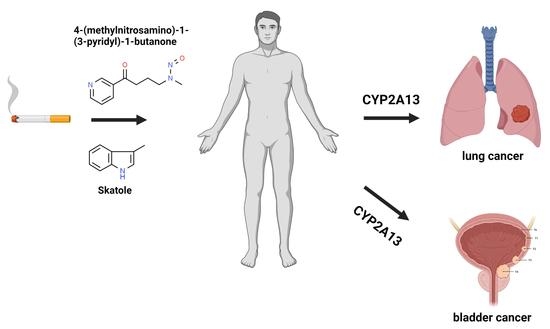
Genetic and Enzymatic Characteristics of CYP2A13 in Relation to Lung Damage
Abstract
1. Introduction
2. Tissue Distribution
3. Substrates and Inhibitors
3.1. AFB1
3.2. NNK
3.3. Skatole
4. Genetic Polymorphisms
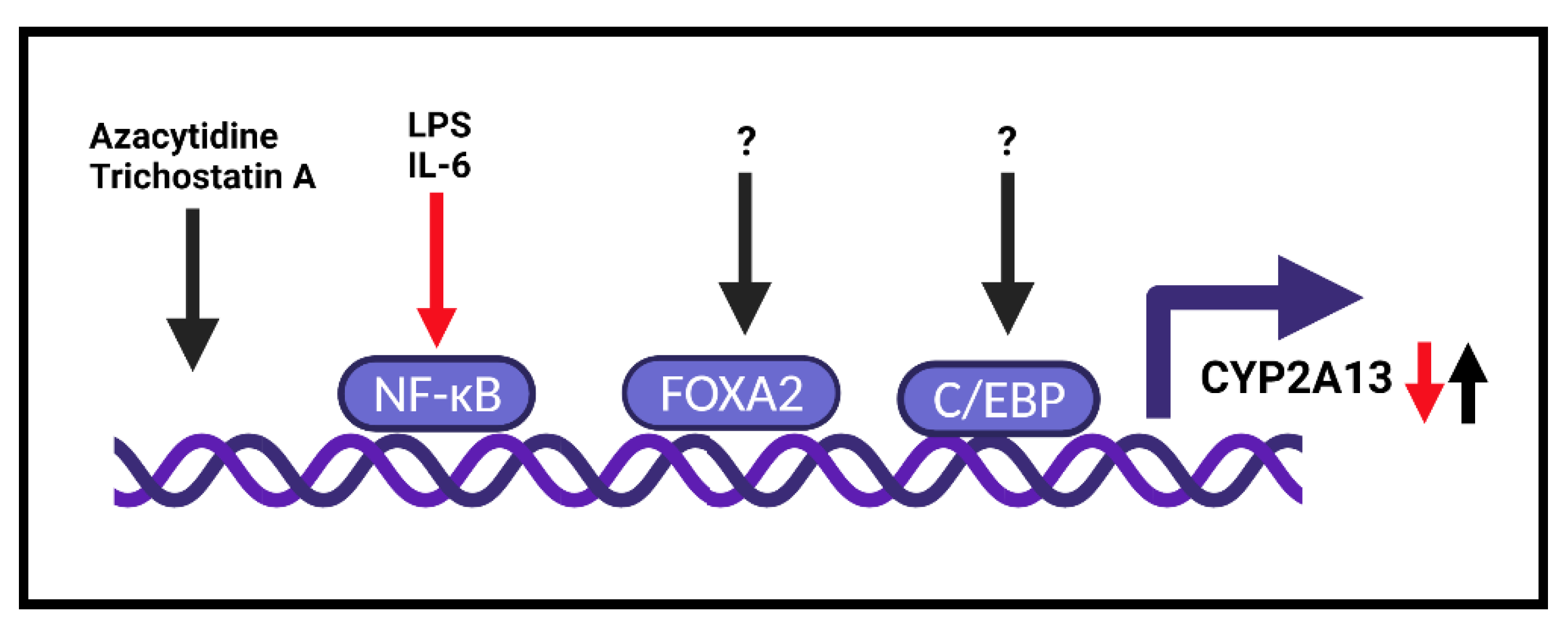
5. Transcription Regulation
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 3MI | 3-methylindole |
| 8-MOP | 8-methoxypsoralen |
| AFB1 | aflatoxin B1 |
| AFG1 | aflatoxin G1 |
| ATM | ATM serine/threonine kinase |
| ATR | ATR serine/threonine kinase |
| BRCA1 | Breast cancer type 1 susceptibility protein |
| ChIP | Chromatin immunoprecipitation |
| Chk2 | Checkpoint kinase 2 |
| FOXA2 | Forkhead box A |
| γH2AX | Phosphorylated H2A.X variant histone |
| Km | Michaelis–Menten constant |
| NF-kB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NNK | 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone |
| p53 | tumor protein P53 |
| TNFα | Tumor necrosis factor alpha |
| Vmax | maximal velocity |
References
- Omura, T. Structural diversity of cytochrome P450 enzyme system. J. Biochem. 2010, 147, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Tralau, T.; Luch, A. The evolution of our understanding of endo-xenobiotic crosstalk and cytochrome P450 regulation and the therapeutic implications. Expert Opin. Drug Metab. Toxicol. 2013, 9, 1541–1554. [Google Scholar] [CrossRef]
- Podust, L.M.; Sherman, D.H. Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 2012, 29, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W.; Dalton, T.P. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat. Rev. Cancer 2006, 6, 947–960. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W.; Wikvall, K.; Miller, W.L. Human cytochromes P450 in health and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120431. [Google Scholar] [CrossRef]
- Lewis, D.F. 57 varieties: The human cytochromes P450. Pharmacogenomics 2004, 5, 305–318. [Google Scholar] [CrossRef]
- Fernandez-Salguero, P.; Hoffman, S.M.; Cholerton, S.; Mohrenweiser, H.; Raunio, H.; Rautio, A.; Pelkonen, O.; Huang, J.D.; Evans, W.E.; Idle, J.R.; et al. A genetic polymorphism in coumarin 7-hydroxylation: Sequence of the human CYP2A genes and identification of variant CYP2A6 alleles. Am. J. Hum. Genet. 1995, 57, 651–660. [Google Scholar] [PubMed]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Su, T.; Bao, Z.; Zhang, Q.Y.; Smith, T.J.; Hong, J.Y.; Ding, X. Human cytochrome P450 CYP2A13: Predominant expression in the respiratory tract and its high efficiency metabolic activation of a tobacco-specific carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 2000, 60, 5074–5079. [Google Scholar]
- Gu, J.; Su, T.; Chen, Y.; Zhang, Q.Y.; Ding, X. Expression of biotransformation enzymes in human fetal olfactory mucosa: Potential roles in developmental toxicity. Toxicol. Appl. Pharmacol. 2000, 165, 158–162. [Google Scholar] [CrossRef]
- Nakajima, M.; Itoh, M.; Sakai, H.; Fukami, T.; Katoh, M.; Yamazaki, H.; Kadlubar, F.F.; Imaoka, S.; Funae, Y.; Yokoi, T. CYP2A13 expressed in human bladder metabolically activates 4-aminobiphenyl. Int. J. Cancer 2006, 119, 2520–2526. [Google Scholar] [CrossRef]
- Borlak, J.; Walles, M.; Levsen, K.; Thum, T. Verapamil: Metabolism in cultures of primary human coronary arterial endothelial cells. Drug Metab. Dispos. 2003, 31, 888–891. [Google Scholar] [CrossRef]
