Garlic Volatile Diallyl Disulfide Induced Cucumber Resistance to Downy Mildew
Abstract
:1. Introduction
2. Results
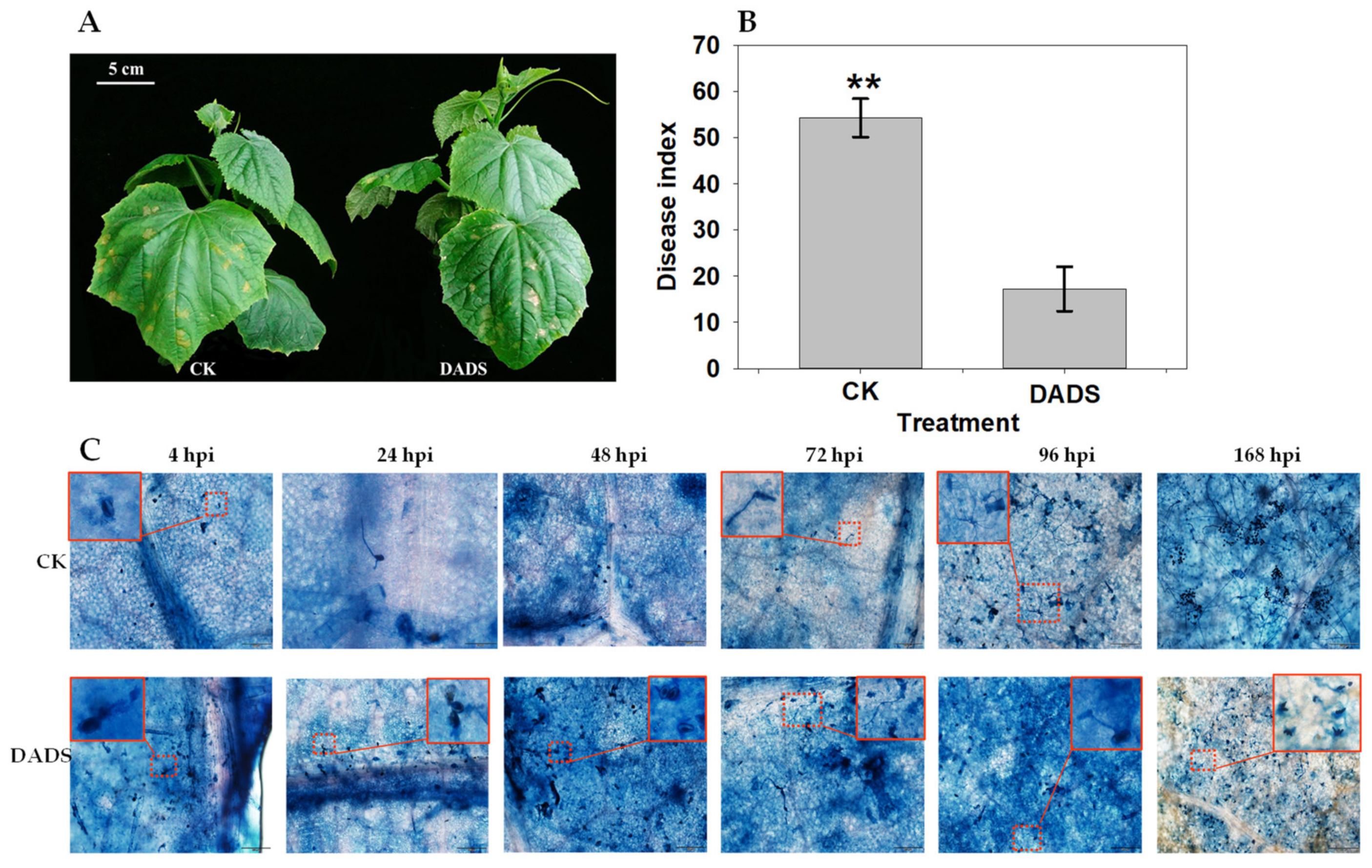
2.1. DADS-Induced Cucumber Resistance to Downy Mildew
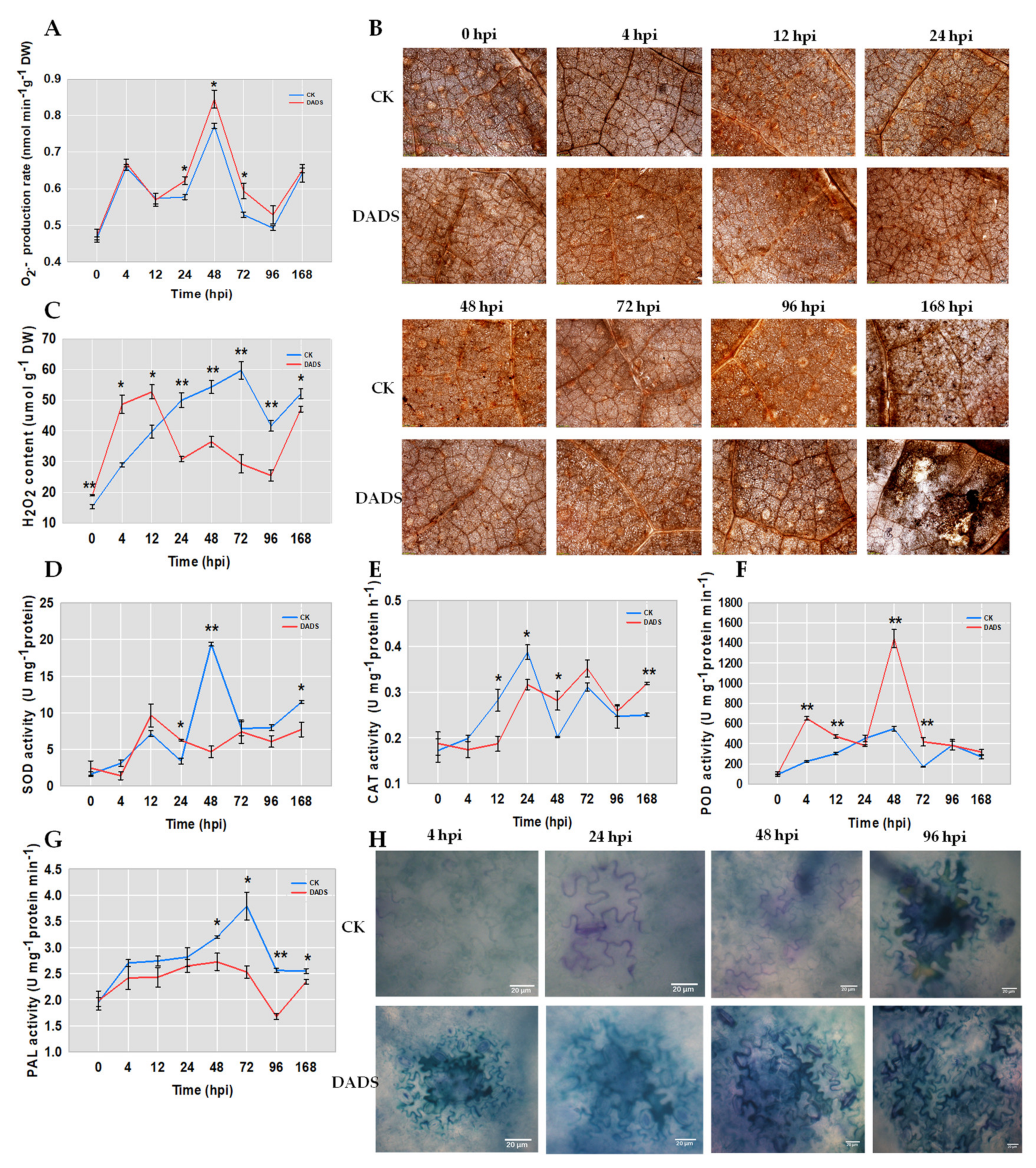
2.2. DADS Induced More ROS and Lignin Accumulations and Defense Enzymes Divergences after P. cubensis Inoculation
2.3. DADS Induced Phytohormone Changes under the Infection of Downy Mildew
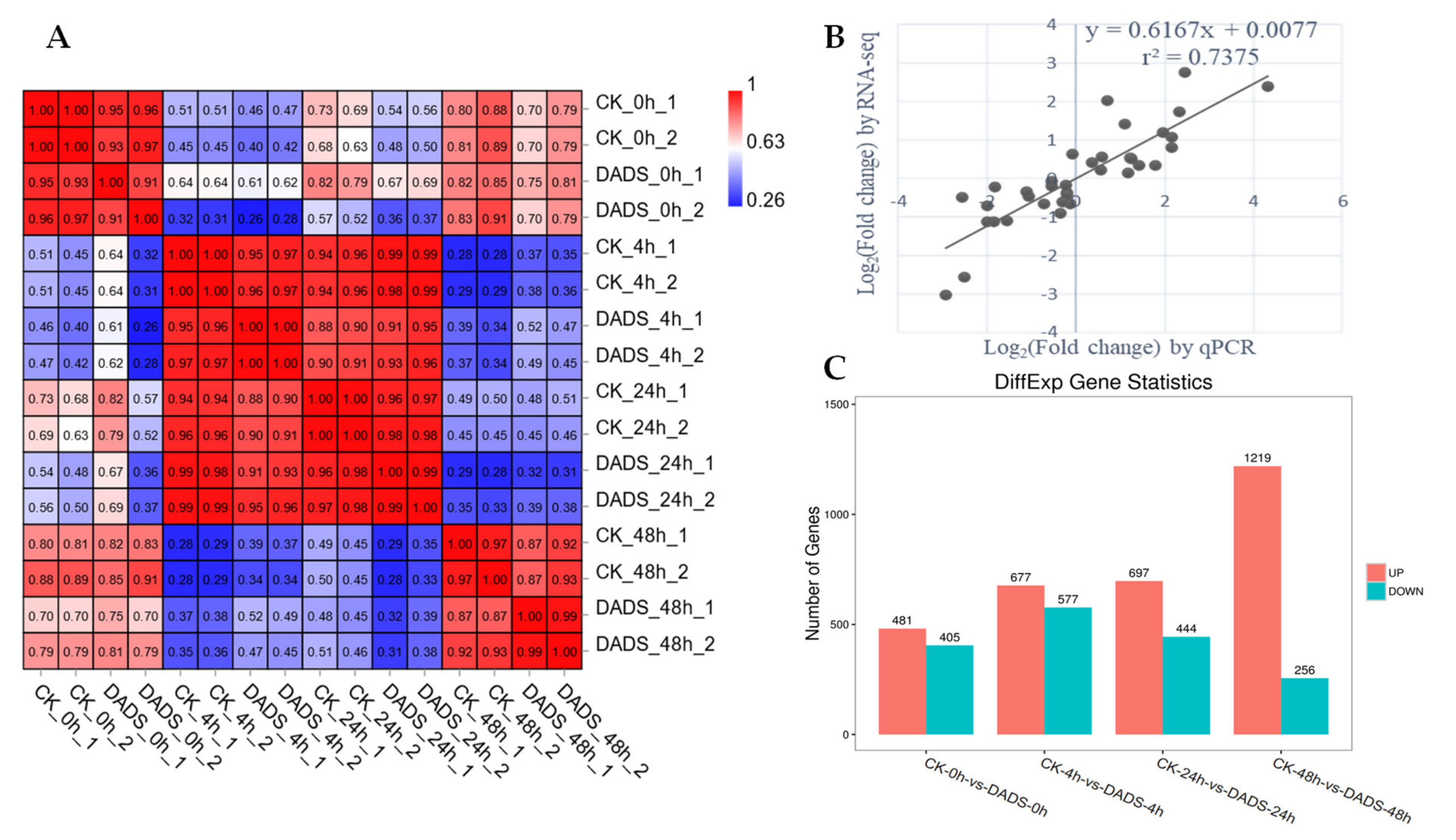
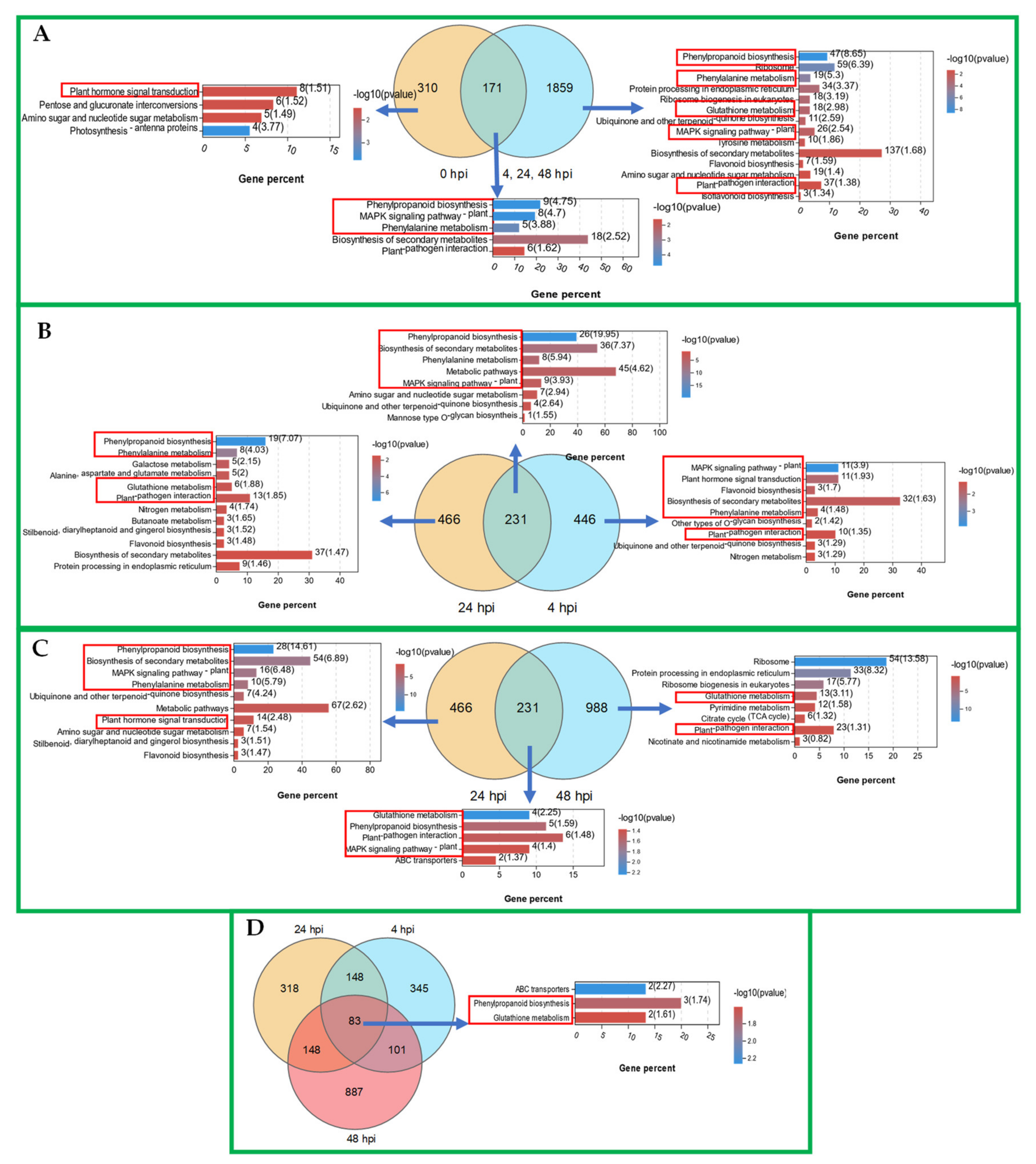
2.4. DADS-Induced Significant DEGs in Cucumber to Improve its Downy Mildew Resistance
2.5. WGCNA Provides Insights into Cucumber Resistance to P. cubensis
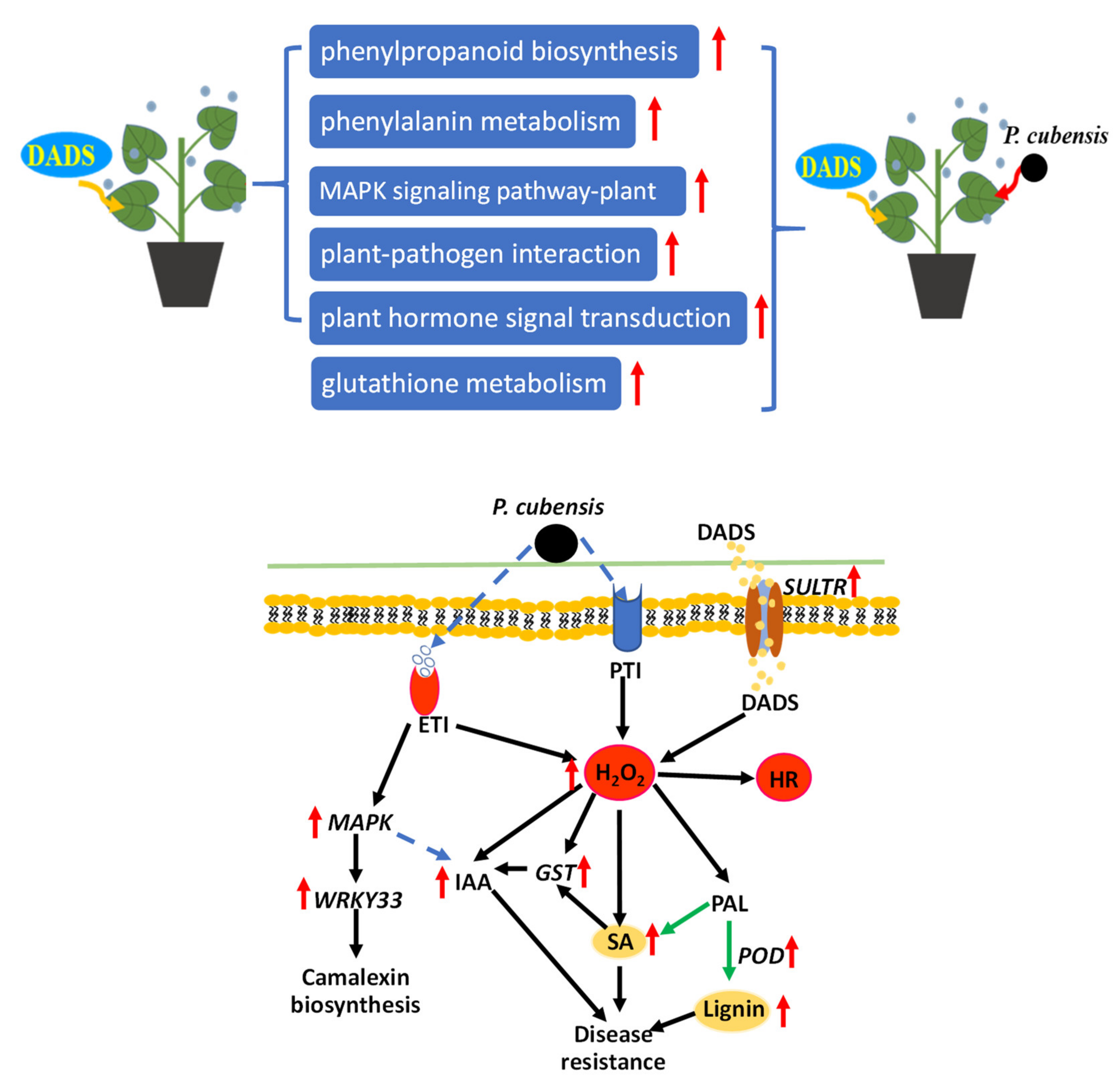
3. Discussion
3.1. Possible PTI and ETI Defense Mechanisms
3.2. DADS Induced H2O2 and Lignin Response to P. cubensis Infection
3.3. DADS Induced GST Response to P. cubensis Infection
3.4. DADS Induced Plant Hormone Response to P. cubensis Infection
3.5. DADS Induced WRKY33 Response to P. cubensis Infection
3.6. Possible Resistance Pathways in DADS Induced Cucumber Resistance to Downy Mildew
4. Materials and Methods
4.1. Cucumber Plant and P. cubensis Preparation
4.2. DADS Treatment and P. cubensis Inoculation
4.3. Histological Observation
4.4. Biochemical Assays
4.5. RNA Extraction, cDNA Library Construction and Sequencing
4.6. Differentially Expressed Genes (DEGs) Analysis
4.7. Expression Analysis of Disease Resistance-Related Genes
4.8. Analysis of KEGG Enrichment of DEGs
4.9. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, F.; Liu, X.; Wang, H.; Deng, R.; Yu, H.; Cheng, Z. Identification and allelopathy of green garlic (Allium sativum L.) volatiles on scavenging of cucumber (Cucumis sativus L.) reactive oxygen species. Molecules 2019, 24, 3263. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.Y.; Wu, C.N.; Cheng, Z.H.; Meng, H.W.; Zhang, M.R.; Zhang, H.J. Soil chemical property changes in eggplant/garlic relay intercropping systems under continuous cropping. PLoS ONE 2014, 9, e111040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, H.Y.; Cheng, Z.H.; Liu, M.L.; Hayat, S.; Feng, H. Garlic exerts allelopathic effects on pepper physiology in a hydroponic co-culture system. Biol. Open 2016, 5, 631–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Cheng, Z.; Meng, H.; Ahmad, I.; Zhao, H. Growth, yield and quality of spring tomato and physicochemical properties of medium in a tomato/garlic intercropping system under plastic tunnel organic medium cultivation. Sci. Hortic. 2014, 170, 159–168. [Google Scholar] [CrossRef]
- Xiao, X.M.; Cheng, Z.H.; Meng, H.W.; Khan, M.A.; Li, H.Z. Intercropping with garlic alleviated continuous cropping obstacle of cucumber in plastic tunnel. Acta Agric. Scand. Sect. B Soil Plant Sci. 2012, 62, 696–705. [Google Scholar] [CrossRef]
- Xiao, X.M.; Cheng, Z.H.; Meng, H.W.; Liu, L.H.; Li, H.Z.; Dong, Y.X. Intercropping of green garlic (Allium sativum L.) induces nutrient concentration changes in the soil and plants in continuously cropped cucumber (Cucumis sativus L.) in a plastic tunnel. PLoS ONE 2013, 8, e62173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, F.; Cheng, Z.H. Research progress on the use of plant allelopathy in agriculture and the physiological and ecological mechanisms of allelopathy. Front. Plant Sci. 2015, 6, 1020. [Google Scholar] [CrossRef]
- Cheng, F.; Cheng, Z.H.; Meng, H.W.; Tang, X.W. The garlic allelochemical diallyl disulfide affects tomato root growth by influencing cell division, phytohormone balance and expansin gene expression. Front. Plant Sci. 2016, 7, 512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, F.; Cheng, Z.H.; Meng, H.W. Transcriptomic insights into the allelopathic effects of the garlic allelochemical diallyl disulfide on tomato roots. Sci. Rep. 2016, 6, 38902. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Ali, M.; Liu, C.; Deng, R.; Cheng, Z. Garlic allelochemical diallyl disulfide alleviates autotoxicity in the root exudates caused by long-term continuous cropping of tomato. J. Agric. Food Chem. 2020, 68, 11684–11693. [Google Scholar] [CrossRef]
- Ren, K.L.; Hayat, S.; Qi, X.F.; Liu, T.; Cheng, Z.H. The garlic allelochemical DADS influences cucumber root growth involved in regulating hormone levels and modulating cell cycling. Plant Physiol. 2018, 230, 51–60. [Google Scholar] [CrossRef]
- El-Saber Batiha, G.; Magdy Beshbishy, A.; Wasef, L.G.; Elewa, Y.H.A.; Al-Sagan, A.A.; Abd El-Hack, M.E.; Taha, A.E.; Abd-Elhakim, Y.M.; Prasad Devkota, H. Chemical constituents and pharmacological activities of garlic (Allium sativum L.): A review. Nutrients 2020, 12, 872. [Google Scholar] [CrossRef] [Green Version]
- Savory, E.A.; Granke, L.L.; Quesada-Ocampo, L.M.; Varbanova, M.; Hausbeck, M.K.; Day, B. The cucurbit downy mildew pathogen Pseudoperonospora cubensis. Mol. Plant Pathol. 2011, 12, 217–226. [Google Scholar] [CrossRef]
- Burkhardt, A.; Day, B. Transcriptome and small RNAome dynamics during a resistant and susceptible interaction between cucumber and downy mildew. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Guo, P.; Wang, Z.; Chen, C.; Ren, Z. Transcriptome profiling reveals response genes for downy mildew resistance in cucumber. Planta 2021, 253, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Tan, J.Y.; Wu, Z.M.; VandenLangenberg, K.; Wehner, T.C.; Wen, C.L.; Zheng, X.Y.; Owens, K.; Thornton, A.; Bang, H.H.; et al. STAYGREEN, STAY HEALTHY: A loss-of-susceptibility mutation in the STAYGREEN gene provides durable, broad-spectrum disease resistances for over 50 years of US cucumber production. New Phytol. 2019, 221, 415–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, Q.; Chen, C.; Liu, M.; Li, Q.; Wang, L.; Ren, Z. CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant Sci. 2019, 279, 59–69. [Google Scholar] [CrossRef]
