Ets1 Promotes the Differentiation of Post-Selected iNKT Cells through Regulation of the Expression of Vα14Jα18 T Cell Receptor and PLZF
Abstract
:1. Introduction
2. Results
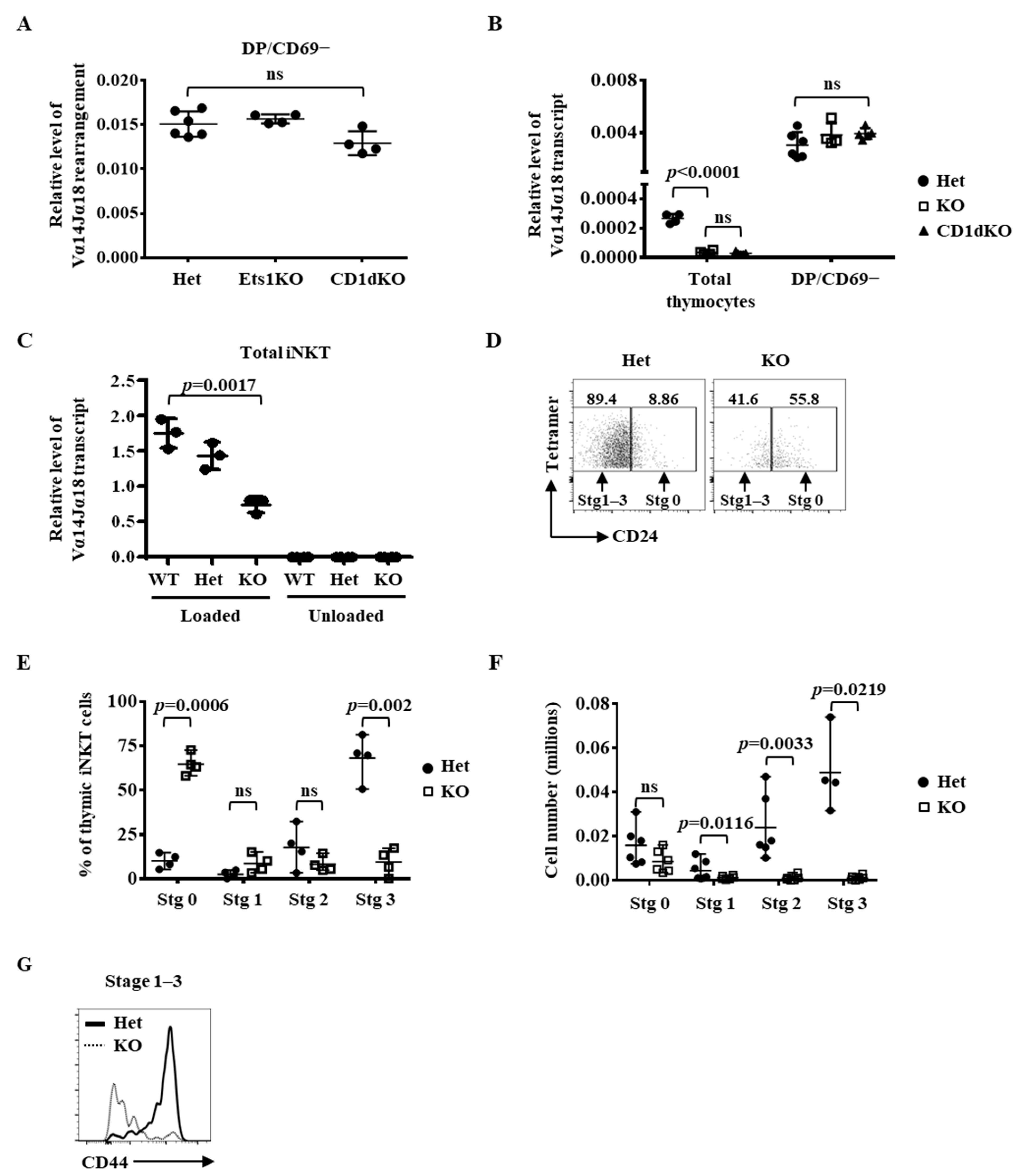
2.1. Regulation of the Immediate Differentiation of Post-Selected iNKT Cells by Ets1
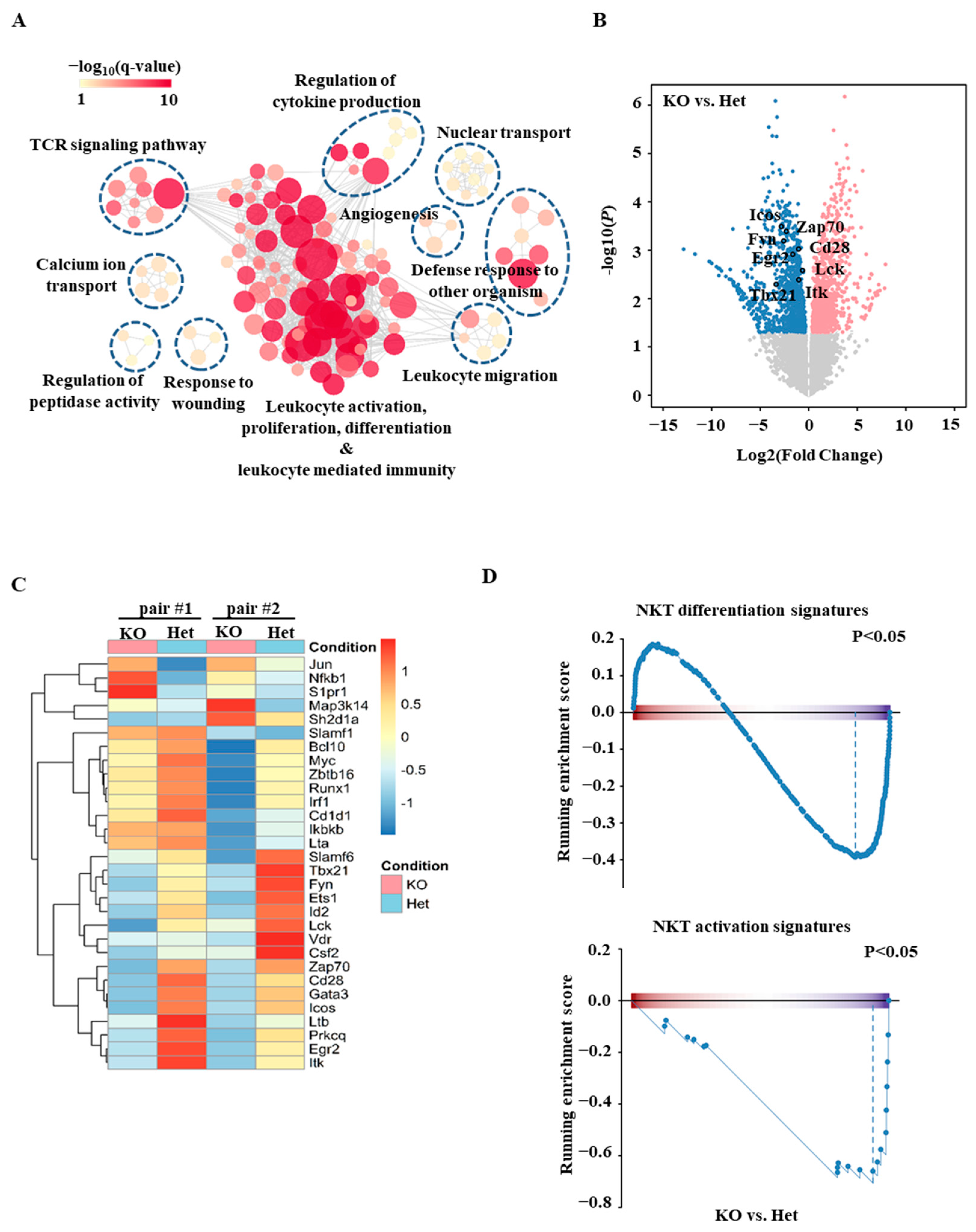
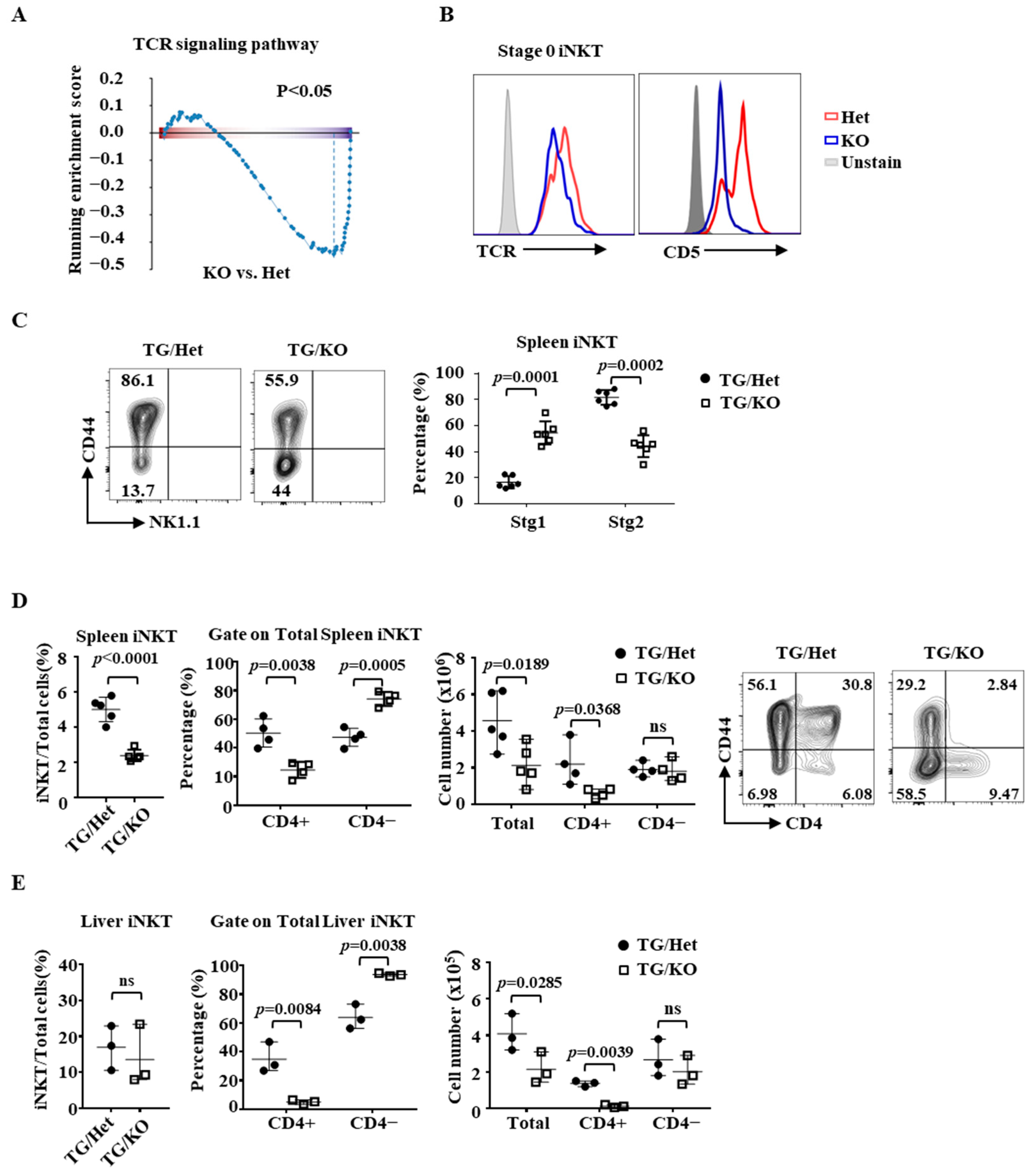
2.2. Transcriptomic Analyses of Ets1KO Thymic iNKT Cells
2.3. Identification of ICOS as a Target Gene of Ets1
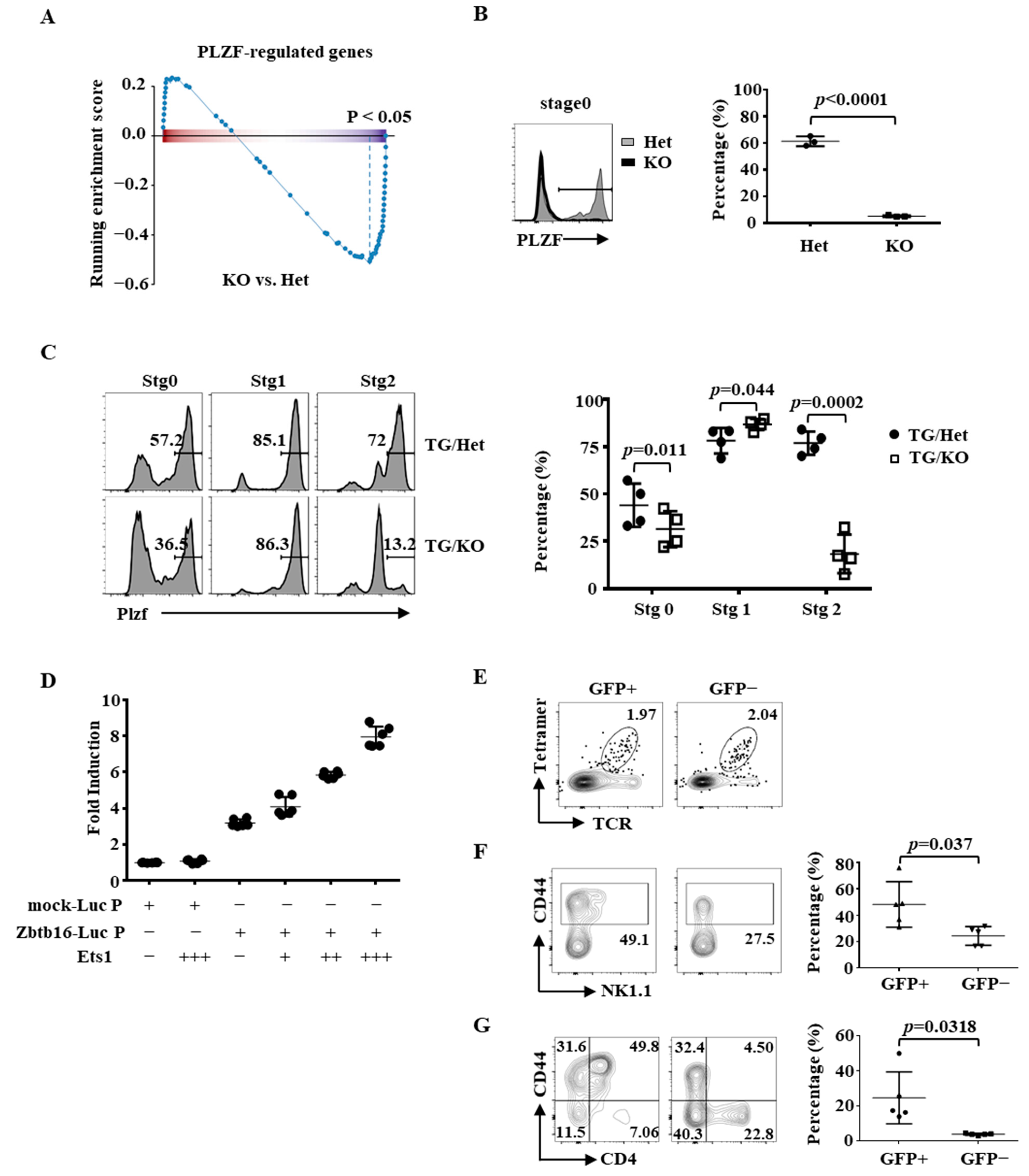
2.4. Partial Rescue of the Differentiation of Ets1KO iNKT Cells by PLZF
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Real-Time PCR
4.3. FACS Analysis and Antibodies
4.4. iNKT Cell Enrichment
4.5. RNA Sequencing of Thymic Stage 0 NKT Cells
4.6. Plasmid, Transfection, and Luciferase Assay
4.7. Preparation of Splenic and Liver iNKT Cells
4.8. Bone Marrow Transfer for PLZF and ICOS Restoration
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godfrey, D.I.; Stankovic, S.; Baxter, A.G. Raising the NKT cell family. Nat. Immunol. 2010, 11, 197–206. [Google Scholar] [CrossRef]
- Bendelac, A.; Savage, P.B.; Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 2007, 25, 297–336. [Google Scholar] [CrossRef] [Green Version]
- Gapin, L.; Matsuda, J.L.; Surh, C.D.; Kronenberg, M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2001, 2, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Bendelac, A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 1995, 182, 2091–2096. [Google Scholar] [CrossRef] [Green Version]
