Phosphate (Pi) Starvation Up-Regulated GmCSN5A/B Participates in Anthocyanin Synthesis in Soybean (Glycine max) Dependent on Pi Availability
Abstract
:1. Introduction
2. Results
2.1. Effects of P Deficiency on Soybean Biomass and Anthocyanin Content
2.2. Effects of P Deficiency on Transcripts of Anthocyanin Synthesis Genes
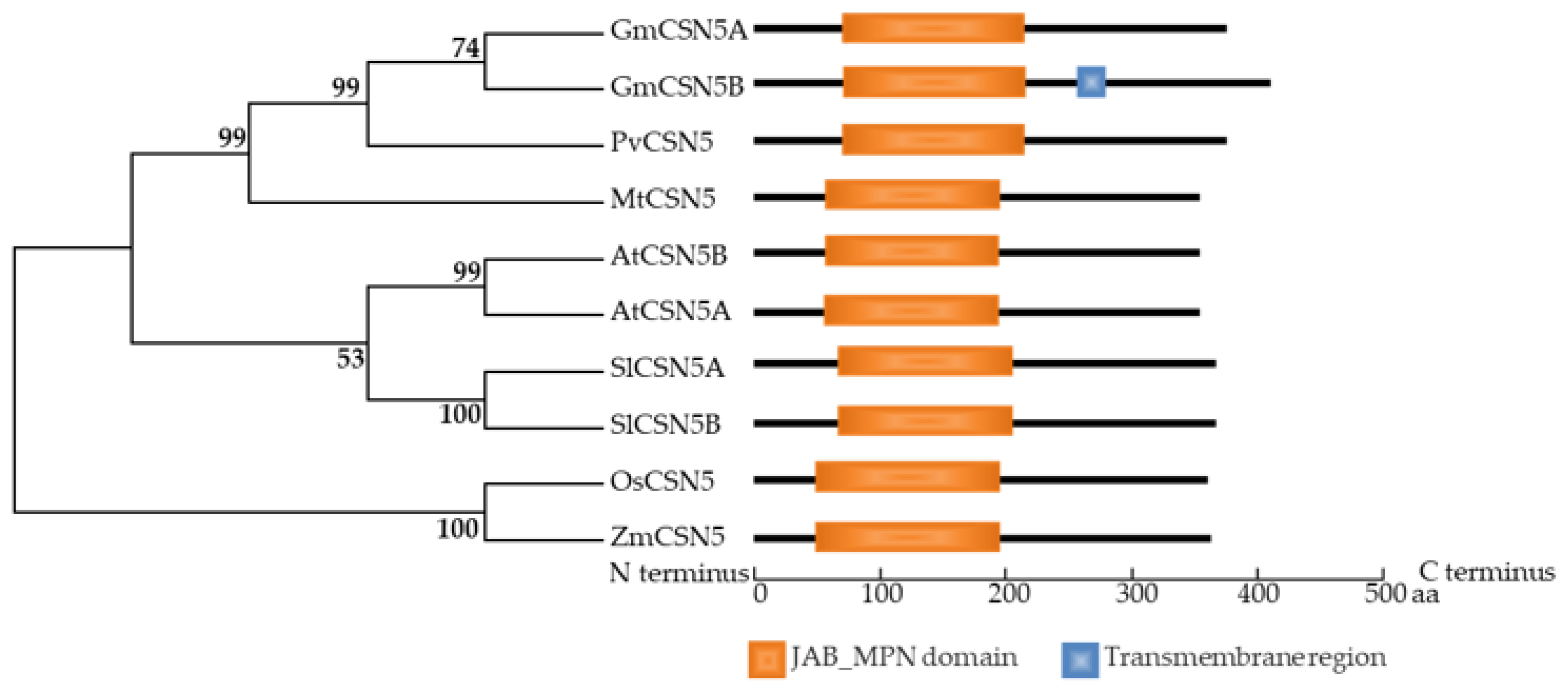
2.3. Identification of GmCSN5A/B in Soybean Genome
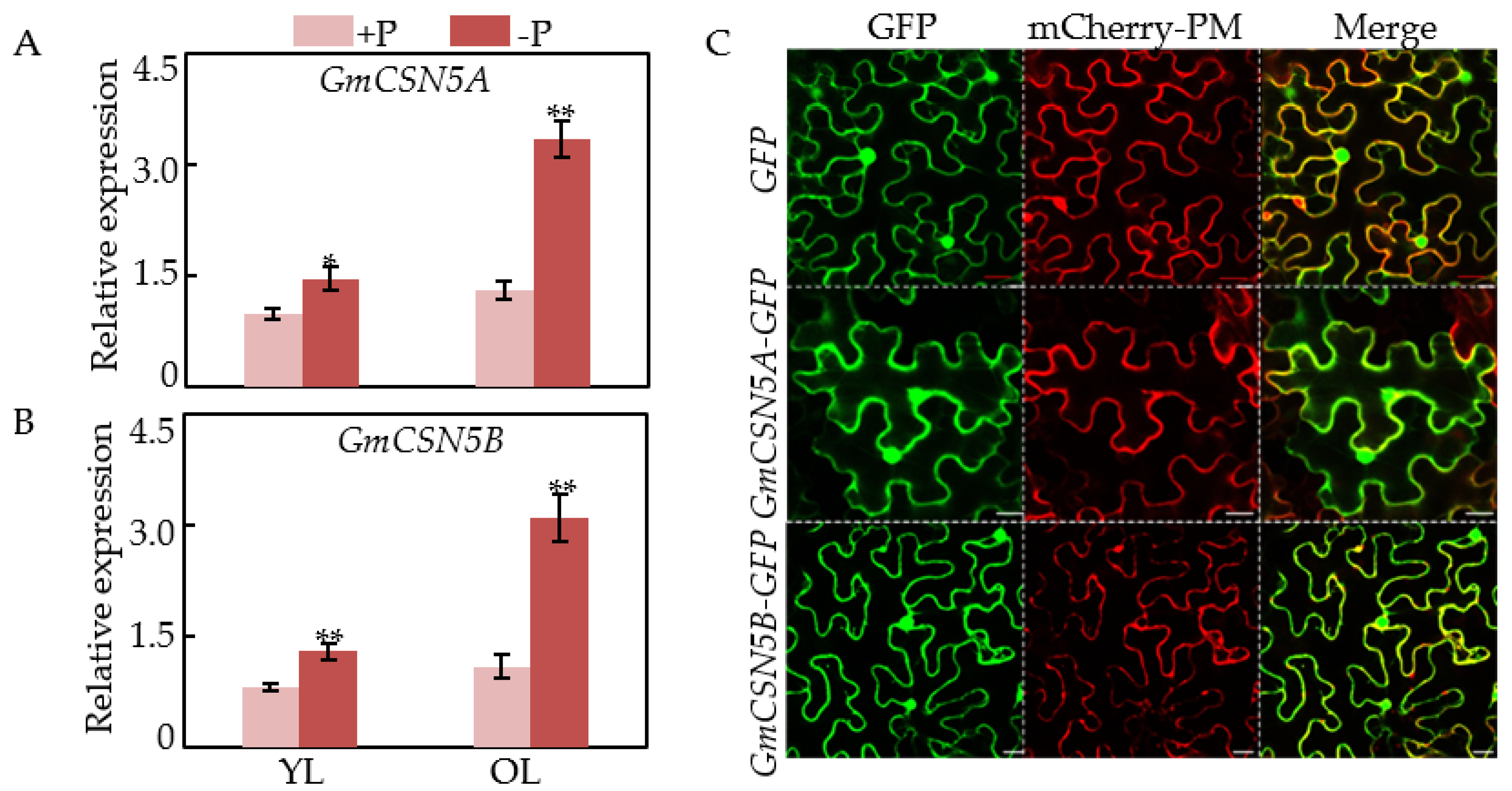
2.4. Expression Patterns and Subcellular Localization of GmCSN5A/B
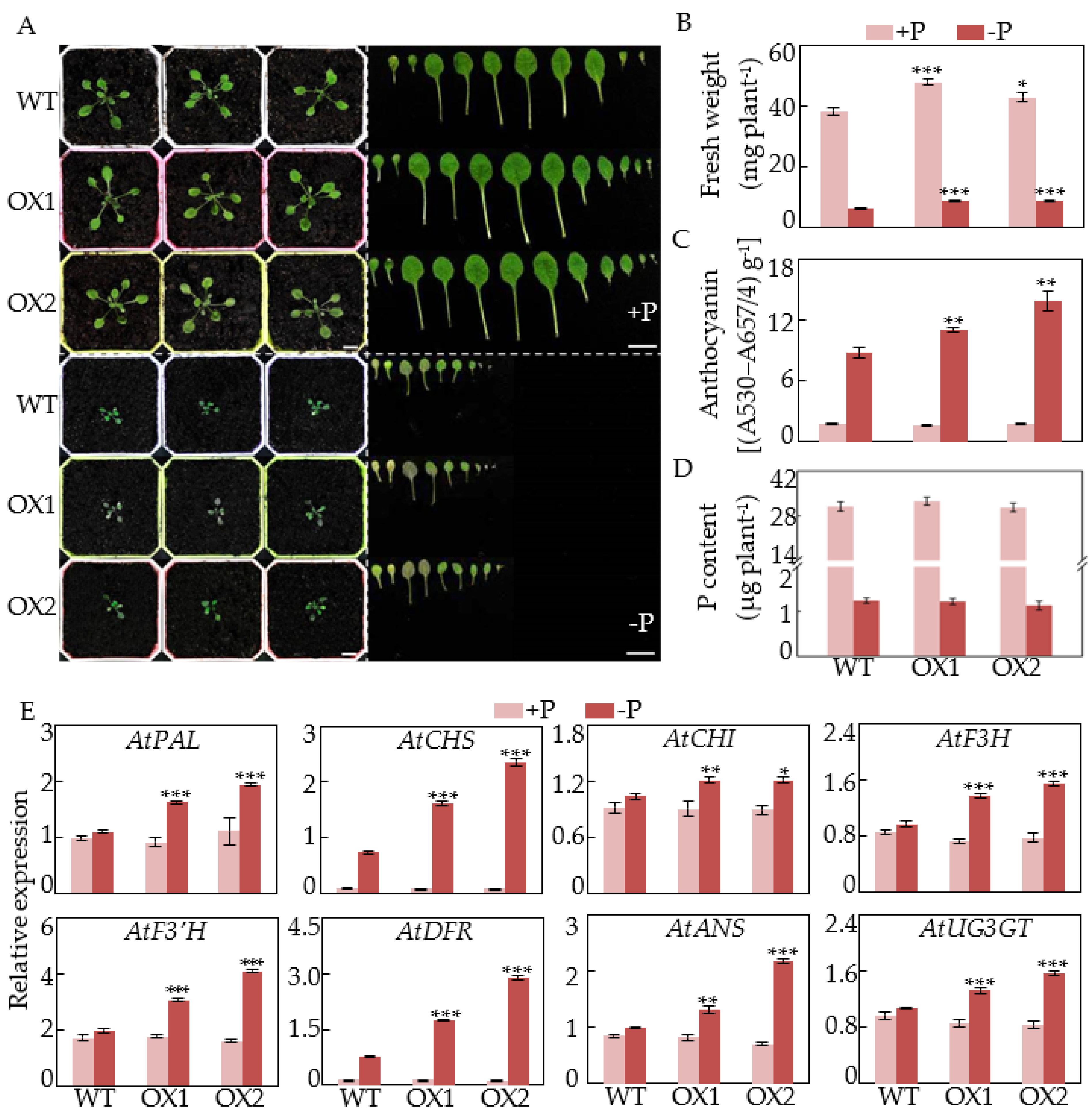
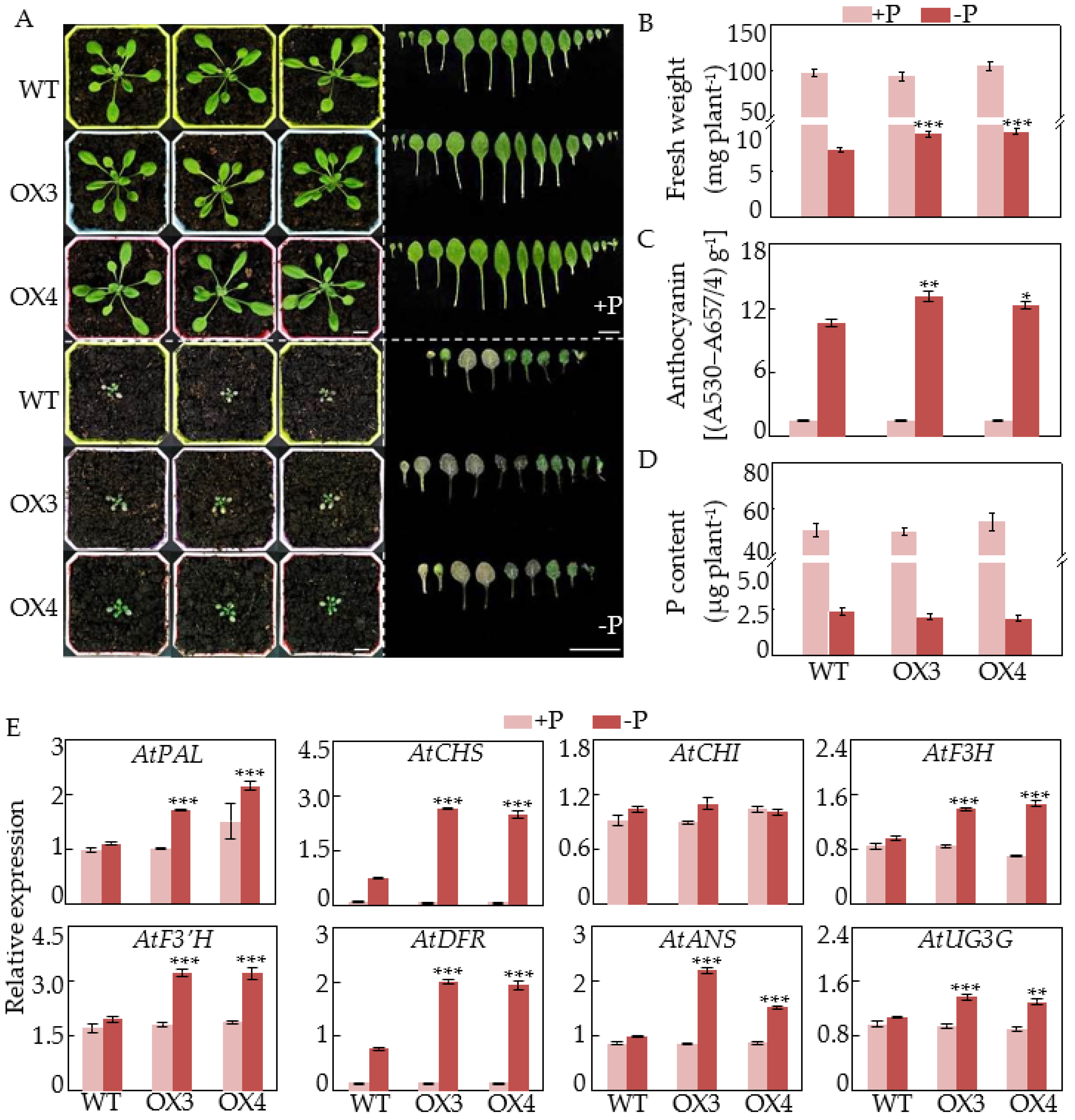
2.5. Overexpression of GmCSN5A/B Increases Anthocyanin Accumulations and Affects Photomorphogenesis in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Analysis of P Concentration and Anthocyanin Content
4.3. Phylogenetic Analysis and Characterization of GmCSN5A/B in Plants
4.4. Subcellular Localization Analysis of GmCSN5A/B
4.5. Functional Characterization of GmCSN5A/B in Arabidopsis
4.6. RNA Extraction and Quantitative RT-PCR Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chiou, T.J.; Lin, S.I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011, 62, 185–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ham, B.K.; Chen, J.; Yan, Y.; Lucas, W.J. Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol. 2018, 49, 1–9. [Google Scholar] [CrossRef]
- Dissanayaka, D.M.S.B.; Ghahremani, M.; Siebers, M.; Wasaki, J.; Plaxton, W.C. Recent insights into the metabolic adaptations of phosphorus-deprived plants. J. Exp. Bot. 2021, 72, 199–223. [Google Scholar] [CrossRef] [PubMed]
- Smith, F.W.; Mudge, S.R.; Rae, A.L.; Glassop, D. Phosphate transport in plants. Plant Soil. 2003, 248, 71–83. [Google Scholar] [CrossRef]