- Cheng, D.D.; Liu, M.J.; Sun, X.B.; Zhao, M.; Chow, W.S.; Sun, G.Y.; Zhang, Z.S.; Hu, Y.B. Light suppresses bacterial population through the accumulation of hydrogen peroxide in tobacco leaves infected with Pseudomonas syringae pv. tabaci. Front. Plant Sci. 2016, 7, 512. [Google Scholar] [CrossRef]
- Gayoso, C.; Pomar, F.; Novo-Uzal, E.; Merino, F.; Martinez de Ilarduya, O. The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 2010, 10, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Kahmann, R. Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front. Plant Sci. 2019, 10, 822. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Song, L.; Jiang, L.; Zhu, Y.; Gao, Q.; Wang, D.; Xie, J.; Lv, M.; Liu, P.; Li, M. The integration of transcriptomic and transgenic analyses reveals the involvement of the SA response pathway in the defense of chrysanthemum against the necrotrophic fungus Alternaria sp. Hortic. Res. 2020, 7, 1744–1757. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yu, J.Q.; Chen, Z. The perplexing role of autophagy in plant innate immune responses. Mol. Plant Pathol. 2014, 15, 637–645. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Ann. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef]
- Wagner, U.; Edwards, R.; Dixon, D.P.; Mauch, F. Probing the diversity of the Arabidopsis glutathione s-transferase gene family. Plant Mol. Biol. 2002, 49, 515–532. [Google Scholar] [CrossRef] [PubMed]
- Demeule, M.; Brossard, M.; Turcotte, S.; Regina, A.; Jodoin, J.; Beliveau, R. Diallyl disulfide, a chemopreventive agent in garlic, induces multidrug resistance-associated protein 2 expression. Biochem. Biophys. Res. Commun. 2004, 324, 937–945. [Google Scholar] [CrossRef]