- Benlagha, K.; Kyin, T.; Beavis, A.; Teyton, L.; Bendelac, A. A thymic precursor to the NK T cell lineage. Science 2002, 296, 553–555. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, J.L.; Gapin, L. Developmental program of mouse Valpha14i NKT cells. Curr. Opin. Immunol. 2005, 17, 122–130. [Google Scholar] [CrossRef]
- Bennstein, S.B. Unraveling Natural Killer T-Cells Development. Front. Immunol. 2017, 8, 1950. [Google Scholar] [CrossRef] [Green Version]
- Cohen, N.R.; Garg, S.; Brenner, M.B. Antigen Presentation by CD1 Lipids, T Cells, and NKT Cells in Microbial Immunity. Adv. Immunol. 2009, 102, 1–94. [Google Scholar] [PubMed]
- Brennan, P.J.; Brigl, M.; Brenner, M.B. Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol. 2013, 13, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhang, H.; Bai, L. NKT cells in liver diseases. Front. Med. 2018, 12, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, S.; Visvabharathy, L.; Wang, C.R. Role of Group 1 CD1-Restricted T Cells in Infectious Disease. Front. Immunol. 2015, 6, 337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Kaer, L.; Wu, L.; Parekh, V.V. Natural killer T cells in multiple sclerosis and its animal model, experimental autoimmune encephalomyelitis. Immunology 2015, 146, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwamura, C.; Nakayama, T. Role of CD1d- and MR1-Restricted T Cells in Asthma. Front. Immunol. 2018, 9, 1942. [Google Scholar] [CrossRef] [Green Version]
