The THO/TREX Complex Active in Alternative Splicing Mediates Plant Responses to Salicylic Acid and Jasmonic Acid
Abstract
:1. Introduction
2. Results
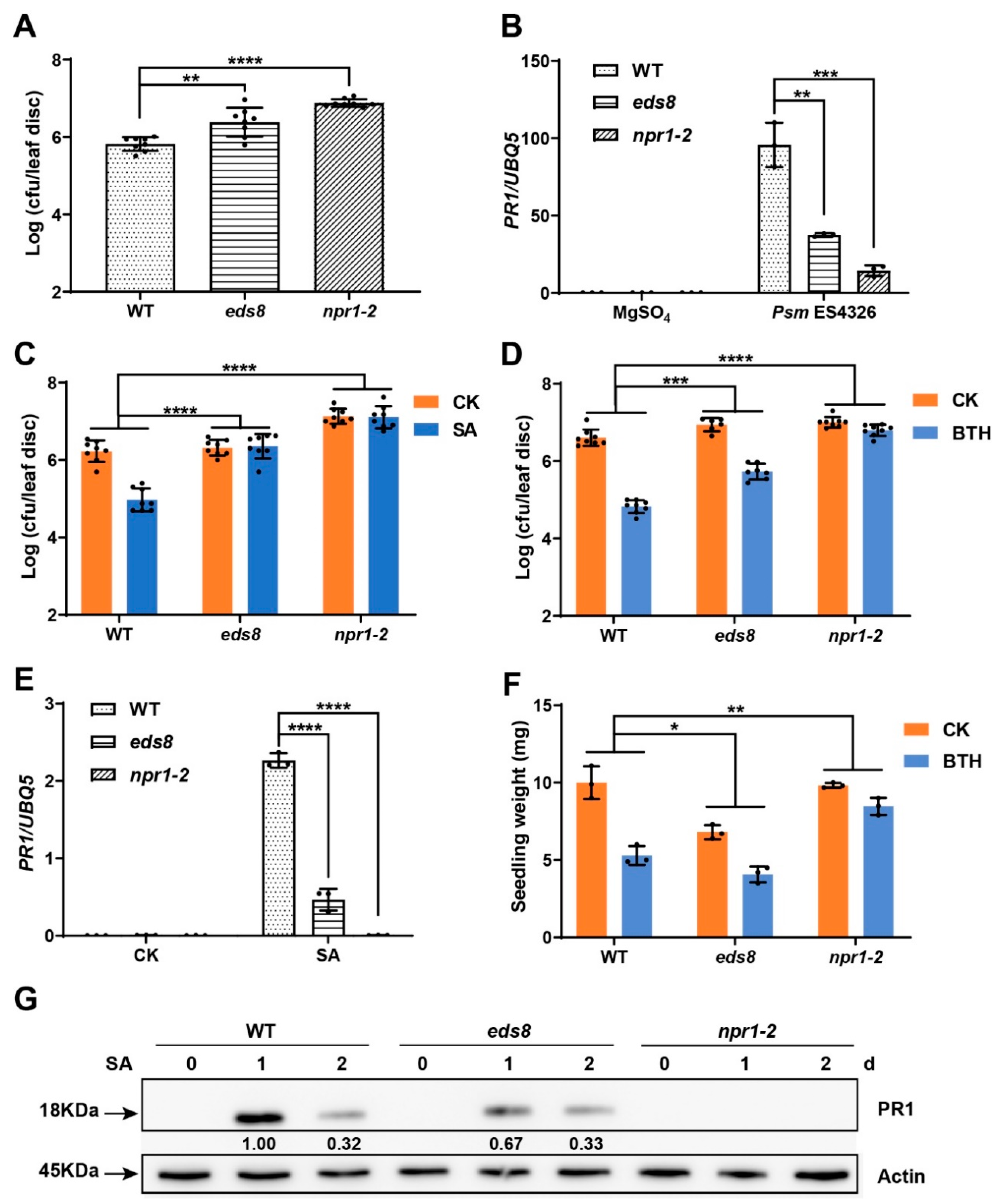
2.1. EDS8 Mutation Compromised Plant Response to SA
2.2. EDS8 Mutation Compromised Plant Response to JA
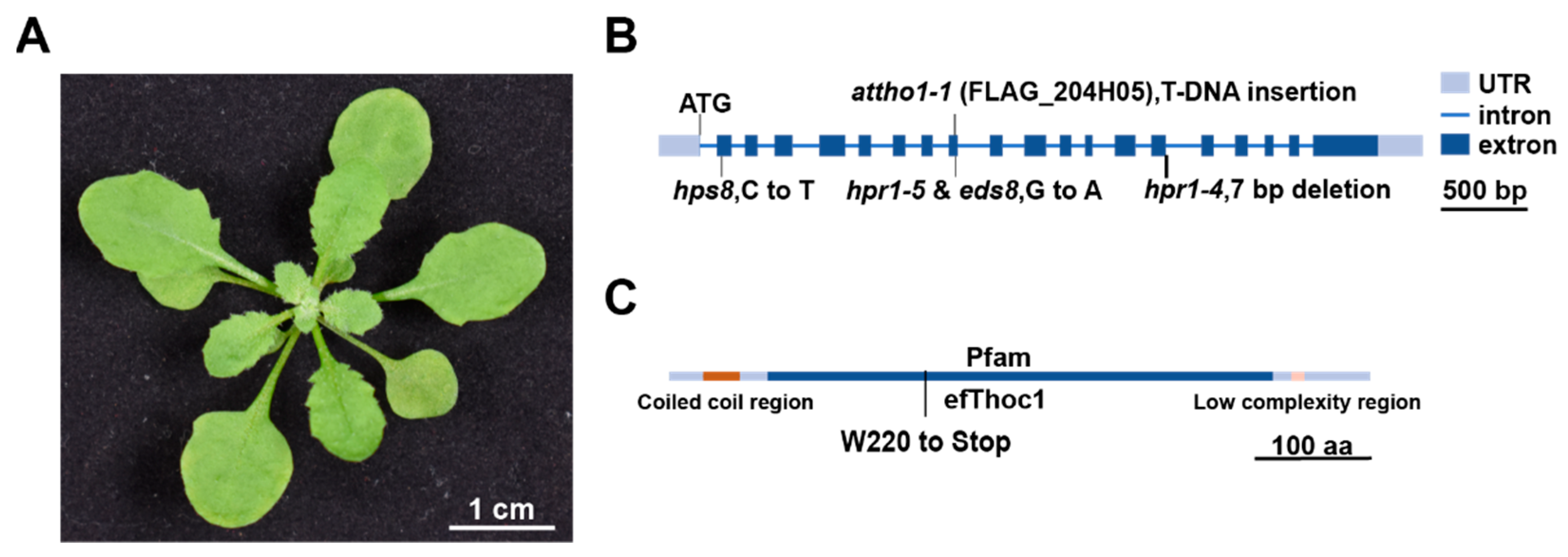
2.3. Identification of EDS8 Gene Using Mapping-by-Sequencing
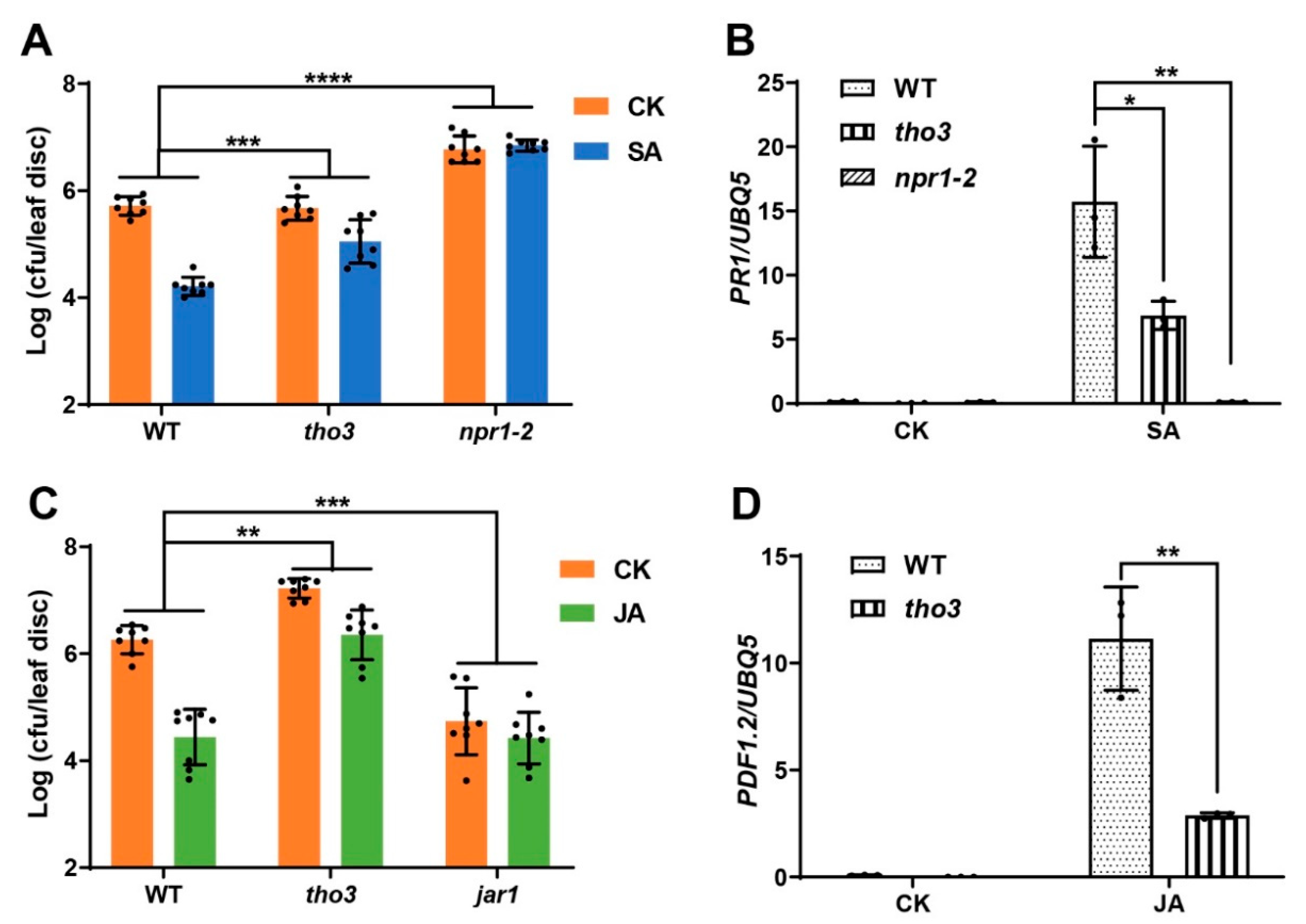
2.4. THO3, Another Subunit of THO/TREX Complex, Positively Regulated Plant Response to SA and JA
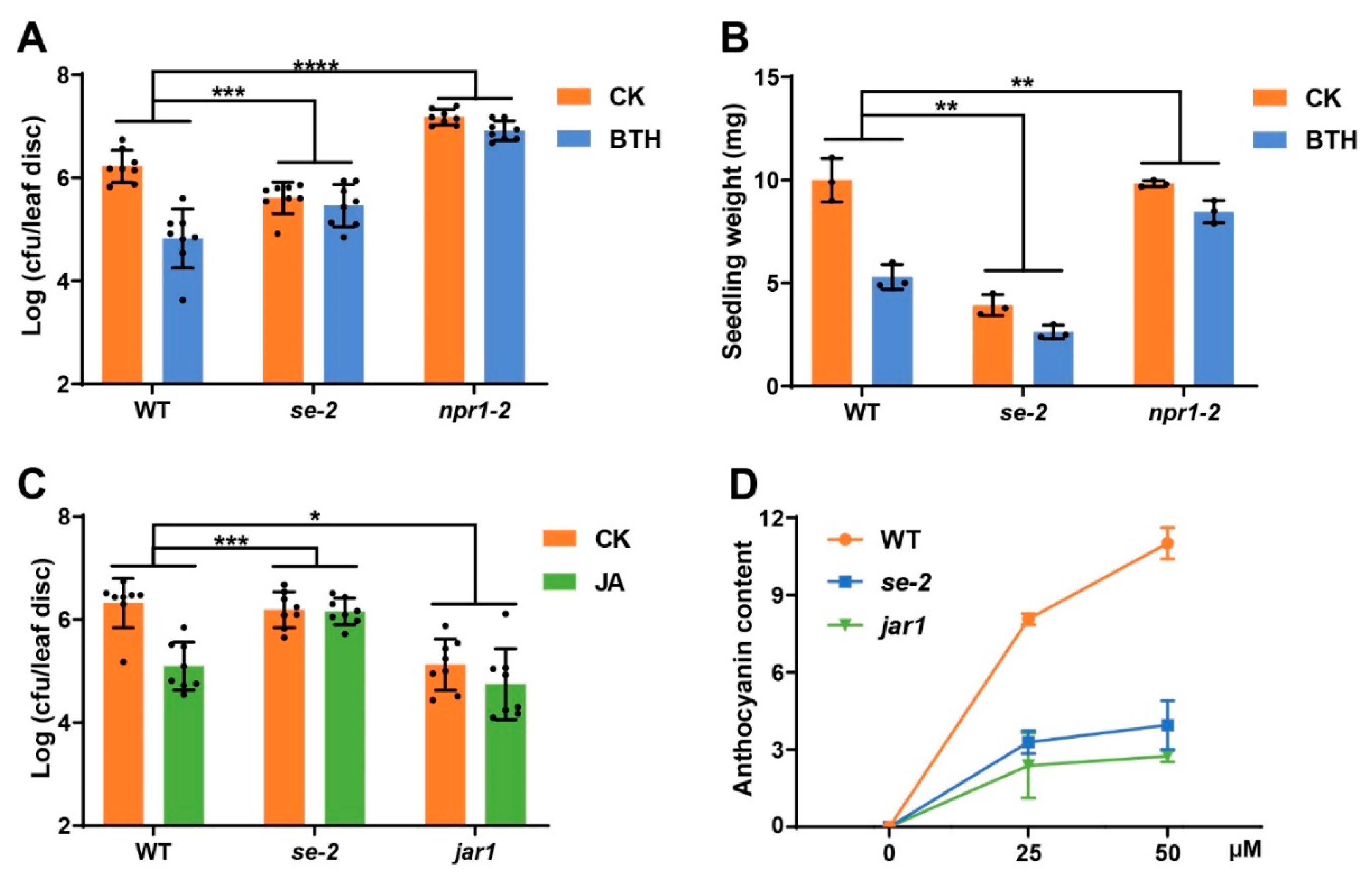
2.5. The THO/TREX Interacting Protein SERRATE Modulated Plant Responses to SA and JA
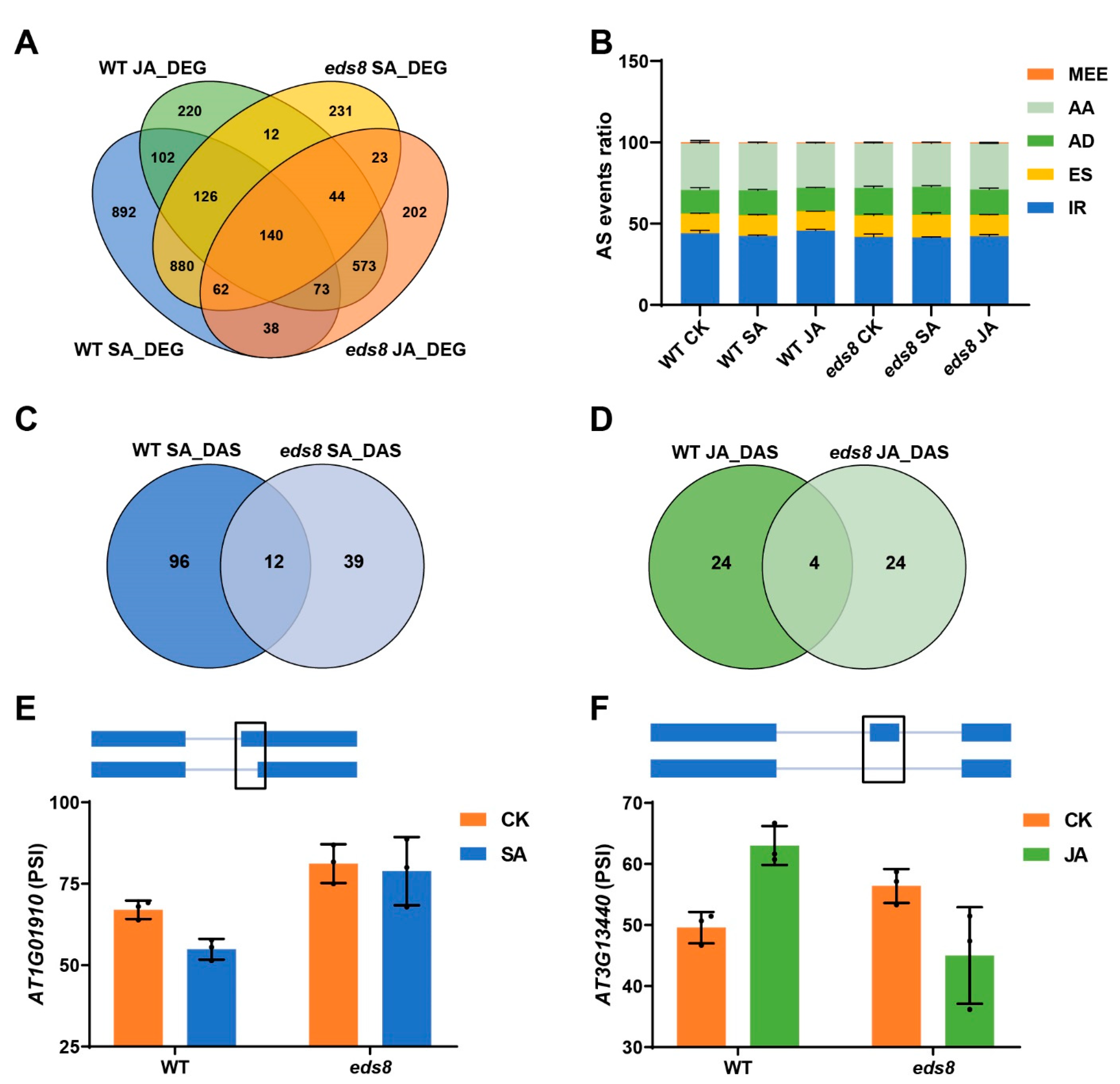
2.6. The SA and JA Induced Different Alternative Splicing Were Dependent on EDS8
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Pathogenic Bacteria Inoculation and Growth Determination
4.3. Botrytis Cinerea Bioassays
4.4. Growth Inhibition Assays Following BTH Treatment
4.5. Anthocyanin Accumulation Assays
4.6. Gene Expression Analyses by qPCR
4.7. Protein Extraction and Immunoblots Analysis
4.8. Mapping-by-Sequencing Analyses
4.9. Transcriptome Sequencing and Alternative Splicing Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ngou, B.P.M.; Jones, J.D.G.; Ding, P. Plant immune networks. Trends Plant Sci. 2021. [Google Scholar] [CrossRef]
- Kim, Y.; Tsuda, K.; Igarashi, D.; Hillmer, R.A.; Sakakibara, H.; Myers, C.L.; Katagiri, F. Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell Host Microbe 2014, 15, 84–94. [Google Scholar] [CrossRef] [Green Version]
- Zhu, F.; Xi, D.H.; Yuan, S.; Xu, F.; Zhang, D.W.; Lin, H.H. Salicylic acid and jasmonic acid are essential for systemic resistance against tobacco mosaic virus in Nicotiana benthamiana. Mol. Plant Microbe Interact. 2014, 27, 567–577. [Google Scholar] [CrossRef] [Green Version]