- Pei, D.; Ma, H.; Zhang, Y.; Ma, Y.; Wang, W.; Geng, H.; Wu, J.; Li, C. Virus-induced gene silencing of a putative glutathione s-transferase gene compromised Ol-1-mediated resistance against powdery mildew in tomato. Plant Mol. Biol. Rep. 2011, 29, 972–978. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Kiraly, L.; Schroeder, P. Glutathione s-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilang, J.; Sturm, A. Cloning and Characterization of a glutathione s-transferase that can be photolabeled with 5-azido-indole-3-acetic acid. Plant Physiol. 1995, 109, 253–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khaleghnezhad, V.; Yousefi, A.R.; Tavakoli, A.; Farajmand, B.; Mastinu, A. Concentrations-dependent effect of exogenous abscisic acid on photosynthesis, growth and phenolic content of Dracocephalum moldavica L. under drought stress. Planta 2021, 253, 127. [Google Scholar] [CrossRef] [PubMed]
- Naservafaei, S.; Sohrabi, Y.; Moradi, P.; Mac Sweeney, E.; Mastinu, A. Biological response of Lallemantia iberica to brassinolide treatment under different watering conditions. Plants 2021, 10, 496. [Google Scholar] [CrossRef] [PubMed]
- Caarls, L.; Pieterse, C.M.; Van Wees, S.C. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef]
- Coelho, J.; Almeida-Trapp, M.; Pimentel, D.; Soares, F.; Reis, P.; Rego, C.; Mithöfer, A.; Fortes, A.M. The study of hormonal metabolism of Trincadeira and Syrah cultivars indicates new roles of salicylic acid, jasmonates, ABA and IAA during grape ripening and upon infection with Botrytis cinerea. Plant Sci. 2019, 283, 266–277. [Google Scholar] [CrossRef]
- Koornneef, A.; Leon-Reyes, A.; Ritsema, T.; Verhage, A.; Den Otter, F.C.; Van Loon, L.C.; Pieterse, C.M. Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 2008, 147, 1358–1368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Ning, K.; Wang, Z.; Liu, X.; Zhong, Y.; Ding, L.; Zi, H.; Cheng, Z.; Li, X.; Shan, H.; et al. CsIVP functions in vasculature development and downy mildew resistance in cucumber. PLoS Biol. 2020, 18, e3000671. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Ulate, B.; Vincenti, E.; Powell, A.L.; Cantu, D. Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea. Front. Plant Sci. 2013, 4, 142. [Google Scholar] [CrossRef] [Green Version]