- Walunas, T.L.; Wang, B.; Wang, C.R.; Leiden, J.M. Cutting edge: The Ets1 transcription factor is required for the development of NK T cells in mice. J. Immunol. 2000, 164, 2857–2860. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.J.; Geng, Y.; Cho, H.; Li, S.; Giri, P.K.; Felio, K.; Wang, C.R. Differential requirements for the Ets transcription factor Elf-1 in the development of NKT cells and NK cells. Blood 2011, 117, 1880–1887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stankovic, S.; Gugasyan, R.; Kyparissoudis, K.; Grumont, R.; Banerjee, A.; Tsichlis, P.; Gerondakis, S.; Godfrey, D.I. Distinct roles in NKT cell maturation and function for the different transcription factors in the classical NF-kappaB pathway. Immunol. Cell Biol. 2011, 89, 294–303. [Google Scholar] [CrossRef]
- Kim, E.Y.; Lynch, L.; Brennan, P.J.; Cohen, N.R.; Brenner, M.B. The transcriptional programs of iNKT cells. Semin. Immunol. 2015, 27, 26–32. [Google Scholar] [CrossRef]
- Lazarevic, V.; Zullo, A.J.; Schweitzer, M.N.; Staton, T.L.; Gallo, E.M.; Crabtree, G.R.; Glimcher, L.H. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat. Immunol. 2009, 10, 306–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsao, H.W.; Tai, T.S.; Tseng, W.; Chang, H.H.; Grenningloh, R.; Miaw, S.C.; Ho, I.C. Ets-1 facilitates nuclear entry of NFAT proteins and their recruitment to the IL-2 promoter. Proc. Natl. Acad. Sci. USA 2013, 110, 15776–15781. [Google Scholar] [CrossRef] [Green Version]
