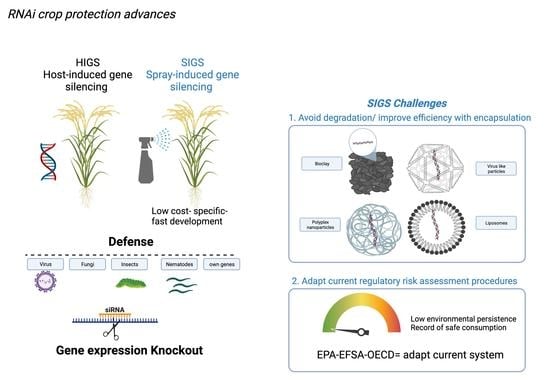
RNAi Crop Protection Advances
Abstract
:1. Introduction
2. How it Works
3. Potential Targets
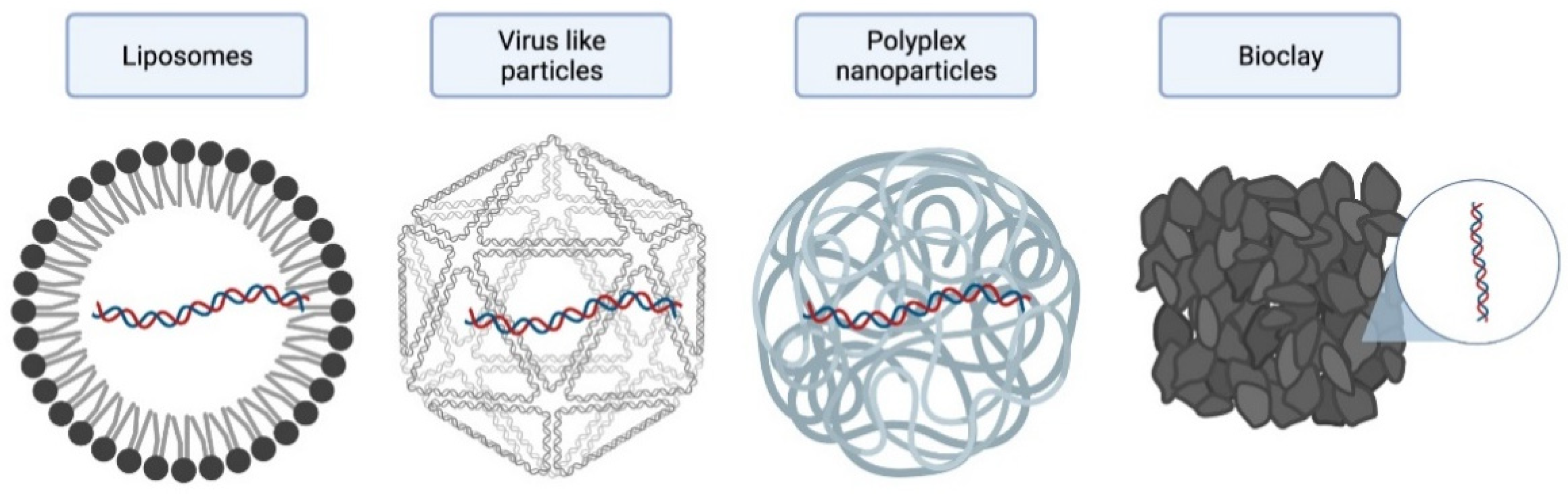
4. Encapsulation Technology to Improve Efficiency
4.1. Liposomes
4.2. Virus Like-Particles (VLPs)
4.3. Polyplex Nanoparticles
4.4. Bioclays
5. Regulatory Approaches
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hernández-Soto, A.; Echeverría-Beirute, F.; Guzmán-Hernández, T. The RNAi as a tool to control tropical pathogens. Agron. Mesoam. 2021, 32, 326–337. [Google Scholar] [CrossRef]
- Yogindran, S.; Rajam, M.V. RNAi for Crop Improvement Sneha. In Plant Biology and Biotechnology; Bahadur, B., Rajam, M.V., Sahijram, L., Krishnamurthy, K.V., Eds.; Springer: New Delhi, India, 2015; Volume II, pp. 623–637. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Lin, F.-M.; Thomma, B.P.H.J.; Huang, H.-D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 1–10. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.; Kogel, K.-H.; Jin, H. Cross-kingdom RNA trafficking and environmental RNAi—Nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 2018, 46, 58–64. [Google Scholar] [CrossRef]
- Castillo-González, C.; Zhang, X. The Trojan Horse of the Plant Kingdom. Cell Host Microbe 2018, 24, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Liang, X.; Zhou, J.-M. Small RNA trafficking at the forefront of plant–pathogen interactions. F1000Research 2018, 7, 1633. [Google Scholar] [CrossRef]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.-M.; Palmquist, J.; Huang, S.-D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiberg, A.; Wang, M.; Lin, F.-M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.-D.; Jin, H. Fungal Small RNAs Suppress Plant Immunity by Hijacking Host RNA Interference Pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [Green Version]
- Giudice, G.; Moffa, L.; Varotto, S.; Cardone, M.F.; Bergamini, C.; De Lorenzis, G.; Velasco, R.; Nerva, L.; Chitarra, W. Novel and emerging biotechnological crop protection approaches. Plant Biotechnol. J. 2021, 19, 1495–1510. [Google Scholar] [CrossRef] [PubMed]
- Taning, C.N.T.; Arpaia, S.; Christiaens, O.; Dietz-Pfeilstetter, A.; Jones, H.; Mezzetti, B.; Sabbadini, S.; Sorteberg, H.; Sweet, J.; Ventura, V.; et al. RNA-based biocontrol compounds: Current status and perspectives to reach the market. Pest Manag. Sci. 2020, 76, 841–845. [Google Scholar] [CrossRef]
- Rodrigues, T.B.; Petrick, J.S. Safety Considerations for Humans and Other Vertebrates Regarding Agricultural Uses of Externally Applied RNA Molecules. Front. Plant Sci. 2020, 11, 407. [Google Scholar] [CrossRef] [Green Version]
- Zotti, M.; dos Santos, E.A.; Cagliari, D.; Christiaens, O.; Taning, C.N.T.; Smagghe, G. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag. Sci. 2018, 74, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Mezzetti, B.; Smagghe, G.; Arpaia, S.; Christiaens, O.; Dietz-Pfeilstetter, A.; Jones, H.; Kostov, K.; Sabbadini, S.; Opsahl-Sorteberg, H.-G.; Ventura, V.; et al. RNAi: What is its position in agriculture? J. Pest Sci. 2020, 93, 1125–1130. [Google Scholar] [CrossRef]
- Rutter, B.; Innes, R.W. Extracellular vesicles as key mediators of plant–microbe interactions. Curr. Opin. Plant Biol. 2018, 44, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Song, X.-S.; Gu, K.-X.; Duan, X.-X.; Xiao, X.-M.; Hou, Y.-P.; Duan, Y.-B.; Wang, J.-X.; Yu, N.; Zhou, M.-G. Secondary amplification of siRNA machinery limits the application of spray-induced gene silencing. Mol. Plant Pathol. 2018, 19, 2543–2560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reagan, B.C.; Ganusova, E.E.; Fernandez, J.C.; McCray, T.N.; Burch-Smith, T.M. RNA on the move: The plasmodesmata perspective. Plant Sci. 2018, 275, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wytinck, N.; Manchur, C.L.; Li, V.H.; Whyard, S.; Belmonte, M.F. dsRNA Uptake in Plant Pests and Pathogens: Insights into RNAi-Based Insect and Fungal Control Technology. Plants 2020, 9, 1780. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Wilson, R.C.; Doudna, J.A. Molecular Mechanisms of RNA Interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef] [Green Version]
