. 2008, 15, 2129–2136. [Google Scholar] [CrossRef]
- Stanilov, N.; Miteva, L.; Dobreva, Z.; Stanilova, S. Colorectal cancer severity and survival in correlation with tumour necrosis factor-alpha. Biotechnol. Biotechnol. Equip. 2014, 28, 911–917. [Google Scholar] [CrossRef]
- Li, J.; Huang, L.; Zhao, H.; Yan, Y.; Lu, J. The Role of Interleukins in Colorectal Cancer. Int. J. Biol. Sci. 2020, 16, 2323–2339. [Google Scholar] [CrossRef]
- Ohchi, T.; Akagi, Y.; Kinugasa, T.; Kakuma, T.; Kawahara, A.; Sasatomi, T.; Gotanda, Y.; Yamaguchi, K.; Tanaka, N.; Ishibashi, Y.; et al. Amphiregulin is a prognostic factor in colorectal cancer. Anticancer Res. 2012, 32, 2315–2321. [Google Scholar]
- Martins, S.F.; A Garcia, E.; Luz, M.A.M.; Pardal, F.; Rodrigues, M.; Filho, A.L. Clinicopathological correlation and prognostic significance of VEGF-A, VEGF-C, VEGFR-2 and VEGFR-3 expression in colorectal cancer. Cancer Genom. Proteom. 2013, 10, 55–67. [Google Scholar]
- Caiado, H.; Conceição, N.; Tiago, D.; Marreiros, A.; Vicente, S.; Enriquez, J.L.; Vaz, A.M.; Antunes, A.; Guerreiro, H.; Caldeira, P.; et al. Data on the evaluation of FGF2 gene expression in Colorectal Cancer. Data Brief 2020, 31, 105765. [Google Scholar] [CrossRef]
- Galzie, Z.; Fernig, D.G.; Smith, J.A.; Poston, G.J.; Kinsella, A.R. Basic fibroblast growth factor and colorectal carcinoma invasion. Biochem. Soc. Trans. 1996, 24, 501S. [Google Scholar] [CrossRef]
- Chen, X.-L.; Chen, Z.-Q.; Zhu, S.-L.; Liu, T.-W.; Wen, Y.; Su, Y.-S.; Xi, X.-J.; Hu, Y.; Lian, L.; Liu, F.-B. Prognostic value of transforming growth factor-beta in patients with colorectal cancer who undergo surgery: A meta-analysis. BMC Cancer 2017, 17, 240. [Google Scholar] [CrossRef]
- Sizemore, S.T.; Sizemore, G.M.; Booth, C.N.; Thompson, C.L.; Silverman, P.; Bebek, G.; Abdul-Karim, F.W.; Avril, S.; Keri, R.A. Hypomethylation of the MMP7 promoter and increased expression of MMP7 distinguishes the basal-like breast cancer subtype from other triple-negative tumors. Breast Cancer Res. Treat. 2014, 146, 25–40. [Google Scholar] [CrossRef]
- Chicoine, É.; Estève, P.-O.; Robledo, O.; Van Themsche, C.; Potworowski, E.F.; St-Pierre, Y. Evidence for the role of promoter methylation in the regulation of MMP-9 gene expression. Biochem. Biophys. Res. Commun. 2002, 297, 765–772. [Google Scholar] [CrossRef]
- Yan, C.; Boyd, D.D. Regulation of matrix metalloproteinase gene expression. J. Cell. Physiol. 2007, 211, 19–26. [Google Scholar] [CrossRef]