- Wagner, S.; Stuttmann, J.; Rietz, S.; Guerois, R.; Brunstein, E.; Bautor, J.; Niefind, K.; Parker, J.E. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 2013, 14, 619–630. [Google Scholar] [CrossRef] [Green Version]
- Ding, P.; Ding, Y. Stories of salicylic acid: A plant defense hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Vasquez, A.; Salinas, P.; Holuigue, L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front. Plant Sci. 2015, 6, 171. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.; Yan, J.; Li, Y.; Jiang, H.; Sun, J.; Chen, Q.; Li, H.; Chu, J.; Yan, C.; Sun, X.; et al. Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol. 2012, 195, 872–882. [Google Scholar] [CrossRef]
- Naseem, M.; Kaltdorf, M.; Dandekar, T. The nexus between growth and defence signalling: Auxin and cytokinin modulate plant immune response pathways. J. Exp. Bot. 2015, 66, 4885–4896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Pajerowska-Mukhtar, K.; Culler, A.H.; Dong, X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 2007, 17, 1784–1790. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Abu Qamar, S.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Maechtel, R.; Ammon, A.; Engelsdorf, T.; Schmitz, J.; Maurino, V.G.; Voll, L.M. Reactive oxygen species dosage in Arabidopsis chloroplasts can improve resistance towards Colletotrichum higginsianum by the induction of WRKY33. New Phytol. 2020, 226, 189–204. [Google Scholar] [CrossRef] [Green Version]
- Tian, M.; Win, J.; Savory, E.; Burkhardt, A.; Held, M.; Brandizzi, F.; Day, B. 454 Genome sequencing of Pseudoperonospora cubensis reveals effector proteins with a QXLR translocation motif. Mol. Plant Microbe Interact. 2011, 24, 543–553. [Google Scholar] [CrossRef] [Green Version]