- Axtell, M.J. Classification and Comparison of Small RNAs from Plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef] [Green Version]
- Muthamilarasan, M.; Prasad, M. Plant innate immunity: An updated insight into defense mechanism. J. Biosci. 2013, 38, 433–449. [Google Scholar] [CrossRef]
- Lichtenberg, S.S.; Tsyusko, O.V.; Palli, S.R.; Unrine, J.M. Uptake and Bioactivity of Chitosan/Double-Stranded RNA Polyplex Nanoparticles in Caenorhabditis elegans. Environ. Sci. Technol. 2019, 53, 3832–3840. [Google Scholar] [CrossRef] [PubMed]
- Rosa, C.; Kuo, Y.-W.; Yan, Z.; Falk, B.W. RNA Interference Mechanisms and Applications in Plant Pathology. Annu. Rev. Phytopathol. 2018, 56, 581–610. [Google Scholar] [CrossRef] [PubMed]
- Worrall, E.; Bravo-Cazar, A.; Nilon, A.T.; Fletcher, S.J.; Robinson, K.E.; Carr, J.; Mitter, N. Exogenous Application of RNAi-Inducing Double-Stranded RNA Inhibits Aphid-Mediated Transmission of a Plant Virus. Front. Plant Sci. 2019, 10, 265. [Google Scholar] [CrossRef] [Green Version]
- Konakalla, N.C.; Kaldis, A.; Berbati, M.; Masarapu, H.; Voloudakis, A.E. Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta 2016, 244, 961–969. [Google Scholar] [CrossRef]
- Niehl, A.; Soininen, M.; Poranen, M.M.; Heinlein, M. Synthetic biology approach for plant protection using dsRNA. Plant Biotechnol. J. 2018, 16, 1679–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLoughlin, A.G.; Wytinck, N.; Walker, P.L.; Girard, I.J.; Rashid, K.Y.; De Kievit, T.; Fernando, W.G.D.; Whyard, S.; Belmonte, M.F. Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-Based Control of Fusarium graminearum Infections through Spraying of Long dsRNAs Involves a Plant Passage and Is Controlled by the Fungal Silencing Machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef]
- Gu, K.-X.; Song, X.-S.; Xiao, X.-M.; Duan, X.-X.; Wang, J.-X.; Duan, Y.-B.; Hou, Y.-P.; Zhou, M.-G. A β-tubulin dsRNA derived from Fusarium asiaticum confers plant resistance to multiple phytopathogens and reduces fungicide resistance. Pestic. Biochem. Physiol. 2019, 153, 36–46. [Google Scholar] [CrossRef]
- Mumbanza, F.M.; Kiggundu, A.; Tusiime, G.; Tushemereirwe, W.K.; Niblett, C.; Bailey, A. In vitro antifungal activity of synthetic dsRNA molecules against two pathogens of banana, Fusarium oxysporumf. sp. cubense and Mycosphaerella fijiensis. Pest Manag. Sci. 2013, 69, 1155–1162. [Google Scholar] [CrossRef]
- Höfle, L.; Biedenkopf, D.; Werner, B.T.; Shrestha, A.; Jelonek, L.; Koch, A. Study on the efficiency of dsRNAs with increasing length in RNA-based silencing of the Fusarium CYP51 genes. RNA Biol. 2020, 17, 463–473. [Google Scholar] [CrossRef]
- Banerjee, S.; Banerjee, A.; Gill, S.; Gupta, O.; Dahuja, A.; Jain, P.K.; Sirohi, A. RNA Interference: A Novel Source of Resistance to Combat Plant Parasitic Nematodes. Front. Plant Sci. 2017, 8, 834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, S.; Jones, M.G.K.; Fosu-Nyarko, J. RNA interference of an orthologue of Dicer of Meloidogyne incognita alludes to the gene’s importance in nematode development. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lilley, C.J.; Davies, L.J.; Urwin, P.E. RNA interference in plant parasitic nematodes: A summary of the current status. Parasitology 2012, 139, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, E.; Di Vito, M.; Jones, J.; De Giorgi, C. Analysis of chitin synthase function in a plant parasitic nematode, Meloidogyne artiellia, using RNAi. Gene 2005, 349, 87–95. [Google Scholar] [CrossRef]
- Kimber, M.J.; McKinney, S.; McMaster, S.; Day, T.A.; Fleming, C.C.; Maule, A.G. flp gene disruption in a parasitic nematode reveals motor dysfunction and unusual neuronal sensitivity to RNA interference. FASEB J. 2007, 21, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Whyard, S.; Vélez, A.M.; Smagghe, G. Double-Stranded RNA Technology to Control Insect Pests: Current Status and Challenges. Front. Plant Sci. 2020, 11, 451. [Google Scholar] [CrossRef]