- Jin, M.-Z.; Jin, W.-L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 2020, 5, 166. [Google Scholar] [CrossRef]
- Caon, I.; D’Angelo, M.; Bartolini, B.; Caravà, E.; Parnigoni, A.; Contino, F.; Cancemi, P.; Moretto, P.; Karamanos, N.; Passi, A.; et al. The Secreted Protein C10orf118 Is a New Regulator of Hyaluronan Synthesis Involved in Tumour-Stroma Cross-Talk. Cancers 2021, 13, 1105. [Google Scholar] [CrossRef]
- Klemm, F.; Joyce, J.A. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol. 2015, 25, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.R.; Fingleton, B.; Rothenberg, M.L.; Matrisian, L.M. Matrix Metalloproteinases: Biologic Activity and Clinical Implications. J. Clin. Oncol. 2000, 18, 1135. [Google Scholar] [CrossRef] [PubMed]
- Juurikka, K.; Butler, G.S.; Salo, T.; Nyberg, P.; Åström, P. The Role of MMP8 in Cancer: A Systematic Review. Int. J. Mol. Sci. 2019, 20, 4506. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Fabián, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argáez, V.; Lara-Riegos, J.; Ramírez-Camacho, M.A.; Alvarez-Sánchez, M.E. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front. Oncol. 2019, 9, 1370. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; Mu, L.; Huang, K.; Hu, Y.; Yan, C.; Zhao, H.; Ma, C.; Li, X.; Tao, D.; Qin, J. Hypoxic colorectal cancer cells promote metastasis of normoxic cancer cells depending on IL-8/p65 signaling pathway. Cell Death Dis. 2020, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhou, R.; Yang, T.; Liu, S.; Cui, Z.; Qiao, Q.; Zhang, J. Hypoxia promotes colorectal cancer cell migration and invasion in a SIRT1-dependent manner. Cancer Cell Int. 2019, 19, 116. [Google Scholar] [CrossRef] [PubMed]
- Collins, H.M.; Morris, T.M.; A Watson, S. Spectrum of matrix metalloproteinase expression in primary and metastatic colon cancer: Relationship to the tissue inhibitors of metalloproteinases and membrane type-1-matrix metalloproteinase. Br. J. Cancer 2001, 84, 1664–1670. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Damodharan, U.; Ganesan, R.; Radhakrishnan, U.C. Expression of MMP2 and MMP9 (Gelatinases A and B) in Human Colon Cancer Cells. Appl. Biochem. Biotechnol. 2011, 165, 1245–1252. [Google Scholar] [CrossRef]