- Zhang, X.; D’Agostino, J.; Wu, H.; Zhang, Q.Y.; von Weymarn, L.; Murphy, S.E.; Ding, X. CYP2A13: Variable expression and role in human lung microsomal metabolic activation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. J. Pharmacol. Exp. Ther. 2007, 323, 570–578. [Google Scholar] [CrossRef]
- Zhu, L.R.; Thomas, P.E.; Lu, G.; Reuhl, K.R.; Yang, G.Y.; Wang, L.D.; Wang, S.L.; Yang, C.S.; He, X.Y.; Hong, J.Y. CYP2A13 in human respiratory tissues and lung cancers: An immunohistochemical study with a new peptide-specific antibody. Drug Metab. Dispos. 2006, 34, 1672–1676. [Google Scholar] [CrossRef]
- Jia, K.; Li, L.; Liu, Z.; Hartog, M.; Kluetzman, K.; Zhang, Q.Y.; Ding, X. Generation and characterization of a novel CYP2A13--transgenic mouse model. Drug Metab. Dispos. 2014, 42, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhu, L.R.; Lu, G.; Wang, H.; Hong, J.Y. Selective expression of CYP2A13 in human pancreatic alpha-islet cells. Drug Metab. Dispos. 2012, 40, 1878–1882. [Google Scholar] [CrossRef] [PubMed]
- Gandini, S.; Botteri, E.; Iodice, S.; Boniol, M.; Lowenfels, A.B.; Maisonneuve, P.; Boyle, P. Tobacco smoking and cancer: A meta-analysis. Int. J. Cancer 2008, 122, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.D.; Sanders, J.L.; Porubsky, P.R.; Lushington, G.H.; Stout, C.D.; Scott, E.E. Structure of the human lung cytochrome P450 2A13. J. Biol. Chem. 2007, 282, 17306–17313. [Google Scholar] [CrossRef] [PubMed]
- He, X.Y.; Shen, J.; Hu, W.Y.; Ding, X.; Lu, A.Y.; Hong, J.Y. Identification of Val117 and Arg372 as critical amino acid residues for the activity difference between human CYP2A6 and CYP2A13 in coumarin 7-hydroxylation. Arch. Biochem. Biophys. 2004, 427, 143–153. [Google Scholar] [CrossRef]
- Von Weymarn, L.B.; Murphy, S.E. CYP2A13-catalysed coumarin metabolism: Comparison with CYP2A5 and CYP2A6. Xenobiotica 2003, 33, 73–81. [Google Scholar] [CrossRef]
- Bao, Z.; He, X.Y.; Ding, X.; Prabhu, S.; Hong, J.Y. Metabolism of nicotine and cotinine by human cytochrome P450 2A13. Drug Metab. Dispos. 2005, 33, 258–261. [Google Scholar] [CrossRef]
- He, X.Y.; Shen, J.; Ding, X.; Lu, A.Y.; Hong, J.Y. Identification of critical amino acid residues of human CYP2A13 for the metabolic activation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco-specific carcinogen. Drug Metab. Dispos. 2004, 32, 1516–1521. [Google Scholar] [CrossRef]
- Von Weymarn, L.B.; Zhang, Q.Y.; Ding, X.; Hollenberg, P.F. Effects of 8-methoxypsoralen on cytochrome P450 2A13. Carcinogenesis 2005, 26, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, J.; Zhang, C.; Yang, B.; Wang, L.; Zhou, J. The inhibition of cytochrome P450 2A13-catalyzed NNK metabolism by NAT, NAB and nicotine. Toxicol. Res. 2016, 5, 1115–1121. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kramlinger, V.M.; von Weymarn, L.B.; Murphy, S.E. Inhibition and inactivation of cytochrome P450 2A6 and cytochrome P450 2A13 by menthofuran, beta-nicotyrine and menthol. Chem.-Biol. Interact. 2012, 197, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Abente, G.; Gonzalez, C.A.; Errezola, M.; Escolar, A.; Izarzugaza, I.; Nebot, M.; Riboli, E. Tobacco smoke inhalation pattern, tobacco type, and bladder cancer in Spain. Am. J. Epidemiol. 1991, 134, 830–839. [Google Scholar] [CrossRef]