- Ticconi, C.A.; Abel, S. Short on phosphate: Plant surveillance and countermeasures. Trends Plant Sci. 2004, 9, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Raghothama, K.G. Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 665–693. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Q.; Zhou, X.; Dong, L.; Guo, J.; Chen, Y.; Zhang, Y.; Wu, L.; Xu, M. iTRAQ-based analysis of the Arabidopsis proteome reveals insights into the potential mechanisms of anthocyanin accumulation regulation in response to phosphate deficiency. J. Proteom. 2018, 184, 39–53. [Google Scholar] [CrossRef]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef] [Green Version]
- Morcuende, R.; Bari, R.; Gibon, Y.; Zheng, W.; Pant, B.D.; Bläsing, O.; Usadel, B.; Czechowski, T.; Udvardi, M.K.; Stitt, M.; et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007, 30, 85–112. [Google Scholar] [CrossRef]
- Li, Y.; Gu, M.; Zhang, X.; Zhang, J.; Fan, H.; Li, P.; Li, Z.; Xu, G. Engineering a sensitive visual-tracking reporter system for real-time monitoring phosphorus deficiency in tobacco. Plant Biotechnol. J. 2014, 12, 674–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manna, M.; Islam, T.; Kaul, T.; Reddy, C.S.; Fartyal, D.; James, D.; Reddy, M.K. A comparative study of effects of increasing concentrations of phosphate and phosphite on rice seedlings. Acta Physiol. Plant. 2015, 37, 258. [Google Scholar] [CrossRef]
- Sun, Y.; Mu, C.; Chen, Y.; Kong, X.; Xu, Y.; Zheng, H.; Zhang, H.; Wang, Q.; Xue, Y.; Li, Z.; et al. Comparative transcript profiling of maize inbreds in response to long-term phosphorus deficiency stress. Plant Physiol. Biochem. 2016, 109, 467–481. [Google Scholar] [CrossRef] [Green Version]