- Langers, A.M.J.; Verspaget, H.W.; Hawinkels, L.J.A.C.; Kubben, F.J.G.M.; Van Duijn, W.; Van Der Reijden, J.J.; Hardwick, J.C.H.; Hommes, D.W.; Sier, C.F.M. MMP-2 and MMP-9 in normal mucosa are independently associated with outcome of colorectal cancer patients. Br. J. Cancer 2012, 106, 1495–1498. [Google Scholar] [CrossRef]
- Salem, N.; Kamal, I.; Al-Maghrabi, J.; Abuzenadah, A.; Peer-Zada, A.A.; Qari, Y.; Al-Ahwal, M.; Al-Qahtani, M.; Buhmeida, A. High expression of matrix metalloproteinases: MMP-2 and MMP-9 predicts poor survival outcome in colorectal carcinoma. Future Oncol. 2016, 12, 323–331. [Google Scholar] [CrossRef]
- Murnane, M.J.; Cai, J.; Shuja, S.; McAneny, D.; Klepeis, V.; Willett, J.B. Active MMP-2 effectively identifies the presence of colorectal cancer. Int. J. Cancer 2009, 125, 2893–2902. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pander, A.; Chinnaiyan, A.M. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef]
- Buttacavoli, M.; Di Cara, G.; D’Amico, C.; Geraci, F.; Pucci-Minafra, I.; Feo, S.; Cancemi, P. Prognostic and Functional Significant of Heat Shock Proteins (HSPs) in Breast Cancer Unveiled by Multi-Omics Approaches. Biology 2021, 10, 247. [Google Scholar] [CrossRef] [PubMed]
- Cancemi, P.; Buttacavoli, M.; DI Cara, G.; Albanese, N.N.; Bivona, S.; Pucci-Minafra, I.; Feo, S. A multiomics analysis of S100 protein family in breast cancer. Oncotarget 2018, 9, 29064–29081. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Sanz-Pamplona, R.; Berenguer, A.; Cordero, D.; Molleví, D.G.; Crous-Bou, M.; Sole, X.; Paré-Brunet, L.; Guino, E.; Salazar, R.; Santos, C.; et al. Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer. Mol. Cancer 2014, 13, 46. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Chisanga, D.; Alessandro, R.; Ang, C.-S.; Askenase, P.; Batagov, A.O.; Martin, A.B.; Camussi, G.; Clayton, A.; et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles 2017, 6, 1321455. [Google Scholar] [CrossRef] [PubMed]
- Buttacavoli, M.; Albanese, N.N.; Roz, E.; Pucci-Minafra, I.; Feo, S.; Cancemi, P. Proteomic Profiling of Colon Cancer Tissues: Discovery of New Candidate Biomarkers. Int. J. Mol. Sci. 2020, 21, 3096. [Google Scholar] [CrossRef] [PubMed]
- Cancemi, P.; Aiello, A.; Accardi, G.; Caldarella, R.; Candore, G.; Caruso, C.; Ciaccio, M.; Cristaldi, L.; Di Gaudio, F.; Siino, V.; et al. The Role of Matrix Metalloproteinases (MMP-2 and MMP-9) in Ageing and Longevity: Focus on Sicilian Long-Living Individuals (LLIs). Mediat. Inflamm. 2020, 2020, 8635158. [Google Scholar] [CrossRef]