- National Center for Chronic Disease Prevention and Health Promotion. The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General; National Center for Chronic Disease Prevention and Health Promotion: Atlanta, GA, USA, 2014. [Google Scholar]
- Barbosa, A.L.A.; Vermeulen, S.; Aben, K.K.; Grotenhuis, A.J.; Vrieling, A.; Kiemeney, L.A. Smoking intensity and bladder cancer aggressiveness at diagnosis. PLoS ONE 2018, 13, e0194039. [Google Scholar]
- Boonruang, S.; Prakobsri, K.; Pouyfung, P.; Srisook, E.; Prasopthum, A.; Rongnoparut, P.; Sarapusit, S. Inhibition of human cytochromes P450 2A6 and 2A13 by flavonoids, acetylenic thiophenes and sesquiterpene lactones from Pluchea indica and Vernonia cinerea. J. Enzyme Inhib. Med. Chem. 2017, 32, 1136–1142. [Google Scholar] [CrossRef] [PubMed]
- Chougnet, A.; Woggon, W.D.; Locher, E.; Schilling, B. Synthesis and in vitro activity of heterocyclic inhibitors of CYP2A6 and CYP2A13, two cytochrome P450 enzymes present in the respiratory tract. ChemBioChem 2009, 10, 1562–1567. [Google Scholar] [CrossRef]
- Shimada, T.; Takenaka, S.; Kakimoto, K.; Murayama, N.; Lim, Y.R.; Kim, D.; Foroozesh, M.K.; Yamazaki, H.; Guengerich, F.P.; Komori, M. Structure-function studies of naphthalene, phenanthrene, biphenyl, and their derivatives in interaction with and Oxidation by CYTOCHROMES P450 2A13 and 2A6. Chem. Res. Toxicol. 2016, 29, 1029–1040. [Google Scholar] [CrossRef]
- Shimada, T.; Takenaka, S.; Murayama, N.; Kramlinger, V.M.; Kim, J.H.; Kim, D.; Liu, J.; Foroozesh, M.K.; Yamazaki, H.; Guengerich, F.P.; et al. Oxidation of pyrene, 1-hydroxypyrene, 1-nitropyrene and 1-acetylpyrene by human cytochrome P450 2A13. Xenobiotica 2016, 46, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Zhang, Z.; Li, N.; Xia, R.; Wang, C.; Yu, Y.; Yao, S.; Shen, J.; Wang, S.L. Identification of 5-hydroxymethylfurfural in cigarette smoke extract as a new substrate metabolically activated by human cytochrome P450 2A13. Toxicol. Appl. Pharmacol. 2018, 359, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Fayyaz, A.; Makwinja, S.; Auriola, S.; Raunio, H.; Juvonen, R.O. Comparison of In Vitro Hepatic Scoparone 7-O-Demethylation between Humans and Experimental Animals. Planta Med. 2018, 84, 320–328. [Google Scholar] [CrossRef] [PubMed]
- He, X.Y.; Tang, L.; Wang, S.L.; Cai, Q.S.; Wang, J.S.; Hong, J.Y. Efficient activation of aflatoxin B1 by cytochrome P450 2A13, an enzyme predominantly expressed in human respiratory tract. Int. J. Cancer 2006, 118, 2665–2671. [Google Scholar] [CrossRef]
- D’Agostino, J.; Zhuo, X.; Shadid, M.; Morgan, D.G.; Zhang, X.; Humphreys, W.G.; Shu, Y.Z.; Yost, G.S.; Ding, X. The pneumotoxin 3-methylindole is a substrate and a mechanism-based inactivator of CYP2A13, a human cytochrome P450 enzyme preferentially expressed in the respiratory tract. Drug Metab. Dispos. 2009, 37, 2018–2027. [Google Scholar] [CrossRef]
- Shimada, T.; Murayama, N.; Tanaka, K.; Takenaka, S.; Guengerich, F.P.; Yamazaki, H.; Komori, M. Spectral modification and catalytic inhibition of human cytochromes P450 1A1, 1A2, 1B1, 2A6, and 2A13 by four chemopreventive organoselenium compounds. Chem. Res. Toxicol. 2011, 24, 1327–1337. [Google Scholar] [CrossRef] [PubMed]
- Pouyfung, P.; Prasopthum, A.; Sarapusit, S.; Srisook, E.; Rongnoparut, P. Mechanism-based inactivation of cytochrome P450 2A6 and 2A13 by Rhinacanthus nasutus constituents. Drug Metab. Pharmacokinet. 2014, 29, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Von Weymarn, L.B.; Chun, J.A.; Knudsen, G.A.; Hollenberg, P.F. Effects of eleven isothiocyanates on P450 2A6- and 2A13-catalyzed coumarin 7-hydroxylation. Chem. Res. Toxicol. 2007, 20, 1252–1259. [Google Scholar] [CrossRef]