- Tominaga-Wada, R.; Masakane, A.; Wada, T. Effect of phosphate deficiency-induced anthocyanin accumulation on the expression of Solanum lycopersicum GLABRA3 (SlGL3) in tomato. Plant Signal. Behav. 2018, 13, e1477907. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.A.; Grayer, R.J. Anthocyanins and other flavonoids. Nat. Prod. Rep. 2004, 21, 539–573. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Butelli, E.; De Stefano, R.; Schoonbeek, H.J.; Magusin, A.; Pagliarani, C.; Wellner, N.; Hill, L.; Orzaez, D.; Granell, A.; et al. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Biol. 2013, 23, 1094–1100. [Google Scholar] [CrossRef] [Green Version]
- Khan, G.A.; Vogiatzaki, E.; Glauser, G.; Poirier, Y. Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol. 2016, 171, 632–644. [Google Scholar] [CrossRef] [Green Version]
- Stewart, A.J.; Chapman, W.; Jenkins, G.I.; Graham, I.; Martin, T.; Crozier, A. The effect of nitrogen and phosphorus deficiency on flavonol accumulation in plant tissues. Plant Cell Environ. 2001, 24, 1189–1197. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Viola, I.L.; Camoirano, A.; Gonzalez, D.H. Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol. 2016, 170, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Solfanelli, C.; Poggi, A.; Loreti, E.; Alpi, A.; Perata, P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006, 140, 637–646. [Google Scholar] [CrossRef] [Green Version]
- Sakuta, M. Diversity in plant red pigments: Anthocyanins and betacyanins. Plant Biotech. Rep. 2014, 8, 37–48. [Google Scholar] [CrossRef]
- Yin, Y.; Borges, G.; Sakuta, M.; Crozier, A.; Ashihara, H. Effect of phosphate deficiency on the content and biosynthesis of anthocyanins and the expression of related genes in suspension-cultured grape (Vitis sp.) cells. Plant Physiol. Biochem. 2012, 55, 77–84. [Google Scholar] [CrossRef]
- Zheng, X.T.; Chen, Y.L.; Zhang, X.H.; Cai, M.L.; Yu, Z.C.; Peng, C.L. ANS-deficient Arabidopsis is sensitive to high light due to impaired anthocyanin photoprotection. Funct. Plant Biol. 2019, 46, 756–765. [Google Scholar] [CrossRef]