- Cancemi, P.; Buttacavoli, M.; Roz, E.; Feo, S. Expression of Alpha-Enolase (ENO1), Myc Promoter-Binding Protein-1 (MBP-1) and Matrix Metalloproteinases (MMP-2 and MMP-9) Reflect the Nature and Aggressiveness of Breast Tumors. Int. J. Mol. Sci. 2019, 20, 3952. [Google Scholar] [CrossRef]
- Coppola, A.; Cancemi, P.; Tomasello, L.; Guarnotta, V.; Pitrone, M.; Failla, V.; Cillino, S.; Feo, S.; Pizzolanti, G.; Giordano, C. Anti-Inflammatory Action of Heterogeneous Nuclear Ribonucleoprotein A2/B1 in Patients with Autoimmune Endocrine Disorders. J. Clin. Med. 2019, 9, 9. [Google Scholar] [CrossRef]
- DI Cara, G.; Marengo, G.; Albanese, N.N.; Marabeti, M.R.; Musso, R.; Cancemi, P.; Pucci-Minafra, I. Proteomic profiling of Trastuzumab (Herceptin(R))-sensitive and -resistant SKBR-3 breast cancer cells. Anticancer Res. 2013, 33, 489–503. [Google Scholar]
- Pucci-Minafra, I.; Albanese, N.N.; Di Cara, G.; Minafra, L.; Marabeti, M.R.; Cancemi, P. Breast Cancer Cells Exhibit Selective Modulation Induced by Different Collagen Substrates. Connect. Tissue Res. 2008, 49, 252–256. [Google Scholar] [CrossRef]
- Raimondi, M.V.; Presentato, A.; Petri, G.L.; Buttacavoli, M.; Ribaudo, A.; De Caro, V.; Alduina, R.; Cancemi, P. New Synthetic Nitro-Pyrrolomycins as Promising Antibacterial and Anticancer Agents. Antibiotics 2020, 9, 292. [Google Scholar] [CrossRef]
- Di Cara, G.; Marabeti, M.R.; Musso, R.; Riili, I.; Cancemi, P.; Minafra, I.P. New Insights into the Occurrence of Matrix Metalloproteases -2 and -9 in a Cohort of Breast Cancer Patients and Proteomic Correlations. Cells 2018, 7, 89. [Google Scholar] [CrossRef]
- Cancemi, P.; Di Falco, F.; Feo, S.; Arizza, V.; Vizzini, A. The gelatinase MMP-9like is involved in regulation of LPS inflammatory response in Ciona robusta. Fish Shellfish Immunol. 2018, 86, 213–222. [Google Scholar] [CrossRef]
- Cancemi, P.; Albanese, N.N.; DI Cara, G.; Marabeti, M.R.; Costantini, F.; Minafra, S.; Pucci-Minafra, I. Multiple Changes Induced by Fibroblasts on Breast Cancer Cells. Connect. Tissue Res. 2009, 51, 88–104. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Coppola, A.; Tomasello, L.; Pizzolanti, G.; Pucci-Minafra, I.; Albanese, N.; DI Cara, G.; Cancemi, P.; Pitrone, M.; Bommarito, A.; Carissimi, E.; et al. In Vitro Phenotypic, Genomic and Proteomic Characterization of a Cytokine-Resistant Murine β-TC3 Cell Line. PLoS ONE 2012, 7, e32109. [Google Scholar] [CrossRef] [PubMed]