- Zho, Y.J.; Su, J.; Shi, L.; Liao, Q.J.; Su, Q. DADS downregulates the Rac1-ROCK1/PAK1-LIMK1-ADF/cofilin signaling pathway, inhibiting cell migration and invasion. Oncol Rep. 2013, 29, 605–612. [Google Scholar] [CrossRef]
- Savory, E.A.; Adhikari, B.N.; Hamilton, J.P.; Brieanne, V.; Robin, B.C.; Brad, D.; Rdam, A.M. mRNA-Seq analysis of the Pseudoperonospora cubensis transcriptome during cucumber (Cucumis sativus L.) infection. PLoS ONE 2012, 7, e35796. [Google Scholar] [CrossRef]
- Zhang, H.C.; Wang, C.F.; Cheng, Y.L.; Chen, X.M.; Han, Q.M.; Huang, L.L.; Wei, G.R.; Kang, Z.S. Histological and cytological characterization of adult plant resistance to wheat stripe rust. Plant Cell Rep. 2012, 31, 2121–2137. [Google Scholar] [CrossRef]
- Han, X.; Mao, L.; Wei, X.; Lu, W. Stimulatory involvement of abscisic acid in wound suberization of postharvest kiwifruit. Sci. Hortic. 2017, 224, 244–250. [Google Scholar] [CrossRef]
- Wang, B.; Zhu, S.J. Pre-storage cold acclimation maintained quality of cold-stored cucumber through differentially and orderly activating ROS scavengers. Postharvest Biol. Technol. 2017, 129, 1–8. [Google Scholar] [CrossRef]
- Gong, H.J.; Randall, D.P.; Flowers, T.J. Silicon deposition in the root reduces sodium uptake in rice (Oryza sativa L.) seedlings by reducing bypass flow. Plant Cell Environ. 2006, 29, 1970–1979. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, J.; Yang, F.; Weng, Y.; Chen, P.; Du, S.; Wei, A.; Li, Y. CsKTN1 for a katanin p60 subunit is associated with the regulation of fruit elongation in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2021, 134, 2429–2441. [Google Scholar] [CrossRef]
- Liu, X.; Pan, Y.; Liu, C.; Ding, Y.; Meng, H. Cucumber fruit size and shape variations explored from the aspects of morphology, histology, and endogenous hormones. Plants 2020, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