- Hsieh, L.C.; Lin, S.I.; Shih, A.C.; Chen, J.W.; Lin, W.Y.; Tseng, C.Y.; Li, W.H.; Chiou, T.J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Zhang, X.; Li, L.; Sun, Z.; Li, J.; Chen, X.; Hong, G. SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorus-status-dependent anthocyanin biosynthesis. New Phytol. 2021, 230, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitayama, M.; et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005, 42, 218–235. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Bhargava, A.; Mansfield, S.D.; Hall, H.C.; Douglas, C.J.; Ellis, B.E. MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol. 2010, 154, 1428–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, S.; Ma, L.; Wang, X.; Xie, D.; Dinesh-Kumar, S.P.; Wei, N.; Deng, X.W. The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 2003, 15, 1083–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gusmaroli, G.; Figueroa, P.; Serino, G.; Deng, X.W. Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant Cell 2007, 19, 564–581. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yu, Y.; Zhang, Z.; Quan, R.; Zhang, H.; Ma, L.; Deng, X.W.; Huang, R. Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 2013, 25, 625–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, D.; Li, B.; Deng, X.W.; Wei, N. Plant COP9 signalosome subunit 5, CSN5. Plant Sci. 2014, 224, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Gusmaroli, G.; Feng, S.; Deng, X.W. The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell 2004, 16, 2984–3001. [Google Scholar] [CrossRef]
- Dohmann, E.M.; Kuhnle, C.; Schwechheimer, C. Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 2005, 17, 1967–1978. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Li, X.; Gruber, M.Y.; Feyissa, B.A.; Amyot, L.; Hannoufa, A. COP9 signalosome subunit 5A affects phenylpropanoid metabolism, trichome formation and transcription of key genes of a regulatory tri-protein complex in Arabidopsis. BMC Plant Biol. 2018, 18, 134. [Google Scholar] [CrossRef]