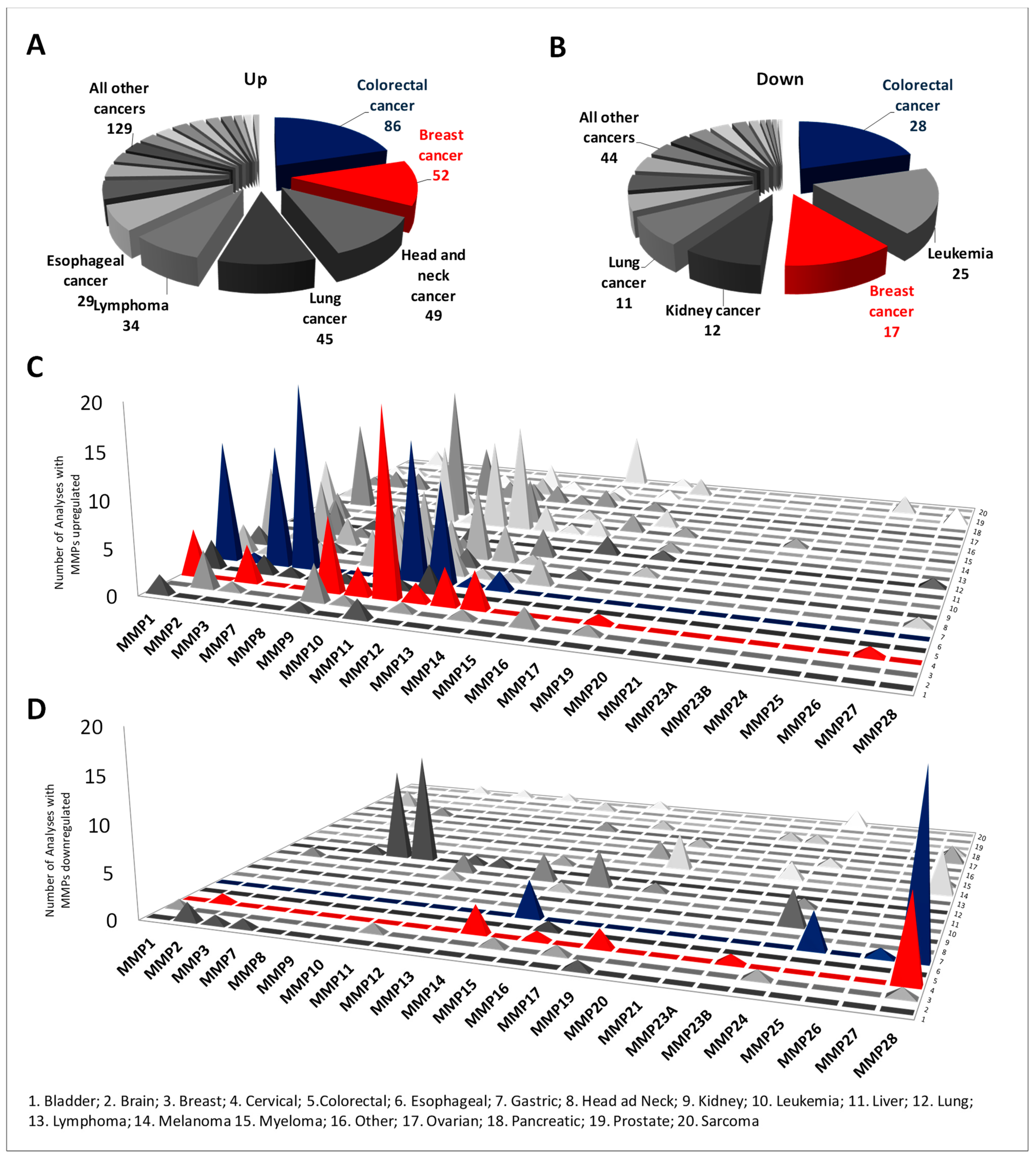

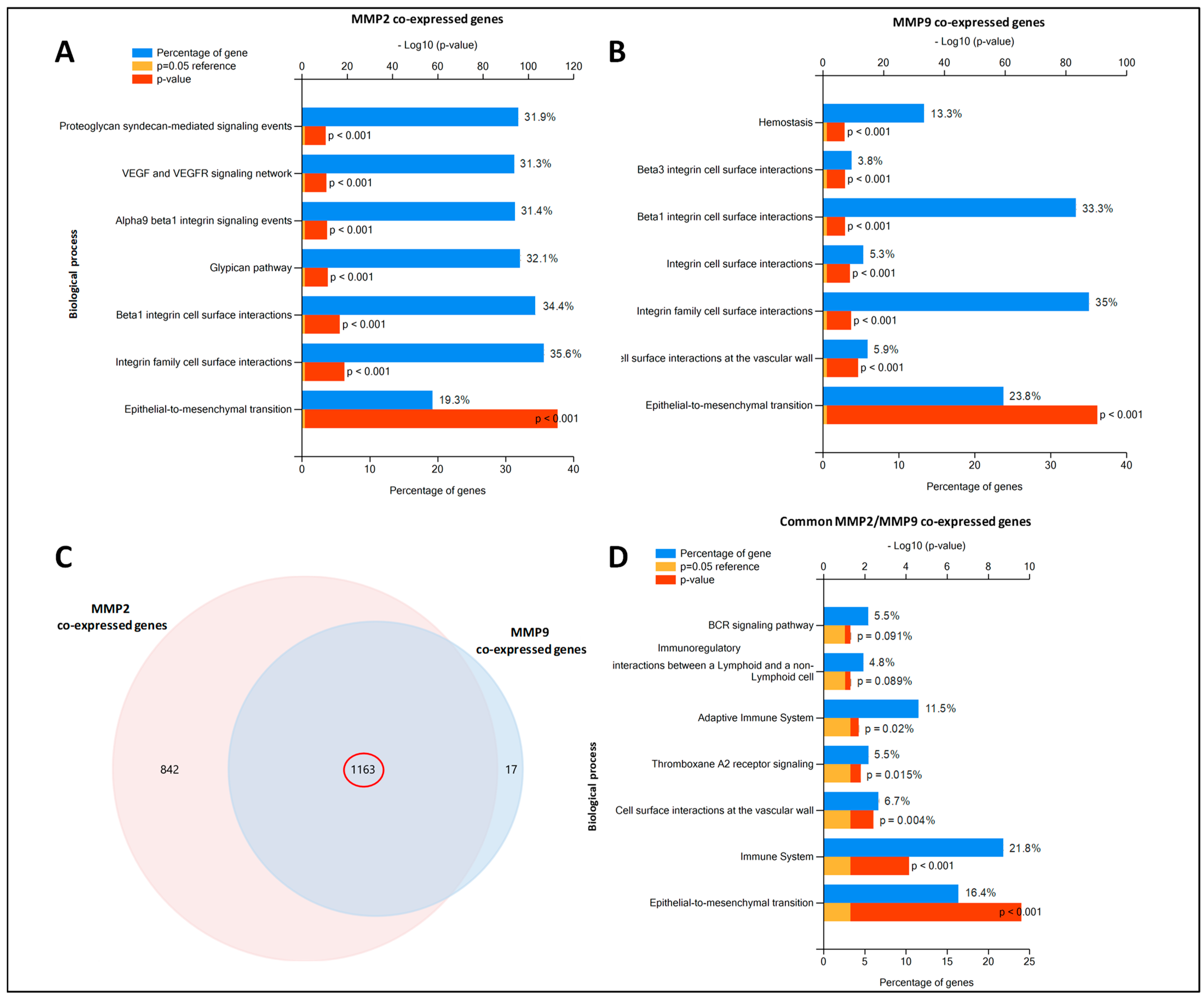
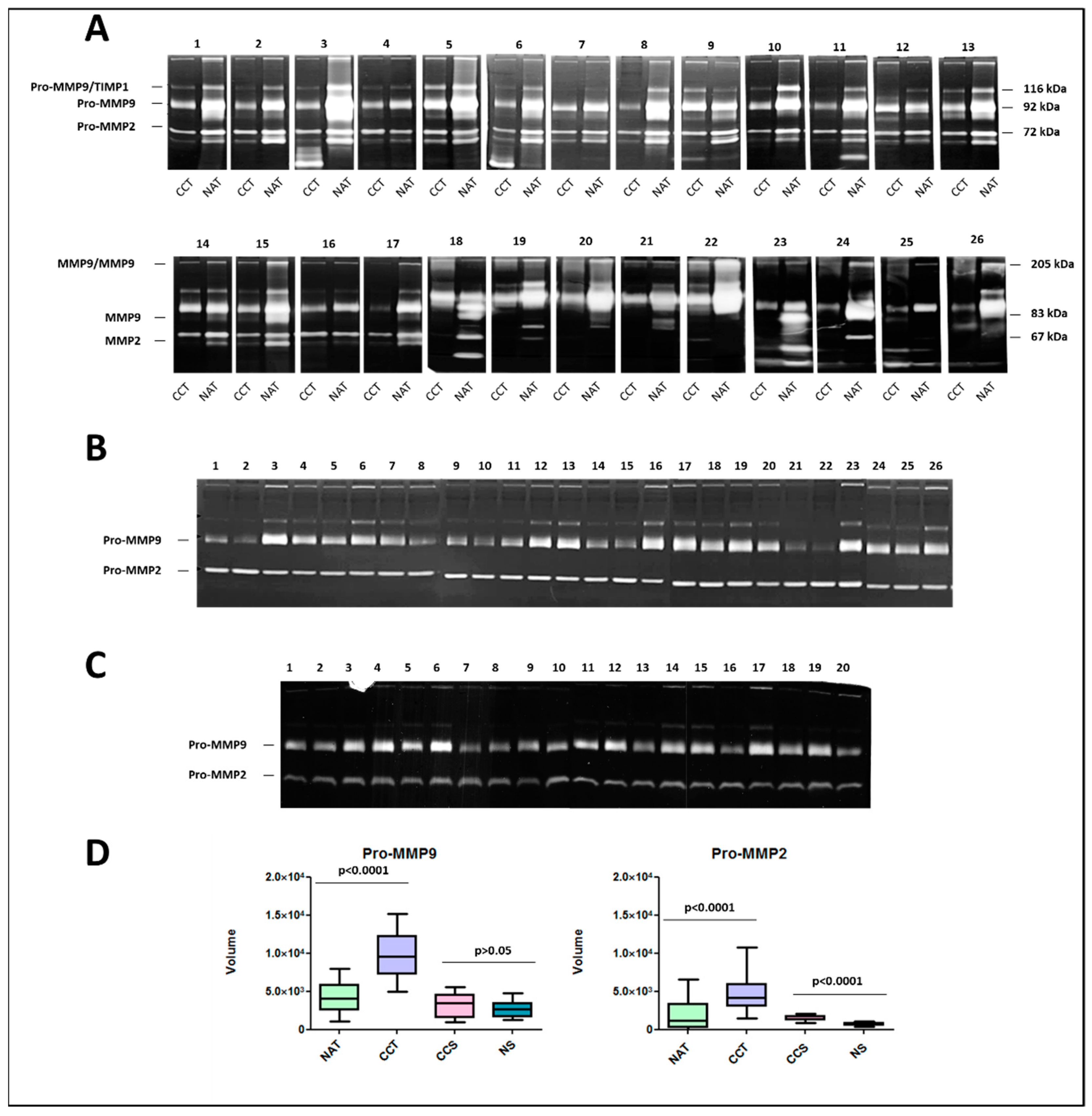
| Gene Name | Chromosomic Localization | Protein Name | Uniprot Access Number | MW (Da)/pI |
|---|---|---|---|---|
| Collagenases | ||||
| MMP1 | 11q22.3 | Interstitial collagenases | P03956 | 54,061/6.97 |
| MMP8 | 11q22.3 | Neutrophil collagenase | P22894 | 53,411/6.86 |
| MMP13 | 11q22.3 | Collagenase-3 | P45452 | 53,819/5.31 |
| Gelatinases | ||||
| MMP2 | 16q13 | 72 kDa type IV collagenase | P08253 | 73,881/5.09 |
| MMP9 | 20q11.2 | Matrix metalloproteinase-9 | P14780 | 78,457/5.91 |
| Stromelysins | ||||
| MMP3 | 11q22.3 | Stromelysin-1 | P08254 | 53,976/6.07 |
| MMP10 | 11q22.3 | Stromelysin-2 | P09238 | 54,150/5.59 |
| MMP11 | 22q11.2 | Stromelysin-3 | P24347 | 54,589/6.87 |
| Matrilysins | ||||
| MMP7 | 11q21 | Matrilysin | P09237 | 29,676/8.09 |
| MMP26 | 11p15 | Matrix metalloproteinase-26 | Q9NRE1 | 29,708/6.45 |
| Membrane-bound/associated MMPs | ||||
| MMP14 | 14q11 | Matrix metalloproteinase-14 | P50281 | 65,893/7.87 |
| MMP15 | 16q13 | Matrix metalloproteinase-15 | P51511 | 75,806/7.49 |
| MMP16 | 8q21 | Matrix metalloproteinase-16 | P51512 | 69,521/8.74 |
| MMP17 | 12q24.3 | Matrix metalloproteinase-17 | Q9ULZ9 | 66,652/6.55 |
| MMP24 | 20q11.2 | Matrix metalloproteinase-24 | Q9Y5R2 | 73,230/9.58 |
| MMP25 | 16p13.3 | Matrix metalloproteinase-25 | Q9NPA2 | 62,553/8.76 |
| Others MMPs | ||||
| MMP12 | 11q22.3 | Macrophage metalloelastase | P39900 | 54,001/8.89 |
| MMP19 | 12q14 | Matrix metalloproteinase-19 | Q99542 | 57,356/7.67 |
| MMP20 | 11q22.3 | Matrix metalloproteinase-20 | O60882 | 54,386/9.08 |
| MMP21 | 10q26.13 | Matrix metalloproteinase-21 | Q8N119 | 65,042/9.42 |
| MMP23A | 1p36.33 | Matrix metalloproteinase-23A | O75900 | 43,934/9.94 |
| MMP23B | 1p36.34 | Matrix metalloproteinase-23B | O75901 | 43,934/10.27 |
| MMP27 | 11q24 | Matrix metalloproteinase-27 | Q9H306 | 59,025/8.95 |
| MMP28 | 17q11 | Matrix metalloproteinase-28 | Q9H239 | 58,938/10.07 |
| UALCAN (T = 286; n = 41) | Colonomics (T = 98; n = 98) | |
|---|---|---|
| MMP1 | p = 1.6 × 10−12 | p < 2 × 10−16 |
| MMP2 | p = 2.1 × 10−3 | - |
| MMP3 | p < 1 × 10−12 | p < 2 × 10−16 |
| MMP7 | p = 1.6 × 10−12 | p < 2 × 10−16 |
| MMP8 | - | p = 7.5 × 10−4 |
| MMP9 | p = 1.4 × 10−10 | p = 3.9 × 10−16 |
| MMP10 | p = 1.8 × 10−6 | p < 2 × 10−16 |
| MMP11 | p = 1.6 × 10−12 | p < 2 × 10−16 |