- Knorr, E.; Fishilevich, E.; Tenbusch, L.; Frey, M.L.F.; Rangasamy, M.; Billion, A.; Worden, S.E.; Gandra, P.; Arora, K.; Lo, W.; et al. Gene silencing in Tribolium castaneum as a tool for the targeted identification of candidate RNAi targets in crop pests. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Willow, J.; Soonvald, L.; Sulg, S.; Kaasik, R.; Silva, A.; Taning, C.; Christiaens, O.; Smagghe, G.; Veromann, E. First Evidence of Bud Feeding-Induced RNAi in a Crop Pest via Exogenous Application of dsRNA. Insects 2020, 11, 769. [Google Scholar] [CrossRef]
- Willow, J.; Soonvald, L.; Sulg, S.; Kaasik, R.; Silva, A.I.; Taning, C.N.T.; Christiaens, O.; Smagghe, G.; Veromann, E. RNAi efficacy is enhanced by chronic dsRNA feeding in pollen beetle. Commun. Biol. 2021, 4, 1–8. [Google Scholar] [CrossRef]
- Nitnavare, R.B.; Bhattacharya, J.; Singh, S.; Kour, A.; Hawkesford, M.J.; Arora, N. Next Generation dsRNA-Based Insect Control: Success So Far and Challenges. Front. Plant Sci. 2021, 12, 673576. [Google Scholar] [CrossRef]
- Mutti, N.S.; Park, Y.; Reese, J.C.; Reeck, G.R. RNAi Knockdown of a Salivary Transcript Leading to Lethality in the Pea Aphid, Acyrthosiphon pisum. J. Insect Sci. 2006, 6, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubrovina, A.S.; Kiselev, K.V. Exogenous RNAs for Gene Regulation and Plant Resistance. Int. J. Mol. Sci. 2019, 20, 2282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitter, N.; Worrall, E.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.; Lu, G.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef] [PubMed]
- Gurusamy, D.; Mogilicherla, K.; Palli, S.R. Chitosan nanoparticles help double-stranded RNA escape from endosomes and improve RNA interference in the fall armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol. 2020, 104, 1–15. [Google Scholar] [CrossRef]
- Killmer, J.; Kumar, A.; McLaughlin, P.D.; Humberto, J.P. Methods and Compositions for Controlling Pests. Patent Application No PCT/US2017/015861, 31 January 2017. [Google Scholar]
- Christiaens, O.; Tardajos, M.G.; Reyna, Z.L.M.; Dash, M.; Dubruel, P.; Smagghe, G. Increased RNAi Efficacy in Spodoptera exigua via the Formulation of dsRNA With Guanylated Polymers. Front. Physiol. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Peng, Y.; Pu, J.; Fu, W.; Wang, J.; Han, Z. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 2016, 77, 1–9. [Google Scholar] [CrossRef]
- Ivashuta, S.; Zhang, Y.; Wiggins, B.E.; Ramaseshadri, P.; Segers, G.C.; Johnson, S.; Meyer, S.E.; Kerstetter, R.A.; McNulty, B.C.; Bolognesi, R.; et al. Environmental RNAi in herbivorous insects. RNA 2015, 21, 840–850. [Google Scholar] [CrossRef] [Green Version]
- Kolliopoulou, A.; Taning, C.N.T.; Smagghe, G.; Swevers, L. Viral Delivery of dsRNA for Control of Insect Agricultural Pests and Vectors of Human Disease: Prospects and Challenges. Front. Physiol. 2017, 8, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Gillet, F.-X.; Garcia, R.A.; Macedo, L.L.P.; Albuquerque, E.; Silva, M.C.M.; Grossi-De-Sa, M.F. Investigating Engineered Ribonucleoprotein Particles to Improve Oral RNAi Delivery in Crop Insect Pests. Front. Physiol. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- Parsons, K.H.; Mondal, M.; McCormick, C.L.; Flynt, A.S. Guanidinium-Functionalized Interpolyelectrolyte Complexes Enabling RNAi in Resistant Insect Pests. Biomacromolecules 2018, 19, 1111–1117. [Google Scholar] [CrossRef] [Green Version]
- Taning, C.N.T.; Christiaens, O.; Berkvens, N.; Casteels, H.; Maes, M.; Smagghe, G. Oral RNAi to control Drosophila suzukii: Laboratory testing against larval and adult stages. J. Pest Sci. 2016, 89, 803–814. [Google Scholar] [CrossRef]
- Taning, C.N.T.; Christiaens, O.; Li, X.; Swevers, L.; Casteels, H.; Maes, M.; Smagghe, G. Engineered Flock House Virus for Targeted Gene Suppression Through RNAi in Fruit Flies (Drosophila melanogaster) In Vitro and In Vivo. Front. Physiol. 2018, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef]
- Castellanos, N.L.; Smagghe, G.; Sharma, R.; E Oliveira, E.; Christiaens, O. Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the Neotropical stink bug Euschistus heros. Pest Manag. Sci. 2019, 75, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, D.; Sandeep, K.; Pandey, D.; Dutta, R.K. Liposomes for Drug Delivery. J. Biotechnol. Biomater. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Maja, L.; Željko, K.; Mateja, P. Sustainable technologies for liposome preparation. J. Supercrit. Fluids 2020, 165, 104984. [Google Scholar] [CrossRef]