- Grenningloh, R.; Kang, B.Y.; Ho, I.C. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J. Exp. Med. 2005, 201, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.G.; Kwon, H.K.; Sahoo, A.; Hwang, W.; So, J.S.; Hwang, J.S.; Chae, C.S.; Kim, G.C.; Kim, J.E.; So, H.S.; et al. Interaction of Ets-1 with HDAC1 represses IL-10 expression in Th1 cells. J. Immunol. 2012, 188, 2244–2253. [Google Scholar] [CrossRef] [Green Version]
- Strempel, J.M.; Grenningloh, R.; Ho, I.C.; Vercelli, D. Phylogenetic and functional analysis identifies Ets-1 as a novel regulator of the Th2 cytokine gene locus. J. Immunol. 2010, 184, 1309–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.J.; Lee, C.G.; Jung, J.Y.; Ghosh, A.; Hasan, S.N.; Hwang, S.M.; Kang, H.; Lee, C.; Kim, G.C.; Rudra, D.; et al. The Transcription Factor Ets1 Suppresses T Follicular Helper Type 2 Cell Differentiation to Halt the Onset of Systemic Lupus Erythematosus. Immunity 2018, 49, 1034–1048 e8. [Google Scholar] [CrossRef] [Green Version]
- Zamisch, M.; Tian, L.; Grenningloh, R.; Xiong, Y.; Wildt, K.F.; Ehlers, M.; Ho, I.C.; Bosselut, R. The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J. Exp. Med. 2009, 206, 2685–2699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grenningloh, R.; Tai, T.S.; Frahm, N.; Hongo, T.C.; Chicoine, A.T.; Brander, C.; Kaufmann, D.E.; Ho, I.C. Ets-1 maintains IL-7 receptor expression in peripheral T cells. J. Immunol. 2011, 186, 969–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; John, S.A.; Clements, J.L.; Percy, D.H.; Barton, K.P.; Garrett-Sinha, L.A. Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease. Int. Immunol. 2005, 17, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Eyquem, S.; Chemin, K.; Fasseu, M.; Bories, J.C. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor beta locus. Proc. Natl. Acad. Sci. USA 2004, 101, 15712–15717. [Google Scholar] [CrossRef] [Green Version]