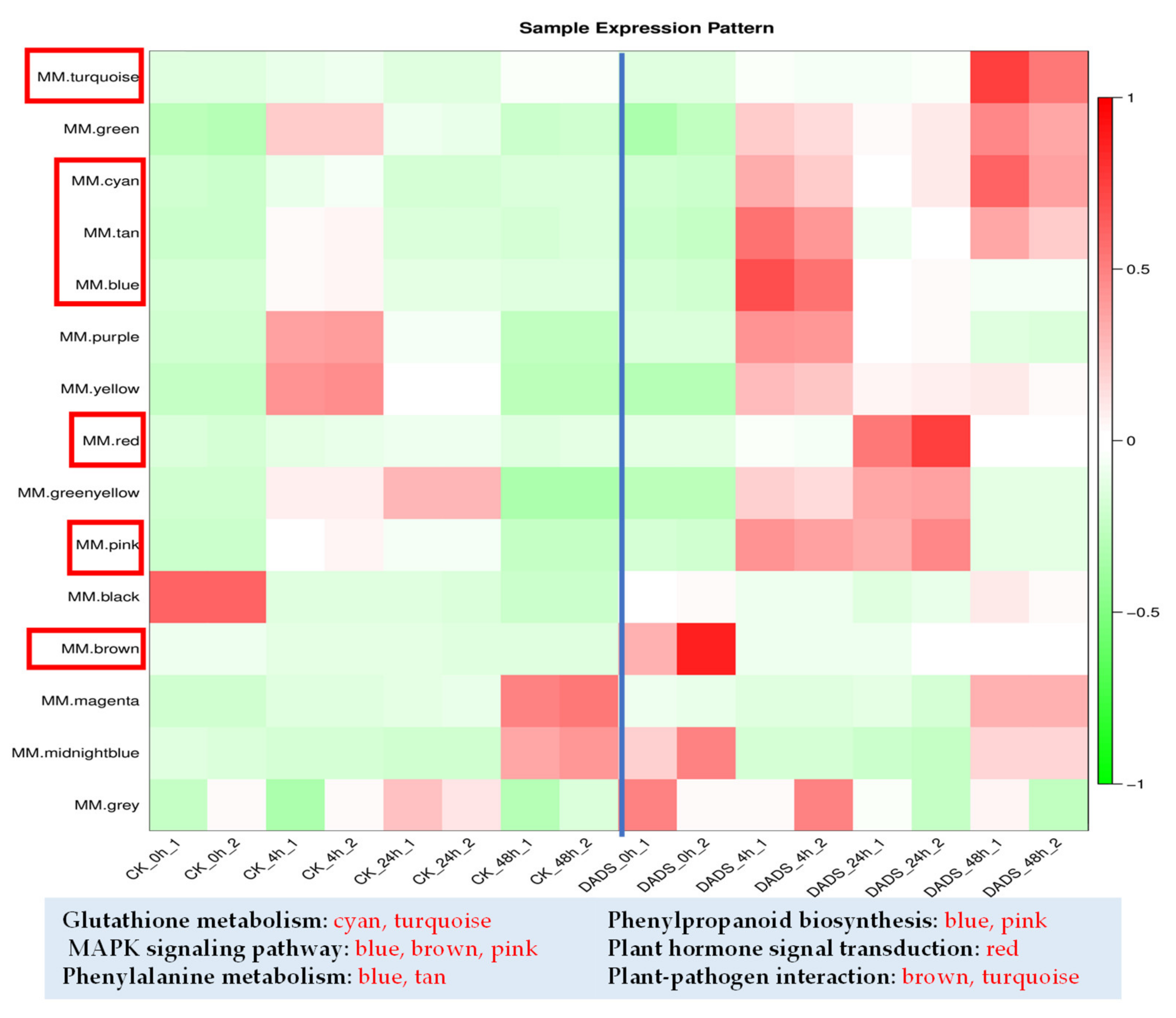
| Gene | Description | Module Colors | All.k within |
|---|---|---|---|
| CsGy4G012800 | Peroxidase | blue | 179.41 |
| CsGy4G000360 | Aminotransferase | blue | 178.46 |
| CsGy6G023900 | Phenylalanine ammonia-lyase | blue | 146.39 |
| CsGy1G004610 | probable WRKY transcription factor 33 | pink | 12.23 |
| CsGy1G004050 | probable glutathione S-transferase | blue | 104.07 |
| CsGy6G010290 | indole-3-acetic acid-amido synthetase GH3.6 | tan | 39.20 |
| CsGy5G008430 | abscisic acid receptor PYL4 | blue | 111.72 |
| CsGy7G006240 | PR1, Cysteine-rich venom protein | turquoise | 27.60 |
| CsGy1G001860 | protein EDS1L-like | cyan | 17.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Wang, H.; Zhi, C.; Chen, B.; Zheng, Y.; Qiao, L.; Gao, J.; Pan, Y.; Cheng, Z. Garlic Volatile Diallyl Disulfide Induced Cucumber Resistance to Downy Mildew. Int. J. Mol. Sci. 2021, 22, 12328. https://doi.org/10.3390/ijms222212328
Yang F, Wang H, Zhi C, Chen B, Zheng Y, Qiao L, Gao J, Pan Y, Cheng Z. Garlic Volatile Diallyl Disulfide Induced Cucumber Resistance to Downy Mildew. International Journal of Molecular Sciences. 2021; 22(22):12328. https://doi.org/10.3390/ijms222212328
Chicago/Turabian StyleYang, Fan, Hui Wang, Chengchen Zhi, Birong Chen, Yujie Zheng, Lijun Qiao, Jingcao Gao, Yupeng Pan, and Zhihui Cheng. 2021. "Garlic Volatile Diallyl Disulfide Induced Cucumber Resistance to Downy Mildew" International Journal of Molecular Sciences 22, no. 22: 12328. https://doi.org/10.3390/ijms222212328
APA StyleYang, F., Wang, H., Zhi, C., Chen, B., Zheng, Y., Qiao, L., Gao, J., Pan, Y., & Cheng, Z. (2021). Garlic Volatile Diallyl Disulfide Induced Cucumber Resistance to Downy Mildew. International Journal of Molecular Sciences, 22(22), 12328. https://doi.org/10.3390/ijms222212328