- Von Weymarn, L.B.; Chun, J.A.; Hollenberg, P.F. Effects of benzyl and phenethyl isothiocyanate on P450s 2A6 and 2A13: Potential for chemoprevention in smokers. Carcinogenesis 2006, 27, 782–790. [Google Scholar] [CrossRef]
- Reddy, B.S.; Upadhyaya, P.; Simi, B.; Rao, C.V. Evaluation of organoselenium compounds for potential chemopreventive properties in colon carcinogenesis. Anticancer Res. 1994, 14, 2509–2514. [Google Scholar]
- Ip, C.; el-Bayoumy, K.; Upadhyaya, P.; Ganther, H.; Vadhanavikit, S.; Thompson, H. Comparative effect of inorganic and organic selenocyanate derivatives in mammary cancer chemoprevention. Carcinogenesis 1994, 15, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Richie, J.P., Jr.; Kleinman, W.; Desai, D.H.; Das, A.; Amin, S.G.; Pinto, J.T.; El-Bayoumy, K. The organoselenium compound 1,4-phenylenebis(methylene)selenocyanate inhibits 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced tumorgenesis and enhances glutathione-related antioxidant levels in A/J mouse lung. Chem.-Biol. Interact. 2006, 161, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Clayson, D.B. Specific aromatic amines as occupational bladder carcinogens. In Carcinogenic and Mutagenic N-substituted Aryl Compounds; National Institutes of Health and National Cancer Institute: Bethesda, MD, USA, 1981; pp. 15–19. [Google Scholar]
- DeVore, N.M.; Smith, B.D.; Wang, J.L.; Lushington, G.H.; Scott, E.E. Key residues controlling binding of diverse ligands to human cytochrome P450 2A enzymes. Drug Metab. Dispos. 2009, 37, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- DeVore, N.M.; Smith, B.D.; Urban, M.J.; Scott, E.E. Key residues controlling phenacetin metabolism by human cytochrome P450 2A enzymes. Drug Metab. Dispos. 2008, 36, 2582–2590. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Murayama, N.; Yamazaki, H.; Tanaka, K.; Takenaka, S.; Komori, M.; Kim, D.; Guengerich, F.P. Metabolic activation of polycyclic aromatic hydrocarbons and aryl and heterocyclic amines by human cytochromes P450 2A13 and 2A6. Chem. Res. Toxicol. 2013, 26, 529–537. [Google Scholar] [CrossRef]
- Kakimoto, K.; Murayama, N.; Takenaka, S.; Nagayoshi, H.; Lim, Y.R.; Kim, V.; Kim, D.; Yamazaki, H.; Komori, M.; Guengerich, F.P.; et al. Cytochrome P450 2A6 and other human P450 enzymes in the oxidation of flavone and flavanone. Xenobiotica 2019, 49, 131–142. [Google Scholar] [CrossRef]
- Li, L.; Carratt, S.; Hartog, M.; Kovalchik, N.; Jia, K.; Wang, Y.; Zhang, Q.Y.; Edwards, P.; Winkle, L.V.; Ding, X. Human CYP2A13 and CYP2F1 Mediate Naphthalene Toxicity in the Lung and Nasal Mucosa of CYP2A13/2F1-Humanized Mice. Environ. Health Perspect. 2017, 125, 067004. [Google Scholar] [CrossRef]
- Toselli, F.; Matthias, A.; Bone, K.M.; Gillam, E.M.; Lehmann, R.P. Metabolism of the major Echinacea alkylamide N-isobutyldodeca-2E,4E,8Z,10Z-tetraenamide by human recombinant cytochrome P450 enzymes and human liver microsomes. Phytother. Res. 2010, 24, 1195–1201. [Google Scholar] [CrossRef]
- Rodu, B.; Cole, P.; Mandel, J.S. Evaluation of the national toxicology program report on carcinogens. Regulat. Toxicol. Pharmacol. 2012, 64, 186–188. [Google Scholar] [CrossRef]