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef] [Green Version]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic Acid: Biosynthesis and Signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 2018, 173, 1454–1467.e15. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.Y.; Liu, Y.; Liu, L.J. Progress on the biosynthesis and signal transduction of phytohormone salicylic acid. Yi Chuan 2020, 42, 858–869. [Google Scholar] [PubMed]
- Liu, H.; Timko, M.P. Jasmonic Acid Signaling and Molecular Crosstalk with Other Phytohormones. Int. J. Mol. Sci. 2021, 22, 2914. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F.; et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 2009, 21, 2220–2236. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhao, D.; Dong, J.; Kong, X.; Zhang, Q.; Li, T.; Meng, Y.; Shan, W. AtRTP5 negatively regulates plant resistance to Phytophthora pathogens by modulating the biosynthesis of endogenous jasmonic acid and salicylic acid. Mol. Plant Pathol. 2020, 21, 95–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mur, L.A.; Kenton, P.; Atzorn, R.; Miersch, O.; Wasternack, C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 2006, 140, 249–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howlader, P.; Bose, S.K.; Jia, X.; Zhang, C.; Wang, W.; Yin, H. Oligogalacturonides induce resistance in Arabidopsis thaliana by triggering salicylic acid and jasmonic acid pathways against Pst DC3000. Int. J. Biol. Macromol. 2020, 164, 4054–4064. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Sonbol, F.M.; Huot, B.; Gu, Y.; Withers, J.; Mwimba, M.; Yao, J.; He, S.Y.; Dong, X. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 2016, 7, 13099. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, V.; Van der Ent, S.; Garcia-Andrade, J.; Coego, A.; Pieterse, C.M.; Vera, P. OCP3 is an important modulator of NPR1-mediated jasmonic acid-dependent induced defenses in Arabidopsis. BMC Plant Biol. 2010, 10, 199. [Google Scholar] [CrossRef] [Green Version]
- Tamaoki, D.; Seo, S.; Yamada, S.; Kano, A.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Jasmonic acid and salicylic acid activate a common defense system in rice. Plant Signal. Behav. 2013, 8, e24260. [Google Scholar] [CrossRef]