- Hou, E.; Luo, Y.; Kuang, Y.; Chen, C.; Lu, X.; Jiang, L.; Luo, X.; Wen, D. Global meta-analysis shows pervasive phosphorus limitation of aboveground plant production in natural terrestrial ecosystems. Nat. Commun. 2020, 11, 637. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yan, X.; Liao, H. Genetic improvement for phosphorus efficiency in soybean: A radical approach. Ann. Bot. 2010, 106, 215–222. [Google Scholar] [CrossRef]
- Liang, C.; Wang, J.; Zhao, J.; Tian, J.; Liao, H. Control of phosphate homeostasis through gene regulation in crops. Curr. Opin. Plant Biol. 2014, 21, 59–66. [Google Scholar] [CrossRef]
- Li, X.; Zeng, R.; Liao, H. Improving crop nutrient efficiency through root architecture modifications. J. Integr. Plant Biol. 2016, 58, 193–202. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Chen, M.; Liang, C.; Xue, Y.; Lin, S.; Tian, J. Characterization of purple acid phosphatase family and functional analysis of GmPAP7a/7b involved in extracellular ATP utilization in soybean. Front. Plant Sci. 2020, 11, 661. [Google Scholar] [CrossRef] [PubMed]
- Truong, H.N.; Thalineau, E.; Bonneau, L.; Fournier, C.; Potin, S.; Balzergue, S.; Van Tuinen, D.; Jeandroz, S.; Morandi, D. The Medicago truncatula hypermycorrhizal B9 mutant displays an altered response to phosphate and is more susceptible to Aphanomyces euteiches. Plant Cell Environ. 2015, 38, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Mahmood, K.; Rothstein, S.J. ROS induces anthocyanin production via late biosynthetic genes and anthocyanin deficiency confers the hypersensitivity to ROS-generating stresses in Arabidopsis. Plant Cell Physiol. 