- Massey, T.E. The 1995 Pharmacological Society of Canada Merck Frosst Award. Cellular and molecular targets in pulmonary chemical carcinogenesis: Studies with aflatoxin B1. Can. J. Physiol. Pharmacol. 1996, 74, 621–628. [Google Scholar] [CrossRef]
- Sorenson, W.G.; Simpson, J.P.; Peach, M.J., 3rd; Thedell, T.D.; Olenchock, S.A. Aflatoxin in respirable corn dust particles. J. Toxicol. Environ. Health 1981, 7, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Hayes, R.B.; van Nieuwenhuize, J.P.; Raatgever, J.W.; ten Kate, F.J. Aflatoxin exposures in the industrial setting: An epidemiological study of mortality. Food Chem. Toxicol. 1984, 22, 39–43. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Z.; Wang, X.; Wang, Y.; Zhang, X.; Lu, H.; Wang, S.L. Cytochrome P450 2A13 enhances the sensitivity of human bronchial epithelial cells to aflatoxin B1-induced DNA damage. Toxicol. Appl. Pharmacol. 2013, 270, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lu, H.; Huan, F.; Meghan, C.; Yang, X.; Wang, Y.; Wang, X.; Wang, X.; Wang, S.L. Cytochrome P450 2A13 mediates the neoplastic transformation of human bronchial epithelial cells at a low concentration of aflatoxin B1. Int. J. Cancer 2014, 134, 1539–1548. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, X.; Wang, Y.; Wang, X.; Lu, H.; Zhang, X.; Xiao, X.; Li, S.; Wang, X.; Wang, S.L. Cytochrome P450 2A13 is an efficient enzyme in metabolic activation of aflatoxin G1 in human bronchial epithelial cells. Arch. Toxicol. 2013, 87, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Megaraj, V.; Zhou, X.; Xie, F.; Liu, Z.; Yang, W.; Ding, X. Role of CYP2A13 in the bioactivation and lung tumorigenicity of the tobacco-specific lung procarcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone: In vivo studies using a CYP2A13-humanized mouse model. Carcinogenesis 2014, 35, 131–137. [Google Scholar] [CrossRef]
- Liu, Z.; Megaraj, V.; Li, L.; Sell, S.; Hu, J.; Ding, X. Suppression of pulmonary CYP2A13 expression by carcinogen-induced lung tumorigenesis in a CYP2A13-humanized mouse model. Drug Metab. Dispos. 2015, 43, 698–702. [Google Scholar] [CrossRef]
- Ibuki, Y.; Shikata, M.; Toyooka, T. Gamma-H2AX is a sensitive marker of DNA damage induced by metabolically activated 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Toxicol. Vitro 2015, 29, 1831–1838. [Google Scholar] [CrossRef]
- Gillam, E.M.; Notley, L.M.; Cai, H.; De Voss, J.J.; Guengerich, F.P. Oxidation of indole by cytochrome P450 enzymes. Biochemistry 2000, 39, 13817–13824. [Google Scholar] [CrossRef]
- Weems, J.M.; Lamb, J.G.; D’Agostino, J.; Ding, X.; Yost, G.S. Potent mutagenicity of 3-methylindole requires pulmonary cytochrome P450-mediated bioactivation: A comparison to the prototype cigarette smoke mutagens B(a)P and NNK. Chemical Res. Toxicol. 2010, 23, 1682–1690. [Google Scholar] [CrossRef]
- Wynder, E.L.; Hoffmann, D. Experimental tobacco carcinogenesis. Science 1968, 162, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Donley, K.M.; Keeney, D.S.; Hoffman, S.M. Organization and evolution of the Cyp2 gene cluster on mouse chromosome 7, and comparison with the syntenic human cluster. Environ. Health Perspect. 2003, 111, 1835–1842. [Google Scholar] [CrossRef] [PubMed]