- Glazebrook, J.; Rogers, E.E.; Ausubel, F.M. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 1996, 143, 973–982. [Google Scholar] [CrossRef]
- Kim, K.C.; Fan, B.; Chen, Z. Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol. 2006, 142, 1180–1192. [Google Scholar] [CrossRef] [Green Version]
- Glazebrook, J.; Chen, W.; Estes, B.; Chang, H.S.; Nawrath, C.; Metraux, J.P.; Zhu, T.; Katagiri, F. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 2003, 34, 217–228. [Google Scholar] [CrossRef]
- Ton, J.; De Vos, M.; Robben, C.; Buchala, A.; Metraux, J.P.; Van Loon, L.C.; Pieterse, C.M. Characterization of Arabidopsis enhanced disease susceptibility mutants that are affected in systemically induced resistance. Plant J. 2002, 29, 11–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.F.; Guo, J.; Zhang, Y.; Huang, C.F. The THO/TREX Complex Component RAE2/TEX1 Is Involved in the Regulation of Aluminum Resistance and Low Phosphate Response in Arabidopsis. Front. Plant Sci. 2021, 12, 698443. [Google Scholar] [CrossRef] [PubMed]
- Canet, J.V.; Dobon, A.; Tornero, P. Non-recognition-of-BTH4, an Arabidopsis mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 2012, 24, 4220–4235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowe, H.C.; Walley, J.W.; Corwin, J.; Chan, E.K.; Dehesh, K.; Kliebenstein, D.J. Deficiencies in jasmonate-mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog. 2010, 6, e1000861. [Google Scholar] [CrossRef] [Green Version]
- Thaler, J.S.; Owen, B.; Higgins, V.J. The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 2004, 135, 530–538. [Google Scholar] [CrossRef] [Green Version]
- Staswick, P.E.; Tiryaki, I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 2004, 16, 2117–2127. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [Green Version]
- James, G.V.; Patel, V.; Nordstrom, K.J.; Klasen, J.R.; Salome, P.A.; Weigel, D.; Schneeberger, K. User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 2013, 14, R61. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Zhang, Y.; Gao, H.; Li, S.; Wang, Z.Y.; Huang, C.F. Mutation of HPR1 encoding a component of the THO/TREX complex reduces STOP1 accumulation and aluminium resistance in Arabidopsis thaliana. New Phytol. 2020, 228, 179–193. [Google Scholar] [CrossRef]