- John, S.A.; Clements, J.L.; Russell, L.M.; Garrett-Sinha, L.A. Ets-1 regulates plasma cell differentiation by interfering with the activity of the transcription factor Blimp-1. J. Biol. Chem. 2008, 283, 951–962. [Google Scholar] [CrossRef] [Green Version]
- Zook, E.C.; Ramirez, K.; Guo, X.; van der Voort, G.; Sigvardsson, M.; Svensson, E.C.; Fu, Y.X.; Kee, B.L. The ETS1 transcription factor is required for the development and cytokine-induced expansion of ILC2. J. Exp. Med. 2016, 213, 687–696. [Google Scholar] [CrossRef]
- Han, J.W.; Zheng, H.F.; Cui, Y.; Sun, L.D.; Ye, D.Q.; Hu, Z.; Xu, J.H.; Cai, Z.M.; Huang, W.; Zhao, G.P.; et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009, 41, 1234–1237. [Google Scholar] [CrossRef]
- Yang, W.; Shen, N.; Ye, D.Q.; Liu, Q.; Zhang, Y.; Qian, X.X.; Hirankarn, N.; Ying, D.; Pan, H.F.; Mok, C.C.; et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010, 6, e1000841. [Google Scholar] [CrossRef] [Green Version]
- Tai, T.S.; Tsao, H.W.; Chuang, W.C.; Liu, C.C.; Huang, Y.W.; Oettgen, P.; Chuang, Y.T.; Ho, I.C. Ets1 regulates the differentiation and function of iNKT cells through both Pointed domain-dependent and domain-independent mechanisms. Cell Mol. Immunol. 2020, 17, 1198–1200. [Google Scholar] [CrossRef]
- Akbari, O.; Stock, P.; Meyer, E.H.; Freeman, G.J.; Sharpe, A.H.; Umetsu, D.T.; DeKruyff, R.H. ICOS/ICOSL interaction is required for CD4+ invariant NKT cell function and homeostatic survival. J. Immunol. 2008, 180, 5448–5456. [Google Scholar] [CrossRef] [Green Version]