- Cauffiez, C.; Pottier, N.; Tournel, G.; Lo-Guidice, J.M.; Allorge, D.; Chevalier, D.; Migot-Nabias, F.; Kenani, A.; Broly, F. CYP2A13 genetic polymorphism in French Caucasian, Gabonese and Tunisian populations. Xenobiotica 2005, 35, 661–669. [Google Scholar] [CrossRef]
- Kim, V.; Yeom, S.; Lee, Y.; Park, H.G.; Cho, M.A.; Kim, H.; Kim, D. In vitro functional analysis of human cytochrome P450 2A13 genetic variants: P450 2A13*2, *3, *4, and *10. J. Toxicol. Environ. Health Pt. A 2018, 81, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Kumondai, M.; Hosono, H.; Maekawa, M.; Yamaguchi, H.; Mano, N.; Oda, A.; Hirasawa, N.; Hiratsuka, M. Functional characterization of 9 CYP2A13 allelic variants by assessment of nicotine C-oxidation and coumarin 7-hydroxylation. Drug Metab. Pharmacokinet. 2018, 33, 82–89. [Google Scholar] [CrossRef]
- Verde, Z.; Santiago, C.; Rodriguez Gonzalez-Moro, J.M.; de Lucas Ramos, P.; Lopez Martin, S.; Bandres, F.; Lucia, A.; Gomez-Gallego, F. ‘Smoking genes’: A genetic association study. PLoS ONE 2011, 6, e26668. [Google Scholar] [CrossRef]
- Tamaki, Y.; Arai, T.; Sugimura, H.; Sasaki, T.; Honda, M.; Muroi, Y.; Matsubara, Y.; Kanno, S.; Ishikawa, M.; Hirasawa, N.; et al. Association between cancer risk and drug-metabolizing enzyme gene (CYP2A6, CYP2A13, CYP4B1, SULT1A1, GSTM1, and GSTT1) polymorphisms in cases of lung cancer in Japan. Drug Metab. Pharmacokinet. 2011, 26, 516–522. [Google Scholar] [CrossRef]
- Kumondai, M.; Hosono, H.; Orikasa, K.; Arai, Y.; Arai, T.; Sugimura, H.; Ozono, S.; Sugiyama, T.; Takayama, T.; Sasaki, T.; et al. CYP2A13 genetic polymorphisms in relation to the risk of bladder cancer in Japanese smokers. Biol. Pharm. Bull. 2016, 39, 1683–1686. [Google Scholar] [CrossRef]
- Sharma, R.; Ahuja, M.; Panda, N.; Khullar, M. Polymorphisms in CYP2A13 and UGT1A7 genes and head and neck cancer susceptibility in North Indians. Oral Dis. 2010, 16, 760–768. [Google Scholar] [CrossRef]
- Schlicht, K.E.; Michno, N.; Smith, B.D.; Scott, E.E.; Murphy, S.E. Functional characterization of CYP2A13 polymorphisms. Xenobiotica 2007, 37, 1439–1449. [Google Scholar] [CrossRef]
- Liu, T.; Hong, Y.; Li, Z.; Hong, J.; Zeng, S.; Zheng, M.; Chen, S. An investigation of the catalytic activity of CYP2A13*4 with coumarin and polymorphisms of CYP2A13 in a Chinese Han population. Drug Metab. Dispos. 2012, 40, 847–851. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tan, W.; Hao, B.; Miao, X.; Zhou, G.; He, F.; Lin, D. Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 2003, 63, 8057–8061. [Google Scholar] [PubMed]
- D’Agostino, J.; Zhang, X.; Wu, H.; Ling, G.; Wang, S.; Zhang, Q.Y.; Liu, F.; Ding, X. Characterization of CYP2A13*2, a variant cytochrome P450 allele previously found to be associated with decreased incidences of lung adenocarcinoma in smokers. Drug Metab. Dispos. 2008, 36, 2316–2323. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, M.N.; Kropp, S.; Sauter, W.; Beckmann, L.; Rosenberger, A.; Illig, T.; Jager, B.; Mittelstrass, K.; Dienemann, H.; Consortium, L.; et al. CYP450 polymorphisms as risk factors for early-onset lung cancer: Gender-specific differences. Carcinogenesis 2009, 30, 1161–1169. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, X.; Ling, G.; D’Agostino, J.; Ding, X. Mechanisms of differential expression of the CYP2A13 7520C and 7520G alleles in human lung: Allelic expression analysis for CYP2A13 heterogeneous nuclear RNA, and evidence for the involvement of multiple cis-regulatory single nucleotide polymorphisms. Pharmacogenet. Genom. 2009, 19, 852–863. [Google Scholar] [CrossRef]