- Pan, H.; Liu, S.; Tang, D. HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. Plant J. 2012, 69, 831–843. [Google Scholar] [CrossRef]
- Pan, H.; Liu, S.; Tang, D. The THO/TREX complex functions in disease resistance in Arabidopsis. Plant Signal. Behav. 2012, 7, 422–424. [Google Scholar] [CrossRef] [Green Version]
- Tao, S.; Zhang, Y.; Wang, X.; Xu, L.; Fang, X.; Lu, Z.J.; Liu, D. The THO/TREX Complex Active in miRNA Biogenesis Negatively Regulates Root-Associated Acid Phosphatase Activity Induced by Phosphate Starvation. Plant Physiol. 2016, 171, 2841–2853. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Zhou, X.; Wen, C.K. HYPER RECOMBINATION1 of the THO/TREX complex plays a role in controlling transcription of the REVERSION-TO-ETHYLENE SENSITIVITY1 gene in Arabidopsis. PLoS Genet. 2015, 11, e1004956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doll, S.; Kuhlmann, M.; Rutten, T.; Mette, M.F.; Scharfenberg, S.; Petridis, A.; Berreth, D.C.; Mock, H.P. Accumulation of the coumarin scopolin under abiotic stress conditions is mediated by the Arabidopsis thaliana THO/TREX complex. Plant J. 2018, 93, 431–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raczynska, K.D.; Stepien, A.; Kierzkowski, D.; Kalak, M.; Bajczyk, M.; McNicol, J.; Simpson, C.G.; Szweykowska-Kulinska, Z.; Brown, J.W.S.; Jarmolowski, A. The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2014, 42, 1224–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajczyk, M.; Lange, H.; Bielewicz, D.; Szewc, L.; Bhat, S.S.; Dolata, J.; Kuhn, L.; Szweykowska-Kulinska, Z.; Gagliardi, D.; Jarmolowski, A. SERRATE interacts with the nuclear exosome targeting (NEXT) complex to degrade primary miRNA precursors in Arabidopsis. Nucleic Acids Res. 2020, 48, 6839–6854. [Google Scholar] [CrossRef] [PubMed]