| MMP12 | p = 2.22 × 10−9 | p < 2 × 10−16 |
| MMP13 | p = 6.2 × 10−9 | p = 8.9 × 10−8 |
| MMP14 | p = 1.62 × 10−12 | p < 2 × 10−16 |
| MMP15 | p = 6 × 10−16 | p < 2 × 10−16 |
| MMP16 | - | - |
| MMP17 | p = 1.9 × 10−5 | p = 3.1 × 10−3 |
| MMP19 | - | - |
| MMP20 | p = 3.6 × 10−5 | - |
| MMP21 | - | - |
| MMP23A | - | - |
| MMP23B | p = 1.2 × 10−3 | - |
| MMP24 | - | - |
| MMP25 | p = 7.6 × 10−7 | p = 1.9 × 10−6 |
| MMP26 | - | - |
| MMP27 | p = 4.7 × 10−12 | p < 2 × 10−16 |
| MMP28 | p < 1 × 10−12 | p < 2 × 10−16 |
| GEPIA (COAD) | GEPIA (READ) | Colonomics (COAD) | UALCAN (COAD) | UCSC Xena COAD COADREAD READ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DFS | OS | DFS | OS | DFS | OS | OS | GDC TCGA OS | TCGA OS | TCGA OS | TCGA OS | |
| MMP1 | - | - | - | - | - | - | - | p = 0.019 | - | - | - |
| MMP2 | p = 0.017 | - | - | - | - | - | - | - | - | - | - |
| MMP3 | - | - | - | - | - | p = 0.046 | - | p = 0.004 | - | - | - |
| MMP7 | - | - | - | - | - | - | - | - | - | - | - |
| MMP8 | p = 0.043 | - | - | - | - | - | - | - | - | - | - |
| MMP9 | - | - | - | - | - | - | - | - | - | - | - |
| MMP10 | - | - | - | - | - | - | - | p = 0.0088 | - | - | - |
| MMP11 | - | - | - | - | - | - | - | - | - | - | - |
| MMP12 | - | - | - | - | - | p = 0.0087 | - | p = 0.027 | - | p = 0.037 | - |
| MMP13 | - | - | - | - | - | - | - | - | - | - | - |
| MMP14 | p = 0.012 | p = 0.013 | p = 0.0037 | - | - | - | - | - | - | p = 0.046 | - |
| MMP15 | - | - | - | - | - | p = 0.014 | p = 0.02 | - | - | - | - |
| MMP16 | - | - | - | - | - | - | - | - | p = 0.016 | p = 0.017 | - |
| MMP17 | - | - | - | - | - | - | - | - | p = 0.011 | - | - |
| MMP19 | - | - | - | - | - | - | - | - | p = 0.002 | p = 0.00033 | - |
| MMP20 | - | - | - | - | - | - | - | - | - | - | - |
| MMP21 | - | - | - | - | - | - | - | - | - | - | - |
| MMP23A | - | - | - | - | - | - | - | - | - | - | - |
| MMP23B | - | p = 0.008 | - | - | - | - | p = 0.0055 | - | - | p = 0.014 | - |
| MMP24 | - | - | - | - | - | - | - | - | - | - | - |
| MMP25 | - | - | - | p = 0.029 | - | - | - | - | - | - | - |
| MMP26 | - | - | - | - | - | - | - | - | - | - | - |
| MMP27 | - | - | - | - | - | - | - | - | - | - | - |
| MMP28 | - | - | - | - | - | - | - | - | - | - | - |
| Purity | B Cell | CD8+ T Cell | CD4+ T Cell | Macrophage | Neutrophil | Dendritic Cell | |
|---|---|---|---|---|---|---|---|
| MMP1 | - | + | + | +++ | ++ | ||
| MMP2 | -- | + | ++ | +++ | +++ | +++ | |