- Sun, L.; Fan, X. Expression of cytochrome P450 2A13 in human non-small cell lung cancer and its clinical significance. J. Biomed. Res. 2013, 27, 202–207. [Google Scholar]
- Chiang, H.C.; Lee, H.; Chao, H.R.; Chiou, Y.H.; Tsou, T.C. Pulmonary CYP2A13 levels are associated with early occurrence of lung cancer-Its implication in mutagenesis of non-small cell lung carcinoma. Cancer Epidemiol. 2013, 37, 653–659. [Google Scholar] [CrossRef]
- Fukami, T.; Nakajima, M.; Matsumoto, I.; Zen, Y.; Oda, M.; Yokoi, T. Immunohistochemical analysis of CYP2A13 in various types of human lung cancers. Cancer Sci. 2010, 101, 1024–1028. [Google Scholar] [CrossRef] [PubMed]
- Ling, G.; Wei, Y.; Ding, X. Transcriptional regulation of human CYP2A13 expression in the respiratory tract by CCAAT/enhancer binding protein and epigenetic modulation. Mol. Pharmacol. 2007, 71, 807–816. [Google Scholar] [CrossRef]
- Wan, H.; Xu, Y.; Ikegami, M.; Stahlman, M.T.; Kaestner, K.H.; Ang, S.L.; Whitsett, J.A. Foxa2 is required for transition to air breathing at birth. Proc. Natl. Acad. Sci. USA 2004, 101, 14449–14454. [Google Scholar] [CrossRef]
- Xiang, C.; Wang, J.; Kou, X.; Chen, X.; Qin, Z.; Jiang, Y.; Sun, C.; Xu, J.; Tan, W.; Jin, L.; et al. Pulmonary expression of CYP2A13 and ABCB1 is regulated by FOXA2, and their genetic interaction is associated with lung cancer. FASEB J. 2015, 29, 1986–1998. [Google Scholar] [CrossRef] [PubMed]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef]
- Li, Z.; Tuteja, G.; Schug, J.; Kaestner, K.H. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell 2012, 148, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Gupta, A.; Wang, Y.; Suzuki, K.; Mirosevich, J.; Orgebin-Crist, M.C.; Matusik, R.J. Foxa1 and Foxa2 interact with the androgen receptor to regulate prostate and epididymal genes differentially. Ann. N. Y. Acad. Sci. 2005, 1061, 77–93. [Google Scholar] [CrossRef]
- Wu, H.; Liu, Z.; Ling, G.; Lawrence, D.; Ding, X. Transcriptional suppression of CYP2A13 expression by lipopolysaccharide in cultured human lung cells and the lungs of a CYP2A13-humanized mouse model. Toxicol. Sci. 2013, 135, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Shao, P.; Guo, N.; Wang, C.; Zhao, M.; Yi, L.; Liu, C.; Kang, L.; Cao, L.; Lv, P.; Xing, L.; et al. Aflatoxin G1 induced TNF-alpha-dependent lung inflammation to enhance DNA damage in alveolar epithelial cells. J. Cell. Physiol. 2019, 234, 9194–9206. [Google Scholar] [CrossRef]
- Sharma, R.; Panda, N.K.; Khullar, M. Hypermethylation of carcinogen metabolism genes, CYP1A1, CYP2A13 and GSTM1 genes in head and neck cancer. Oral Dis. 2010, 16, 668–673. [Google Scholar] [CrossRef]
| Compound | Relation to CYP2A13 | Parameter | Reference |
|---|---|---|---|
| Coumarin | Substrate | Km = 2.21 ± 0.63 or 0.48 ± 0.07 µM, Vmax = 0.69 ± 0.16 or 0.15 ± 0.006 | [19,30] |
| Testosterone | Substrate | Km = 13 ± 3 µM, Vmax = 1.7 ± 0.11 | [23] |
| 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) | Substrate | Km = 10.4 µM, Vmax = 3.6 | [21] |
| Nicotine | Substrate/Inhibitor # | Km = 20.2 µM, Vmax = 8.7, Ki = 6.57−25.01 µM ## | [21,24] |
| Cotinine | Substrate | Km = 45.2 µM, Vmax = 0.7 | [21] |
| 4-aminobiphenyl | Substrate | Km = 38.5 ± 0.6 µM, Vmax = 7.8 ± 0.00 | [11] |