- Furumizu, C.; Tsukaya, H.; Komeda, Y. Characterization of EMU, the Arabidopsis homolog of the yeast THO complex member HPR1. RNA 2010, 16, 1809–1817. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, B.B.; Ehrnsberger, H.F.; Esposito, S.; Pfab, A.; Bruckmann, A.; Hauptmann, J.; Meister, G.; Merkl, R.; Schubert, T.; Langst, G.; et al. The Arabidopsis THO/TREX component TEX1 functionally interacts with MOS11 and modulates mRNA export and alternative splicing events. Plant Mol. Biol. 2017, 93, 283–298. [Google Scholar] [CrossRef]
- Dong, Z.; Han, M.H.; Fedoroff, N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. USA 2008, 105, 9970–9975. [Google Scholar] [CrossRef] [Green Version]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [Green Version]
- Kouzai, Y.; Kimura, M.; Watanabe, M.; Kusunoki, K.; Osaka, D.; Suzuki, T.; Matsui, H.; Yamamoto, M.; Ichinose, Y.; Toyoda, K.; et al. Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon. New Phytol. 2018, 217, 771–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, C.; Tsai, C.J.; Unsicker, S.B.; Xue, L.; Reichelt, M.; Gershenzon, J.; Hammerbacher, A. Salicylic acid activates poplar defense against the biotrophic rust fungus Melampsora larici-populina via increased biosynthesis of catechin and proanthocyanidins. New Phytol. 2019, 221, 960–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courbier, S.; Snoek, B.L.; Kajala, K.; Li, L.; van Wees, S.C.M.; Pierik, R. Mechanisms of far-red light-mediated dampening of defense against Botrytis cinerea in tomato leaves. Plant Physiol. 2021, 187, 1250–1266. [Google Scholar] [CrossRef]
- Selig, P.; Keough, S.; Nalam, V.J.; Nachappa, P. Jasmonate-dependent plant defenses mediate soybean thrips and soybean aphid performance on soybean. Arthropod Plant Interact. 2016, 10, 273–282. [Google Scholar] [CrossRef]
- Jauvion, V.; Elmayan, T.; Vaucheret, H. The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell 2010, 22, 2697–2709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, G.A.; Deforges, J.; Reis, R.S.; Hsieh, Y.F.; Montpetit, J.; Antosz, W.; Santuari, L.; Hardtke, C.S.; Grasser, K.D.; Poirier, Y. The transcription and export complex THO/TREX contributes to transcription termination in plants. PLoS Genet. 