| MMP3 | - | - | + | ++ | + | ||
| MMP7 | - | + | + | ||||
| MMP8 | - | - | + | ++ | ++ | ++ | |
| MMP9 | - | + | ++ | ++ | +++ | +++ | |
| MMP10 | - | + | + | ||||
| MMP11 | - | - | ++ | ++ | + | ||
| MMP12 | - | + | ++ | + | ++ | +++ | +++ |
| MMP13 | - | + | + | ++ | ++ | ++ | |
| MMP14 | -- | + | +++ | +++ | +++ | +++ | |
| MMP15 | - | + | - | ||||
| MMP16 | - | + | ++ | +++ | ++ | ++ | |
| MMP17 | - | ++ | + | + | + | ||
| MMP19 | -- | + | + | +++ | ++ | ++ | ++ |
| MMP20 | |||||||
| MMP21 | - | + | + | + | + | + | |
| MMP23A | - | ||||||
| MMP23B | - | ++ | + | + | + | ||
| MMP24 | - | + | - | - | |||
| MMP25 | -- | + | + | ++ | + | +++ | +++ |
| MMP26 | - | - | - | ||||
| MMP27 | - | + | + | + | + | + | |
| MMP28 | - | + | + | + | + | + |
| MMP2 | MMP9 | ||||
|---|---|---|---|---|---|
| Markers | None | Purity | None | Purity | |
| r | r | r | r | ||
| CD8+ T cell | CD8A | (++) | (--) | (++) | (--) |
| CD8B | (+) | (-) | (+) | (-) | |
| CD3D | (++) | (--) | (++) | (--) | |
| T cell (general) | CD3E | (++) | (--) | (++) | (--) |
| CD2 | (++) | (--) | (++) | (--) | |
| CD19 | (++) | (--) | (+) | (--) | |
| B cell | CD79A | (++) | (--) | (++) | (--) |
| CD86 | (+++) | (--) | (+++) | (--) | |
| Monocyte | CSF1R | (+++) | (--) | (+++) | (--) |
| CCL2 | (+++) | (--) | (+++) | (--) | |
| TAM | CD68 | (+++) | (--) | (+++) | (--) |
| IL10 | (+++) | (--) | (++) | (--) | |
| NOS2 | (-) | (-) | |||
| M1 macrophage | IRF5 | (++) | (++) | ||
| PTGS2 | (++) | (-) | (+) | (-) | |
| CD163 | (+++) | (--) | (+++) | (--) | |
| M2 macrophage | VSIG4 | (+++) | (--) | (+++) | (--) |
| MS4A4A | (+++) | (--) | (+++) | (--) | |
| CEACAM8 | (-) | (-) | (-) | ||
| Neutrophils | ITGAM | (+++) | (--) | (+++) | (--) |
| CCR7 | (++) | (--) | (++) | (--) | |
| KIR2DL1 | (+) | (-) | (+) | (-) | |
| KIR2DL3 | (+) | (-) | (+) | (-) | |
| KIR2DL4 | (+) | (--) | (+) | (--) | |
| Natural killer cells | KIR3DL1 | (+) | (-) | (+) | (-) |
| KIR3DL2 | (+) | (-) | (+) | (-) | |
| KIR3DL3 | (+) | ||||
| KIR2DS4 | (+) | (-) | (+) | (-) | |
| HLA-DPB1 | (+++) | (--) | (+++) | (--) | |
| HLA-DQB1 | (++) | (--) | (++) | (--) | |
| HLA-DRA | (+++) | (--) | (++) | (--) | |
| Dendritic cell | HLA-DPA1 | (+++) | (--) | (+++) | (--) |
| CD1C | (++) | (--) | (++) | (--) | |
| NRP1 | (+++) | (--) | (+++) | (--) | |
| ITGAX | (+++) | (--) | (+++) | (--) | |
| TBX21 | (++) | (--) | (++) | (--) | |
| STAT4 | (++) | (--) | (++) | (--) | |