- Temidayo, O.B.; Olusanya, I.D.; Ahmad, R.R.H.; Ibegbu, D.M.; Smith, J.R.; Elkordy, A.A. Liposomal drug delivery systems and anticancer drugs. Molecules 2018, 23, 907. [Google Scholar] [CrossRef] [Green Version]
- Wagner, A.; Vorauer-Uhl, K. Liposome Technology for Industrial Purposes. J. Drug Deliv. 2011, 2011, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Maherani, B.; Arab-Tehrany, E.; Mozafari, M.R.; Gaiani, C.; Linder, M. Liposomes: A Review of Manufacturing Techniques and Targeting Strategies. Curr. Nanosci. 2011, 7, 436–452. [Google Scholar] [CrossRef]
- Alghuthaymi, M.; Ahmad, A.; Khan, Z.; Khan, S.; Ahmed, F.; Faiz, S.; Nepovimova, E.; Kuča, K.; Abd-Elsalam, K. Exosome/Liposome-like Nanoparticles: New Carriers for CRISPR Genome Editing in Plants. Int. J. Mol. Sci. 2021, 22, 7456. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Matamoros, M.F.; A Villanueva, M.; Islas-Flores, T. Genetic transformation of cell-walled plant and algae cells: Delivering DNA through the cell wall. Briefings Funct. Genom. 2018, 17, 26–33. [Google Scholar] [CrossRef]
- Karny, A.; Zinger, A.; Kajal, A.; Shainsky-Roitman, J.; Schroeder, A. Therapeutic nanoparticles penetrate leaves and deliver nutrients to agricultural crops. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zepeda-Cervantes, J.; Ramírez-Jarquín, J.O.; Vaca, L. Interaction Between Virus-Like Particles (VLPs) and Pattern Recognition Receptors (PRRs) From Dendritic Cells (DCs): Toward Better Engineering of VLPs. Front. Immunol. 2020, 11, 1100. [Google Scholar] [CrossRef]
- Tagliamonte, M.; Tornesello, M.L.; Buonaguro, F.M.; Buonaguro, L. Virus-Like Particles. In Micro and Nanotechnology: Micro and Nanotechnology in Vaccine Development: Chapter 11; Skwarczynski, M., Toth, I., Eds.; William Andrew Publishing: Oxford, UK, 2017; pp. 205–219. [Google Scholar] [CrossRef]
- Biabanikhankahdani, R.; Alitheen, N.B.M.; Ho, K.L.; Tan, W.S. pH-responsive Virus-like Nanoparticles with Enhanced Tumour-targeting Ligands for Cancer Drug Delivery. Sci. Rep. 2016, 6, 37891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeltins, A. Construction and Characterization of Virus-Like Particles: A Review. Mol. Biotechnol. 2012, 53, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Chackerian, B. Virus-like particles: Flexible platforms for vaccine development. Expert Rev. Vaccines 2007, 6, 381–390. [Google Scholar] [CrossRef]
- Karandikar, S.; Mirani, A.; Waybhase, V.; Patravale, V.B.; Patankar, S. Nanovaccines for Oral Delivery-Formulation Strategies and Challenges. In Micro and Nanotechnology: Nanostructures for Oral Medicine: Chapter 10; Andronescu, E., Mihai, A.v., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 263–293. [Google Scholar] [CrossRef]
- Domingo-Espin, J.; Unzueta, U.; Saccardo, P.; Rodríguez-Carmona, E.; Corchero, J.L.; Vazquez, E.; Ferrer-Miralles, N. Engineered Biological Entities for Drug Delivery and Gene Therapy. Eng. Biol. Entities Drug Deliv. Gene Ther. Protein Nanoparticles 2011, 104, 247–298. [Google Scholar] [CrossRef]
- Le, D.; Müller, K. In Vitro Assembly of Virus-Like Particles and Their Applications. Life 2021, 11, 334. [Google Scholar] [CrossRef]
- Yan, D.; Wei, Y.-Q.; Guo, H.-C.; Sun, S.-Q. The application of virus-like particles as vaccines and biological vehicles. Appl. Microbiol. Biotechnol. 2015, 99, 10415–10432. [Google Scholar] [CrossRef]
- Tsanova, T.; Stefanova, L.; Topalova, L.; Atanasov, A.; Pantchev, I. DNA-free gene editing in plants: A brief overview. Biotechnol. Biotechnol. Equip. 2021, 35, 131–138. [Google Scholar] [CrossRef]
- Reineke, T.M.; Davis, M.E. Nucleic Acid Delivery via Polymer Vehicles. In Polymer Science: A Comprehensive Reference: Volumen 09: Polymers in Biology and Medicine; Matyjaszewski, K., Möller, M.v., Eds.; Elsevier B.V.: Amsterdam, The Netherlands, 2012; Volume 09, pp. 497–527. [Google Scholar] [CrossRef]