2020, 16, e1008732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yelina, N.E.; Smith, L.M.; Jones, A.M.; Patel, K.; Kelly, K.A.; Baulcombe, D.C. Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 13948–13953. [Google Scholar] [CrossRef] [Green Version]
- Germain, H.; Qu, N.; Cheng, Y.T.; Lee, E.; Huang, Y.; Dong, O.X.; Gannon, P.; Huang, S.; Ding, P.; Li, Y.; et al. MOS11: A new component in the mRNA export pathway. PLoS Genet. 2010, 6, e1001250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Wang, J.; Tung, J.; Liu, D.; Zhou, Y.; He, S.; Du, Y.; Baker, B.; Li, F. A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 2018, 14, e1006756. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Lee, J.H.; Kim, J.Y.; Park, C.M. Alternative RNA Splicing Expands the Developmental Plasticity of Flowering Transition. Front. Plant Sci. 2019, 10, 606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, X.; Jiao, F.; Xu, H.; Jiao, Q.; Zhang, X.; Zhang, Y.; Du, S.; Xi, M.; Wang, A.; Chen, J.; et al. The role of RNA-binding protein, microRNA and alternative splicing in seed germination: A field need to be discovered. BMC Plant Biol. 2021, 21, 194. [Google Scholar] [CrossRef] [PubMed]
- Rigo, R.; Bazin, J.R.M.; Crespi, M.; Charon, C.L. Alternative Splicing in the Regulation of Plant-Microbe Interactions. Plant Cell Physiol. 2019, 60, 1906–1916. [Google Scholar] [CrossRef] [PubMed]
- Laloum, T.; Martin, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.X.; Chen, M.X.; Zhu, F.Y.; Fan, T.; Zhang, J.H.; Lo, C. Alternative splicing is a Sorghum bicolor defense response to fungal infection. Planta 2020, 251, 14. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Q.; Chen, X.J.; Liang, X.X.; Zhou, X.G.; Yang, F.; Liu, J.; He, S.Y.; Guo, Z.J. Alternative Splicing of Rice WRKY62 and WRKY76 Transcription Factor Genes in Pathogen Defense. Plant Physiol. 2016, 171, 1427–1442. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Liu, H.; Zhuang, H.F.; Zhao, C.X.; Xu, Y.X.; Wu, S.B.; Qi, J.F.; Li, J.; Hettenhausen, C.; Wu, J.Q. Transcriptomics and Alternative Splicing Analyses Reveal Large Differences between Maize Lines B73 and Mo17 in Response to Aphid Rhopalosiphum padi Infestation. Front. Plant Sci. 2017, 8, 1738. [Google Scholar] [CrossRef] [Green Version]