| Naphthalene | Substrate | Km = N.D., Vmax = 6.1 ± 0.88 | [31] |
| Phenanthrene | Substrate | Km = N.D., Vmax = 3.14 ± 0.35 | [31] |
| Biphenyl | Substrate | Km = N.D., Vmax = 3.1 ± 0.19 | [31] |
| Pyrene | Substrate | Km = 1.2 ± 0.2 µM, Vmax = 2.0 ± 0.06 | [32] |
| 5-hydroxymethyfurfural | Substrate | Km = 50.9 ± 8.3 µM, Vmax = 2.7 ± 0.2 | [33] |
| Scoparone | Substrate | Km = 10.1 µM, Vmax = 22 µmol/min/g | [34] |
| Aflatoxin B1 | Substrate | Km = N.D., Vmax = 1.7–6.2 ## | [35] |
| 3-methylindole (skatole) | Substrate/Inhibitor # | Km = 14.3−14.8 µM, Vmax = 1.5−1.9 ## Ki = 10 µM | [36] |
| (R,S)-N-nitrosoanatabine (NAT) | Inhibitor # | Ki = 0.21−0.71 µM ## | [24] |
| (R,S)-N-nitrosoanabasine (NAB) | Inhibitor # | Ki = 0.23−0.87 µM ## | [24] |
| 1-methyl-4-(3-pyridinyl) pyrrole (beta-nicotyrine) | Inhibitor * | Ki = 0.17 µM | [25] |
| Menthofuran | Inhibitor * | Ki = 1.24 µM | [25] |
| (-)-menthol | Inhibitor * | Ki = 8.2 µM | [25] |
| 8-methoxypsoralen (8-MOP) | Inhibitor * | Ki = 0.11 µM | [23] |
| Benzyl selenocyanate (BSC) | Inhibitor * | IC50 = 1.2 ± 0.19 µM | [37] |
| 1,2-phenylenebis(methylene)selenocyanate (o-XSC) | Inhibitor * | IC50 = 1.2 ± 0.13 µM | [37] |
| 1,3-phenylenebis(methylene)selenocyanate (m-XSC) | Inhibitor * | IC50 = 0.22 ± 0.03 µM | [37] |
| 1,4-phenylenebis(methylene)selenocyanate (p-XSC) | Inhibitor * | IC50 = 1.4 ± 0.21 µM | [37] |
| Apigenin | Inhibitor * | IC50 = 0.05 ± 0.01 µM | [29] |
| Luteolin | Inhibitor * | IC50 = 0.18 ± 0.02 µM | [29] |
| Chrysoeriol | Inhibitor * | IC50 = 0.82 ± 0.05 µM | [29] |
| Quercetin | Inhibitor * | IC50 = 0.80 ± 0.01 µM | [29] |
| 2-(penta-1,3-diyn-1-yl)-5-(4-acetoxy-3-hydroxybuta-1-yn-1-yl) thiophene | Inhibitor * | IC50 = 6.18 ± 0.28 µM | [29] |
| 2-(prop-1-inyl)-5-(6-acetoxy-5-hydroxyhexa-1,3-diinyl) thiophene | Inhibitor * | IC50 = 2.94 ± 0.01 µM | [29] |
| 2-(prop-1-inyl)-5-(5, 6-dihydroxyhexa-1,3-diinyl) thiophene | Inhibitor * | IC50 = 2.40 ± 0.33 µM | [29] |
| Rhinacanthin-A | Inhibitor * | IC50 = 1.42 ± 0.05 µM | [38] |
| Rhinacanthin-B | Inhibitor * | IC50 = 1.58 ± 0.17 µM | [38] |
| Rhinacanthin-C | Inhibitor * | IC50 = 7.1 ± 0.81 µM | [38] |
| Rhinacanthin-H/I | Inhibitor * | IC50 = 6.5 ± 1.4 µM | [38] |
| Phenylpropyl isothiocyanate (PPITC) | Inhibitor * | Ki = 0.14 µM | [39] |
| Phenylhexyl isothiocyanate (PHITC) | Inhibitor * | Ki = 1.1 µM | [39] |
| 1-hexyl-1H imidazole B | Inhibitor * | IC50 = 2.1 ± 0.1 µM | [30] |
| Benzyl isothiocyanate (BITC) | Inhibitor * | Ki = 1.3 µM | [40] |
| Phenethyl isothiocyanate (PEITC) | Inhibitor * | Ki = 0.03 µM | [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vrzal, R. Genetic and Enzymatic Characteristics of CYP2A13 in Relation to Lung Damage. Int. J. Mol. Sci. 2021, 22, 12306. https://doi.org/10.3390/ijms222212306
Vrzal R. Genetic and Enzymatic Characteristics of CYP2A13 in Relation to Lung Damage. International Journal of Molecular Sciences. 2021; 22(22):12306. https://doi.org/10.3390/ijms222212306
Chicago/Turabian StyleVrzal, Radim. 2021. "Genetic and Enzymatic Characteristics of CYP2A13 in Relation to Lung Damage" International Journal of Molecular Sciences 22, no. 22: 12306. https://doi.org/10.3390/ijms222212306
APA StyleVrzal, R. (2021). Genetic and Enzymatic Characteristics of CYP2A13 in Relation to Lung Damage. International Journal of Molecular Sciences, 22(22), 12306. https://doi.org/10.3390/ijms222212306