- Rosenkranz, A.; Sobolev, A.S. Polyethylenimine-based polyplex nanoparticles and features of their behavior in cells and tissues. Russ. Chem. Bull. 2015, 64, 2749–2755. [Google Scholar] [CrossRef]
- Wirth, T.; Ylä-Herttuala, S. Gene transfer vectors (DNA vehicles) and their incorporation into biomaterials for bone repair. Biomater. Bone Regen. Novel Tech. Appl. 2014, 374–405. [Google Scholar] [CrossRef]
- Aggarwal, R.; Targhotra, M.; Kumar, B.; Sahoo, P.; Chauhan, M.K. Polyplex: A Promising Gene Delivery System. Int. J. Pharm. Sci. Nanotechnol. 2019, 12, 4681–4686. [Google Scholar] [CrossRef]
- Lin, W.J.; Hsu, W.Y. Pegylation effect of chitosan based polyplex on DNA transfection. Carbohydr. Polym. 2015, 120, 7–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasile, C. Polymeric Nanomaterials: Recent Developments, Properties and Medical Applications. In Micro and Nano Technologies: Polymeric Nanomaterials In Nanotherapeutics: Chapter 01; Vasile, C., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 1–66. [Google Scholar] [CrossRef]
- Zhang, Q.-F.; Yi, W.-J.; Wang, B.; Zhang, J.; Ren, L.; Chen, Q.-M.; Guo, L.; Yu, X.-Q. Linear polycations by ring-opening polymerization as non-viral gene delivery vectors. Biomaterials 2013, 34, 5391–5401. [Google Scholar] [CrossRef]
- Hall, A.; Lächelt, U.; Bartek, J.; Wagner, E.; Moghimi, S.M. Polyplex Evolution: Understanding Biology, Optimizing Performance. Mol. Ther. 2017, 25, 1476–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Firoozi, B.; Nasser, Z.; Sofalian, O.; Sheikhzade-Mosadegh, P. Enhancement of the transfection efficiency of DNA into Crocus sativus L. cells via PEI nanoparticles. J. Integr. Agric. 2018, 17, 1768–1778. [Google Scholar] [CrossRef]
- Zolghadrnasab, M.; Mousavi, A.; Farmany, A.; Arpanaei, A. Ultrasound-mediated gene delivery into suspended plant cells using polyethyleneimine-coated mesoporous silica nanoparticles. Ultrason. Sonochemistry 2021, 73, 105507. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Dzhambazova, T.; Kostov, K.; Arpaia, S.; Joga, M.R.; Urru, I.; Sweet, J.; Smagghe, G. Literature review of baseline information on RNAi to support the environmental risk assessment of RNAi-based GM plants. EFSA Support. Publ. 2018, 15, 173. [Google Scholar] [CrossRef]
- Dhandapani, R.K.; Gurusamy, D.; Howell, J.L.; Palli, S.R. Development of CS-TPP-dsRNA nanoparticles to enhance RNAi efficiency in the yellow fever mosquito, Aedes aegypti. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Kausar, A. Flame Retardant Potential of Clay Nanoparticles. In Micro and Nano Technologies: Clay Nanoparticles: Properties and Applications: Chapter 07; Cavallaro, G., Fakhrullin, R., Pasbakhsh, P., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 169–184. [Google Scholar] [CrossRef]
- Peña-Parás, L.; Sánchez-Fernández, J.A.; Vidaltamayo, R. Nanoclays for Biomedical Applications. In Handbook of Ecomaterials: Volumen 05; Martínez, L., Kharissova, O., Kharisov, B., Eds.; Springer Nature: Cham, Switzerland, 2019; pp. 3453–3471. [Google Scholar] [CrossRef]
- Villanova-de-Benavent, C. Geological Overview of Clay Nanoparticles. In Micro and Nano Technologies: Clay Nanoparticles: Properties and Applications: Chapter 01; Cavallaro, G., Fakhrullin, R., Pasbakhsh, P., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 3–36. [Google Scholar] [CrossRef]
- Massaro, M.; Cavallaro, G.; Lazzara, G.; Riela, S. Covalently Modified Nanoclays: Synthesis, Properties and Applications. In Micro and Nano Technologies: Clay Nanoparticles: Properties and Applications: Chapter 13; Cavallaro, G., Fakhrullin, R., Pasbakhsh, P., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 305–333. [Google Scholar] [CrossRef]
- Cavallaro, G.; Lazzara, G.; Rozhina, E.; Konnova, S.; Kryuchkova, M.; Khaertdinov, N.; Fakhrullin, R. Organic-nanoclay composite materials as removal agents for environmental decontamination. RSC Adv. 2019, 9, 40553–40564. [Google Scholar] [CrossRef] [Green Version]
- Lisuzzo, L.; Cavallaro, G.; Parisi, F.; Riela, S.; Milioto, S.; Lazzara, G. Colloidal Stability and Self-Assembling Behavior of Nanoclays. In Micro and Nano Technologies: Clay Nanoparticles: Properties and Applications: Chapter 04; Cavallaro, G., Fakhrullin, R., Pasbakhsh, P., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 95–116. [Google Scholar] [CrossRef]