2017, 58, 1364–1377. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Sun, L.; Wei, L.; Yuan, J.; Kong, F.; Zhang, Y.; Miao, X.; Xia, G.; Liu, S. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress. Plant J. 2021, 105, 1010–1025. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Hussain, S.; Khan, S.; Geng, M. Seed priming improved antioxidant defense system and alleviated Ni-induced adversities in rice seedlings under N, P, or K deprivation. Front. Plant Sci. 2020, 11, 565647. [Google Scholar] [CrossRef]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; He, J. Protective role of anthocyanins in plants under low nitrogen stress. Biochem. Biophys. Res. Commun. 2018, 498, 946–953. [Google Scholar] [CrossRef]
- Peng, W.; Wu, W.; Peng, J.; Li, J.; Lin, Y.; Wang, Y.; Tian, J.; Sun, L.; Liang, C.; Liao, H. Characterization of the soybean GmALMT family genes and the function of GmALMT5 in response to phosphate starvation. J. Integr. Plant Biol. 2018, 60, 216–231. [Google Scholar] [CrossRef] [Green Version]
- Mo, X.; Zhang, M.; Liang, C.; Cai, L.; Tian, J. Integration of metabolome and transcriptome analyses highlights soybean roots responding to phosphorus deficiency by modulating phosphorylated metabolite processes. Plant Physiol. Biochem. 2019, 139, 697–706. [Google Scholar] [CrossRef]
- Murphy, J.; Riley, J. A modifed single solution method for the determination of phosphate in natural water. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, H.; Ma, Z.; Huang, J. DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol. Plant 2016, 9, 711–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.B.; Xiao, B.X.; Zhu, S.N.; Mo, X.H.; Liang, C.Y.; Tian, J.; Liao, H.; Miriam, G. GmPHR25, a GmPHR member up-regulated by phosphate starvation, controls phosphate homeostasis in soybean. J. Exp. Bot. 2017, 68, 4951–4967. [Google Scholar] [CrossRef] [Green Version]