- Nwugo, C.C.; Sengoda, V.G.; Tian, L.; Lin, H. Characterization of physiological and molecular processes associated with potato response to Zebra chip disease. Hortic. Res. Engl. 2017, 4, 17069. [Google Scholar] [CrossRef] [Green Version]
- Bedre, R.; Irigoyen, S.; Schaker, P.D.C.; Monteiro-Vitorello, C.B.; Da Silva, J.A.; Mandadi, K.K. Genome-wide alternative splicing landscapes modulated by biotrophic sugarcane smut pathogen. Sci. Rep. UK 2019, 9, 8876. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Mao, R.; Wang, Y.Z.; Zhang, L.; Wang, C.Y.; Lv, S.K.; Liu, X.L.; Wang, Y.J.; Ji, W.Q. Transcriptome-wide alternative splicing modulation during plant-pathogen interactions in wheat. Plant Sci. 2019, 288, 110160. [Google Scholar] [CrossRef]
- Bazin, J.; Mariappan, K.; Jiang, Y.H.; Blein, T.; Voelz, R.; Crespi, M.; Hirt, H. Role of MPK4 in pathogen-associated molecular pattern-triggered alternative splicing in Arabidopsis. PLoS Pathog. 2020, 16, e1008401. [Google Scholar] [CrossRef]
- Dressano, K.; Weckwerth, P.R.; Poretsky, E.; Takahashi, Y.; Villarreal, C.; Shen, Z.X.; Schroeder, J.I.; Briggs, S.P.; Huffaker, A. Dynamic regulation of Pep-induced immunity through post-translational control of defence transcript splicing. Nat. Plants 2020, 6, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.B.; Liu, Y.A.; Ding, P.T.; Li, Y.; Kong, Q.; Zhang, Y.L. Splicing of Receptor-Like Kinase-Encoding SNC4 and CERK1 is Regulated by Two Conserved Splicing Factors that Are Required for Plant Immunity. Mol. Plant 2014, 7, 1766–1775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, H.S.; Cooke, T.F.; Depew, C.L.; Patel, L.C.; Ogawa, N.; Kobayashi, Y.; Howe, G.A. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 2010, 63, 613–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Deng, L.; Zhai, Q.; Zhao, J.; Chen, Q.; Li, C. Mediator Subunit MED25 Couples Alternative Splicing of JAZ Genes with Fine-Tuning of Jasmonate Signaling. Plant Cell 2020, 32, 429–448. [Google Scholar] [CrossRef] [Green Version]