- Hazarika, A.; Maji, T.K. Strain sensing behavior and dynamic mechanical properties of carbon nanotubes/nanoclay reinforced wood polymer nanocomposite. Chem. Eng. J. 2014, 247, 33–41. [Google Scholar] [CrossRef]
- Mandal, M.; Halim, Z.; Maji, T.K. Mechanical, moisture absorption, biodegradation and physical properties of nanoclay-reinforced wood/plant oil composites. SN Appl. Sci. 2020, 2, 250. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; White, J.C.; Wang, Z.; Xing, B. Nano-enabled fertilizers to control the release and use efficiency of nutrients. Curr. Opin. Environ. Sci. Health 2018, 6, 77–83. [Google Scholar] [CrossRef]
- Bernardo, M.P.; Guimarães, G.G.F.; Majaron, V.F.; Ribeiro, C. Controlled Release of Phosphate from Layered Double Hydroxide Structures: Dynamics in Soil and Application as Smart Fertilizer. ACS Sustain. Chem. Eng. 2018, 6, 5152–5161. [Google Scholar] [CrossRef]
- Songkhum, P.; Wuttikhun, T.; Chanlek, N.; Khemthong, P.; Laohhasurayotin, K. Controlled release studies of boron and zinc from layered double hydroxides as the micronutrient hosts for agricultural application. Appl. Clay Sci. 2018, 152, 311–322. [Google Scholar] [CrossRef]
- Nourizadeh, H.; Bakhshayesh, A. Nanoclay-Based Products Across Global Markets. In StatNano Applied and Industrial Series: Nanoclay-Based Products Across Global Markets: Applications and Properties; Nourizadeh, H., Bakhshayesh, A., Eds.; StatNano Publications: Lund, Sweden, 2020; pp. 3–33. [Google Scholar] [CrossRef]
- Parker, K.M.; Borrero, V.B.; Van Leeuwen, D.M.; Lever, M.A.; Mateescu, B.; Sander, M. Environmental Fate of RNA Interference Pesticides: Adsorption and Degradation of Double-Stranded RNA Molecules in Agricultural Soils. Environ. Sci. Technol. 2019, 53, 3027–3036. [Google Scholar] [CrossRef]
- Bachman, P.; Fischer, J.; Song, Z.; Urbanczyk-Wochniak, E.; Watson, G. Environmental Fate and Dissipation of Applied dsRNA in Soil, Aquatic Systems, and Plants. Front. Plant Sci. 2020, 11, 11. [Google Scholar] [CrossRef]
- EPA; United States Environmental Protection Agency. RNAi Technology: Program Formulation for Human Health and Ecological Risk Assessment. FIFRA Sci. Advis. Panel Meet. 2014, 1–77. [Google Scholar]

| Target | Experimental Evidence | Target Genes | Reference |
|---|---|---|---|
| Virus | dsRNA + clay resulted in BCMV virus resistance for 20 d | Nib and CP genes of BCMV | [24] |
| TMV Tobacco virus resistance for 7–20 d | CP, P126, RP of TMV | [25,26] | |
| Fungi | Inhibits Botrytis cinerea disease | DCL1, DCL2 of Botrytis cinerea | [27] |
| Efficiently inhibited Fusarium graminearum | CYP51A, CYP51B, CYP51C of F. graminearum | [28] | |
| Sclerotinia sclerotiorum/Botrytis cinerea | mRNA splicing, ribosome biogenesis, protein disulfide oxidoreductase, peroxisomal protein | [27] | |
| Fusarium asiaticum, Botrytis cinerea, Magnaporthe oryzae, Colletotrichum truncatum | β2-tubulin | [29] | |
| Fusarium oxysporum f. sp. cubense and Mycosphaerella fijiensi, Fusarium | adenylate cyclase, DNA polymerase alpha subunit/delta subunit/CYP51 | [30,31] | |
| Nematodes | Caenorhabditis elegans, Radopholus similis, Meloidogyne artiellia, Meloidogyne incognita, Globodera pallida | Several genes such as 16D10 peptide, chitin synthase, xylanase, glutathione-S- transferase, FMRF-like peptides | [18,32,33,34,35,36] |
| Insects | Coleopterans are highly sensitive, Hemiptera, Orthoptera, Diptera, Hymenoptera, and Lepidoptera have different responses. | Several genes such as salivary protein C002, 16D10, αCOP, Cytochrome P450, Acetylcholinesterase, ABC transporter, β-actin, chitin synthase B | [37,38,39,40,41,42] |
| Endogenous plant genes | Arabidopsis, Tobacco, poplar, rice | Transgenes/CHS/EPSPS/STM/WER/MYB1/WRKY23 | [43] |
| Encapsulation System | Potential Crop Protection Application | Strategy | Reference |
|---|---|---|---|