- Bhadouria, J.; Singh, A.P.; Mehra, P.; Verma, L.; Srivastawa, R.; Parida, S.K.; Giri, J. Identification of purple acid phosphatases in chickpea and potential roles of CaPAP7 in seed phytate accumulation. Sci. Rep. 2017, 7, 11012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Sun, L.; Liu, P.; Liu, G.; Tian, J.; Liao, H. Malate synthesis and secretion mediated by a manganese-enhanced malate dehydrogenase confers superior manganese tolerance in Stylosanthes guianensis. Plant Physiol. 2015, 167, 176–188. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mo, X.; Zhang, M.; Zhang, Z.; Lu, X.; Liang, C.; Tian, J. Phosphate (Pi) Starvation Up-Regulated GmCSN5A/B Participates in Anthocyanin Synthesis in Soybean (Glycine max) Dependent on Pi Availability. Int. J. Mol. Sci. 2021, 22, 12348. https://doi.org/10.3390/ijms222212348
Mo X, Zhang M, Zhang Z, Lu X, Liang C, Tian J. Phosphate (Pi) Starvation Up-Regulated GmCSN5A/B Participates in Anthocyanin Synthesis in Soybean (Glycine max) Dependent on Pi Availability. International Journal of Molecular Sciences. 2021; 22(22):12348. https://doi.org/10.3390/ijms222212348
Chicago/Turabian StyleMo, Xiaohui, Mengke Zhang, Zeyu Zhang, Xing Lu, Cuiyue Liang, and Jiang Tian. 2021. "Phosphate (Pi) Starvation Up-Regulated GmCSN5A/B Participates in Anthocyanin Synthesis in Soybean (Glycine max) Dependent on Pi Availability" International Journal of Molecular Sciences 22, no. 22: 12348. https://doi.org/10.3390/ijms222212348
APA StyleMo, X., Zhang, M., Zhang, Z., Lu, X., Liang, C., & Tian, J. (2021). Phosphate (Pi) Starvation Up-Regulated GmCSN5A/B Participates in Anthocyanin Synthesis in Soybean (Glycine max) Dependent on Pi Availability. International Journal of Molecular Sciences, 22(22), 12348. https://doi.org/10.3390/ijms222212348