- Savage, A.K.; Constantinides, M.G.; Han, J.; Picard, D.; Martin, E.; Li, B.; Lantz, O.; Bendelac, A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 2008, 29, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.Y.; Petryniak, B.; Ho, I.C.; Thompson, C.B.; Leiden, J.M. Evolutionarily conserved Ets family members display distinct DNA binding specificities. J. Exp. Med. 1992, 175, 1391–1399. [Google Scholar] [CrossRef] [Green Version]
- Muthusamy, N.; Barton, K.; Leiden, J.M. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature 1995, 377, 639–642. [Google Scholar] [CrossRef]
- Kreslavsky, T.; Savage, A.K.; Hobbs, R.; Gounari, F.; Bronson, R.; Pereira, P.; Pandolfi, P.P.; Bendelac, A.; von Boehmer, H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc. Natl. Acad. Sci. USA 2009, 106, 12453–12458. [Google Scholar] [CrossRef] [Green Version]
- Mao, A.P.; Ishizuka, I.E.; Kasal, D.N.; Mandal, M.; Bendelac, A. A shared Runx1-bound Zbtb16 enhancer directs innate and innate-like lymphoid lineage development. Nat. Commun. 2017, 8, 863. [Google Scholar] [CrossRef] [Green Version]
- Tani-ichi, S.; Shimba, A.; Wagatsuma, K.; Miyachi, H.; Kitano, S.; Imai, K.; Hara, T.; Ikuta, K. Interleukin-7 receptor controls development and maturation of late stages of thymocyte subpopulations. Proc. Natl. Acad. Sci. USA 2013, 110, 612–617. [Google Scholar] [CrossRef] [Green Version]