| Guanylated 2-(aminoethyl) methacrylate (AEMA)/dsRNA polyplex nanoparticles. | Insecticide induces decreased feeding in Lepidopteran larvae (Spodoptera exigua), then promoting weight loss, developmental halt, and mortality. | Increases RNAi efficiency in targeting the essential gene chitin synthase B (ChSB), while preventing the degradation of dsRNA in the alkaline gut of insects and enhancing its cellular uptake in midgut cells. | [47] |
| poly-[N-(3-guanidinopropyl) methacrylamide] (pGPMA)/dsRNA interpolyelectrolyte nanocomplex. | Ingestion insecticide regulates gene silencing in Lepidopteran larvae (Spodoptera frugiperda), increasing mortality from starvation and growth stunting. | Increased internalization and protection of dsRNA in insect cells, decreasing the accumulation of target mRNA due to the knockdown of genes related to vital functions such as nutrient absorption (sfVATPase), intracellular transport (sfKIF), and cell division (sfCDC27). | [52] |
| Chitosan/dsRNA polyplex nanoparticles | Nematicide can homogeneously enter the nematode’s body (Caenorhabditis elegans) through noncanonical endocytotic pathways and attack specific genes. The combined effect decreases the development of the nematode by the action of the chitosan vehicle. | Increases RNAi efficiency of gene knockdown throughout the whole body of the nematode by introducing intact dsRNA through the Clathrin-mediated endocytosis pathway, which is different from the canonical pathway (sid-1 and sid-2) in the study model. Furthermore, chitosan was shown to effectively decrease myosin gene expression, which is critical for the growth and reproduction of the model nematode. | [22] |
| Chitosan/dsRNA polyplex nanoparticles | Insecticide against Lepidopteran larvae (Spodoptera frugiperda) acts on genes related to the apoptosis pathway, inducing growth impairment and larval mortality. | Improves RNAi efficiency through the protection of dsRNA from degradation by intracellular and intercellular RNases. It also reduces the accumulation of dsRNA in the endosome while favoring its transport to the cytoplasm, where the formation of siRNAs is promoted, producing knockdown of apoptosis-related genes (iap). | [45] |
| Layered double hydroxide (LDH) clay nanosheets/dsRNA | Develop a topical product that induces viral resistance in plants (against PMMoV and CMV) using dsRNA absorption technology in clay nanosheets (Bio-Clay). | Increased persistence of the topical treatment due to strong adhesion of dsRNA in the vehicle (LDH) and with the leaves. It also allows the controlled release of the biomolecule and confers protection against environmental degradation while favoring the internalization of dsRNA in the plant. | [44] |
| Lipofectamine 2000 liposomes/dsRNA. | Insecticide against Diptera of the genus Drosophila (D. melanogaster, D. sechellia, D. yakuba, and D. pseudoobscura) acting by ingestion. It attacks essential genes of development through knockdown management. | Promotion of dsRNA internalization in insects through encapsulation protection, increasing silencing efficiency by promoting more significant RNAi accumulation in larvae. Knockdown of the genes of the VATPase (gut lumen pH stabilizer associated with nutrient uptake) and gTub23C (mitosis-related g-tubulin protein, essential for microtubule organization). | [55] |
| Lipofectamine 2000 liposomes/dsRNA. | Specific insecticide against larvae and adults of Drosophila suzukii combining synergic effect of multiple gene knockdown. Oral administration route. | It facilitates uptake in the insect’s gut. It causes significant mortality in larvae and adults by the reduction in transcript levels of essential genes rps13 (housekeeping), alpha COP (coatomer subunit for trans-organelles transport), and vha26 (subunit of the vacuolar ATPase). The synergistic action of knockdown of the rps13 and alpha COP genes significantly increases mortality in the insect. | [53] |
| Liposomes/dsRNA | Oral insecticide for the control of nymphs of Euschistus heros (hemiptera: pentatomidae), which is one of the main soybean pests in the field. | Protection of dsRNA against degradation promoted by the ribonuclease action of insect saliva. Enhanced silencing activity of target genes vATPaseA (V-type proton ATPase catalytic subunit A) and act-2 (muscle actin). | [56] |