- Feng, G.; Yoo, M.J.; Davenport, R.; Boatwright, J.L.; Koh, J.; Chen, S.; Barbazuk, W.B. Jasmonate induced alternative splicing responses in Arabidopsis. Plant Direct 2020, 4, e00245. [Google Scholar] [CrossRef]
- Zhang, J.K.; Jiao, P.; Zhang, C.; Tong, X.L.; Wei, Q.P.; Xu, L.F. Apple NPR1 homologs and their alternative splicing forms may contribute to SA and disease responses. Tree Genet. Genomes 2016, 12, 92. [Google Scholar] [CrossRef]
- Liu, Y.; Li, M.; Li, T.; Chen, Y.; Zhang, L.; Zhao, G.; Zhuang, J.; Zhao, W.; Gao, L.; Xia, T. Airborne fungus-induced biosynthesis of anthocyanins in Arabidopsis thaliana via jasmonic acid and salicylic acid signaling. Plant Sci. 2020, 300, 110635. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, N.; Kong, X.; Liu, Y.; Gong, T.; Gu, X.; Liu, L. The THO/TREX Complex Active in Alternative Splicing Mediates Plant Responses to Salicylic Acid and Jasmonic Acid. Int. J. Mol. Sci. 2021, 22, 12197. https://doi.org/10.3390/ijms222212197
Sun N, Kong X, Liu Y, Gong T, Gu X, Liu L. The THO/TREX Complex Active in Alternative Splicing Mediates Plant Responses to Salicylic Acid and Jasmonic Acid. International Journal of Molecular Sciences. 2021; 22(22):12197. https://doi.org/10.3390/ijms222212197
Chicago/Turabian StyleSun, Nengxu, Xiangjiu Kong, Yueyan Liu, Tingting Gong, Xiaoyong Gu, and Lijing Liu. 2021. "The THO/TREX Complex Active in Alternative Splicing Mediates Plant Responses to Salicylic Acid and Jasmonic Acid" International Journal of Molecular Sciences 22, no. 22: 12197. https://doi.org/10.3390/ijms222212197
APA StyleSun, N., Kong, X., Liu, Y., Gong, T., Gu, X., & Liu, L. (2021). The THO/TREX Complex Active in Alternative Splicing Mediates Plant Responses to Salicylic Acid and Jasmonic Acid. International Journal of Molecular Sciences, 22(22), 12197. https://doi.org/10.3390/ijms222212197