- Griewank, K.; Borowski, C.; Rietdijk, S.; Wang, N.; Julien, A.; Wei, D.G.; Mamchak, A.A.; Terhorst, C.; Bendelac, A. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity 2007, 27, 751–762. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chuang, Y.-T.; Chuang, W.-C.; Liu, C.-C.; Liu, C.-W.; Huang, Y.-W.; Yang, H.-Y.; Ho, I.-C.; Tai, T.-S. Ets1 Promotes the Differentiation of Post-Selected iNKT Cells through Regulation of the Expression of Vα14Jα18 T Cell Receptor and PLZF. Int. J. Mol. Sci. 2021, 22, 12199. https://doi.org/10.3390/ijms222212199
Chuang Y-T, Chuang W-C, Liu C-C, Liu C-W, Huang Y-W, Yang H-Y, Ho I-C, Tai T-S. Ets1 Promotes the Differentiation of Post-Selected iNKT Cells through Regulation of the Expression of Vα14Jα18 T Cell Receptor and PLZF. International Journal of Molecular Sciences. 2021; 22(22):12199. https://doi.org/10.3390/ijms222212199
Chicago/Turabian StyleChuang, Ya-Ting, Wan-Chu Chuang, Chih-Chun Liu, Chia-Wei Liu, Yu-Wen Huang, Huang-Yu Yang, I-Cheng Ho, and Tzong-Shyuan Tai. 2021. "Ets1 Promotes the Differentiation of Post-Selected iNKT Cells through Regulation of the Expression of Vα14Jα18 T Cell Receptor and PLZF" International Journal of Molecular Sciences 22, no. 22: 12199. https://doi.org/10.3390/ijms222212199
APA StyleChuang, Y.-T., Chuang, W.-C., Liu, C.-C., Liu, C.-W., Huang, Y.-W., Yang, H.-Y., Ho, I.-C., & Tai, T.-S. (2021). Ets1 Promotes the Differentiation of Post-Selected iNKT Cells through Regulation of the Expression of Vα14Jα18 T Cell Receptor and PLZF. International Journal of Molecular Sciences, 22(22), 12199. https://doi.org/10.3390/ijms222212199