| Recombinant Flock House Virus FHV/dsRNA | Recombinant insecticide based on a viral vehicle transporting dsRNA silencers of essential genes in Drosophila melanogaster. For potential massive application in other species susceptible to FHV infection. | Use of the insect cell machinery to assemble infective recombinant FHV virions that carry target sequences for the production of dsRNA when replicating in cells. Thus, virions protect the sequences responsible for silencing the rps13 (housekeeping), alpha COP (coatomer subunit for trans-organelle transport), and vha26 (subunit of the vacuolar ATPase) genes while at the same time favoring dispersal in insects. It simulates natural viral infection. | [54] |
| Virus-Like Particles (VLP)/dsRNA | Oral insecticide for the control of ants of several genera (Solenopsis invicta (fire ants), Camponotus pennsylvanicus and Camponotus floridanus (carpenter ants), Linepithema humile (Argentine ants), Tapinoma sessile (odorous ants), Tetramorium caespitum (pavementom ants), and Monstrous ants) pharaonis (pharaoh ants); inducing the silencing of physiological genes required for the survival of the colony. | Recombinant production in E. coli, which through specific plasmids manufactures capsid proteins of bacteriophages Qß and MS2 and inducible RNAi precursor sequences. The packaging of dsRNAs in VLPs protects them from degradation by nonspecific environmental organisms and the intestinal RNases of the target organism. It also favors its absorption by lining the gut cells. The silenced genes are related to the viability of the colony, for example, the induction of sterility and individual mortality. VLP carrying dsRNA is sprayed on the ground for spot application or incorporated into the bait. Target genes include VgR (vitellogenin receptor protein), TVXl (telomerase variant XI protein), PBAN (pheromone biosynthesis activating neuropeptide), PBANR (pheromone biosynthetic activating neuropeptide receptor), WLS (wntless protein), MEGF10 (multiple epidermal growth factor-like domain proteins 10), CHCP (clatherin heavy chain protein), CDC7 (cell division cycle 7-related protein), Cep89 (centrosomal protein 89 kdal), PSMBl (beta subunit of the type-1 proteosome), A5C (actin 5C protein), ATPSD (ATP synthase delta subunit); as well others related with anamorsin, beta actin, and Csp9 proteins | [46] WO2017/136353Al for APSE RNA Containers (ARCs) |
| Ribonucleoprotein particle (RNP)/dsRNA | Insecticide for control of cotton boll weevil (Anthonomus grandis) adults. | Developing a protection and stability system for dsRNA avoids degradation by nucleases in the insect’s gut and favors rapid cellular incorporation. The above is based on a chimeric protein PTD-DRBD (peptide transduction domain–dsRNA binding domain) combined with dsRNA. This type of resulting protein is known as cell-penetrating peptide (CPP). | [51] |
| Regulatory Agency | Proposal | Reference |
|---|---|---|
| EPA | Propose using Problem Formulation- Risk assessment | [102] |
| European Food Safety Authority (EFSA) | Do not directly address the spray products, but a literature review focused on RNAi-based GM plants and Risk Assessment | [86] |
| OECD | Proposed using risk assessment to evaluate the toxicity profile and exposure by using the current regulatory framework for small-molecule agrochemicals as a general framework for dsRNA-based agricultural products. Proposed using the experience with the review of dsRNA-based GE crops | [11] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Soto, A.; Chacón-Cerdas, R. RNAi Crop Protection Advances. Int. J. Mol. Sci. 2021, 22, 12148. https://doi.org/10.3390/ijms222212148
Hernández-Soto A, Chacón-Cerdas R. RNAi Crop Protection Advances. International Journal of Molecular Sciences. 2021; 22(22):12148. https://doi.org/10.3390/ijms222212148
Chicago/Turabian StyleHernández-Soto, Alejandro, and Randall Chacón-Cerdas. 2021. "RNAi Crop Protection Advances" International Journal of Molecular Sciences 22, no. 22: 12148. https://doi.org/10.3390/ijms222212148
APA StyleHernández-Soto, A., & Chacón-Cerdas, R. (2021). RNAi Crop Protection Advances. International Journal of Molecular Sciences, 22(22), 12148. https://doi.org/10.3390/ijms222212148