| Th1 | STAT1 | (++) | (-) | (++) | (-) |
| IFNG | (+) | (-) | (+) | (-) | |
| TNF | (++) | (-) | (++) | (-) | |
| GATA3 | (+++) | (--) | (++) | (--) | |
| Th2 | STAT6 | ||||
| STAT5A | (++) | (-) | (++) | (-) | |
| IL13 | (++) | (-) | (+) | (-) | |
| Tfh | BCL6 | (+++) | (--) | (+++) | (--) |
| STAT3 | (++) | (-) | (++) | (-) | |
| Th17 | IL17A | ||||
| FOXP3 | (+++) | (--) | (+++) | (--) | |
| CCR8 | (+++) | (--) | (+++) | (--) | |
| T reg | STAT5B | (++) | (+) | ||
| TGFB1 | (+++) | (--) | (+++) | (--) | |
| PDCD1 | (++) | (--) | (++) | (--) | |
| CTLA4 | (+++) | (--) | (++) | (--) | |
| T cell exhaustion | LAG3 | (++) | (--) | (++) | (--) |
| HAVCR2 | (+++) | (--) | (+++) | (--) | |
| GZMB | (+) | ||||
| CD14 | (+++) | (--) | (+++) | (--) | |
| FERMT3 | (+++) | (--) | (+++) | (--) | |
| GPSM3 | (+++) | (--) | (+++) | (--) | |
| Myeloid-derived suppressor cells | IL18BP | (+++) | (--) | (+++) | (--) |
| PSAP | (+++) | (--) | (+++) | (--) | |
| PTGES2 | (-) | (-) | |||
| CFD | (+) | (-) | (+) | (-) | |
| MBL2 | (+) | (-) | (+) | (-) | |
| C2 | (+) | (-) | (+) | (-) | |
| C5 | (+) | ||||
| Complement | C8G | (-) | |||
| MASP2 | |||||
| C3 | (+++) | (--) | (+++) | (--) | |
| C1S | (+++) | (--) | (+++) | (--) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buttacavoli, M.; Di Cara, G.; Roz, E.; Pucci-Minafra, I.; Feo, S.; Cancemi, P. Integrated Multi-Omics Investigations of Metalloproteinases in Colon Cancer: Focus on MMP2 and MMP9. Int. J. Mol. Sci. 2021, 22, 12389. https://doi.org/10.3390/ijms222212389
Buttacavoli M, Di Cara G, Roz E, Pucci-Minafra I, Feo S, Cancemi P. Integrated Multi-Omics Investigations of Metalloproteinases in Colon Cancer: Focus on MMP2 and MMP9. International Journal of Molecular Sciences. 2021; 22(22):12389. https://doi.org/10.3390/ijms222212389
Chicago/Turabian StyleButtacavoli, Miriam, Gianluca Di Cara, Elena Roz, Ida Pucci-Minafra, Salvatore Feo, and Patrizia Cancemi. 2021. "Integrated Multi-Omics Investigations of Metalloproteinases in Colon Cancer: Focus on MMP2 and MMP9" International Journal of Molecular Sciences 22, no. 22: 12389. https://doi.org/10.3390/ijms222212389
APA StyleButtacavoli, M., Di Cara, G., Roz, E., Pucci-Minafra, I., Feo, S., & Cancemi, P. (2021). Integrated Multi-Omics Investigations of Metalloproteinases in Colon Cancer: Focus on MMP2 and MMP9. International Journal of Molecular Sciences, 22(22), 12389. https://doi.org/10.3390/ijms222212389






