Relevance of BRAF Subcellular Localization and Its Interaction with KRAS and KIT Mutations in Skin Melanoma
Abstract
1. Introduction
2. Materials and Methods
2.1. In Silico Analysis of Public Databases Regarding SKMs
2.1.1. Gene Expression and Survival Analysis of BRAF, KRAS, and KIT in SKMs
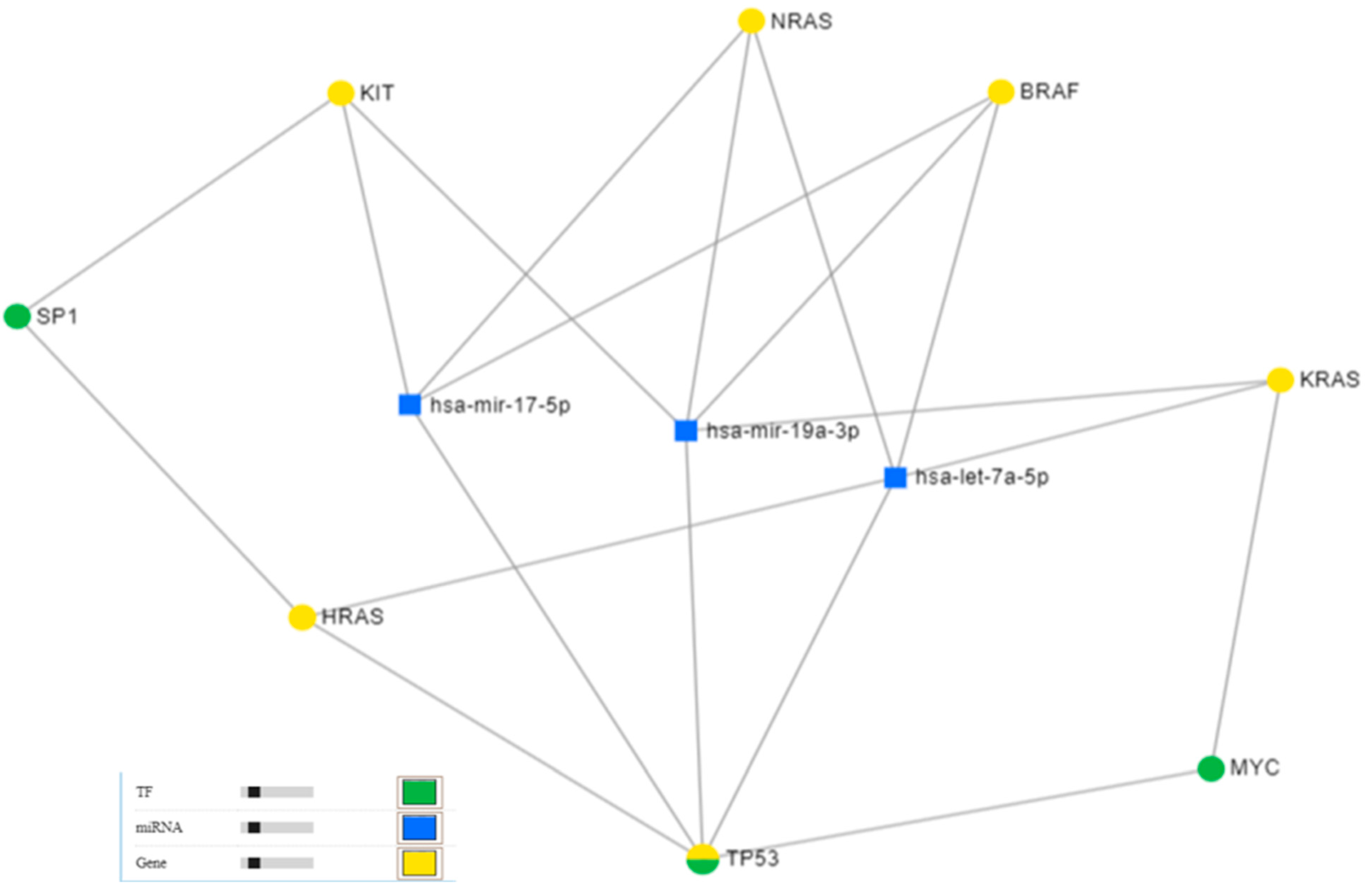
2.1.2. mRNA–miRNA Network Interaction
2.2. Protein Expression Levels of BRAF, KIT, and KRAS
2.3. Statistical Analysis and Survival Curves
3. Results
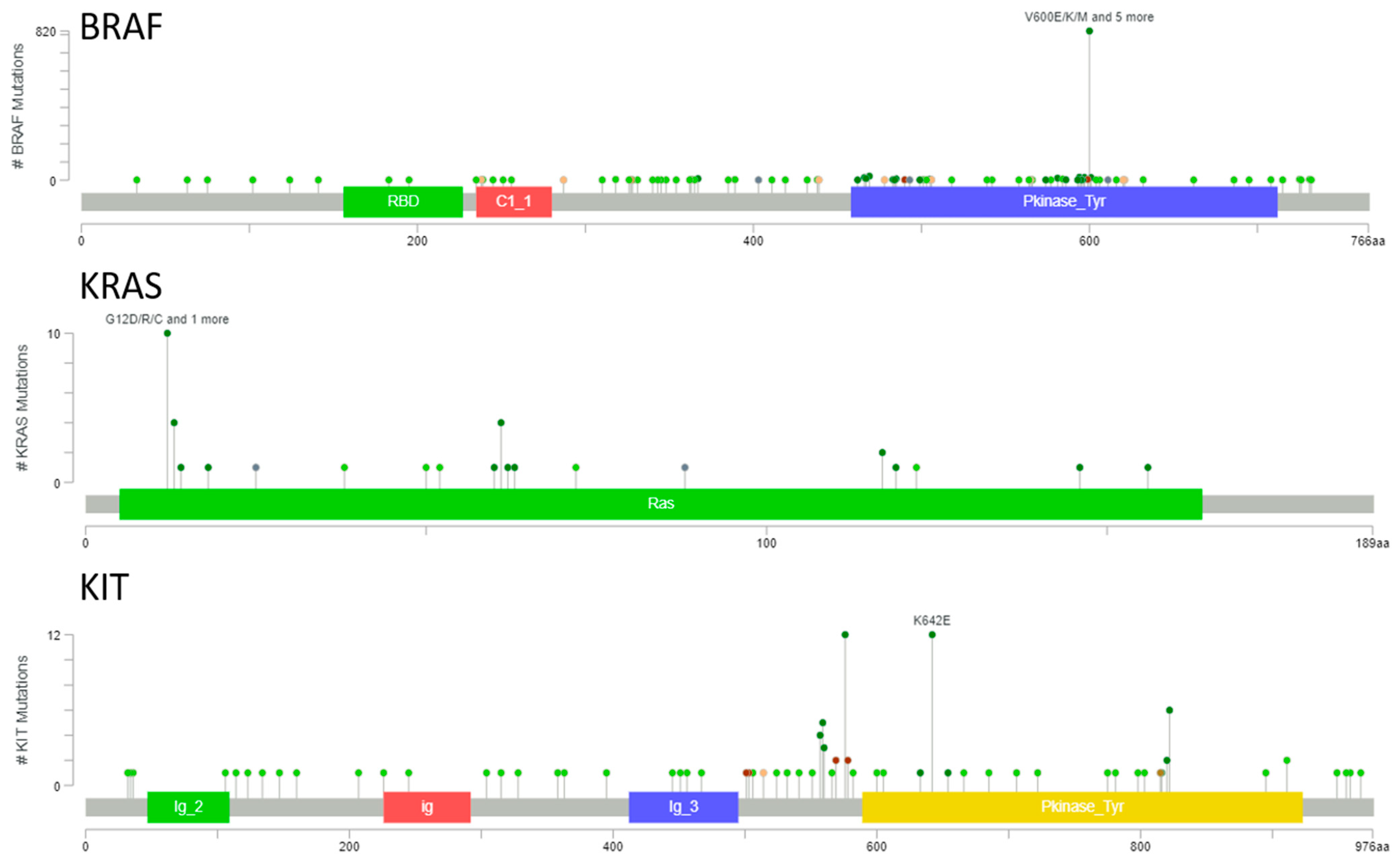
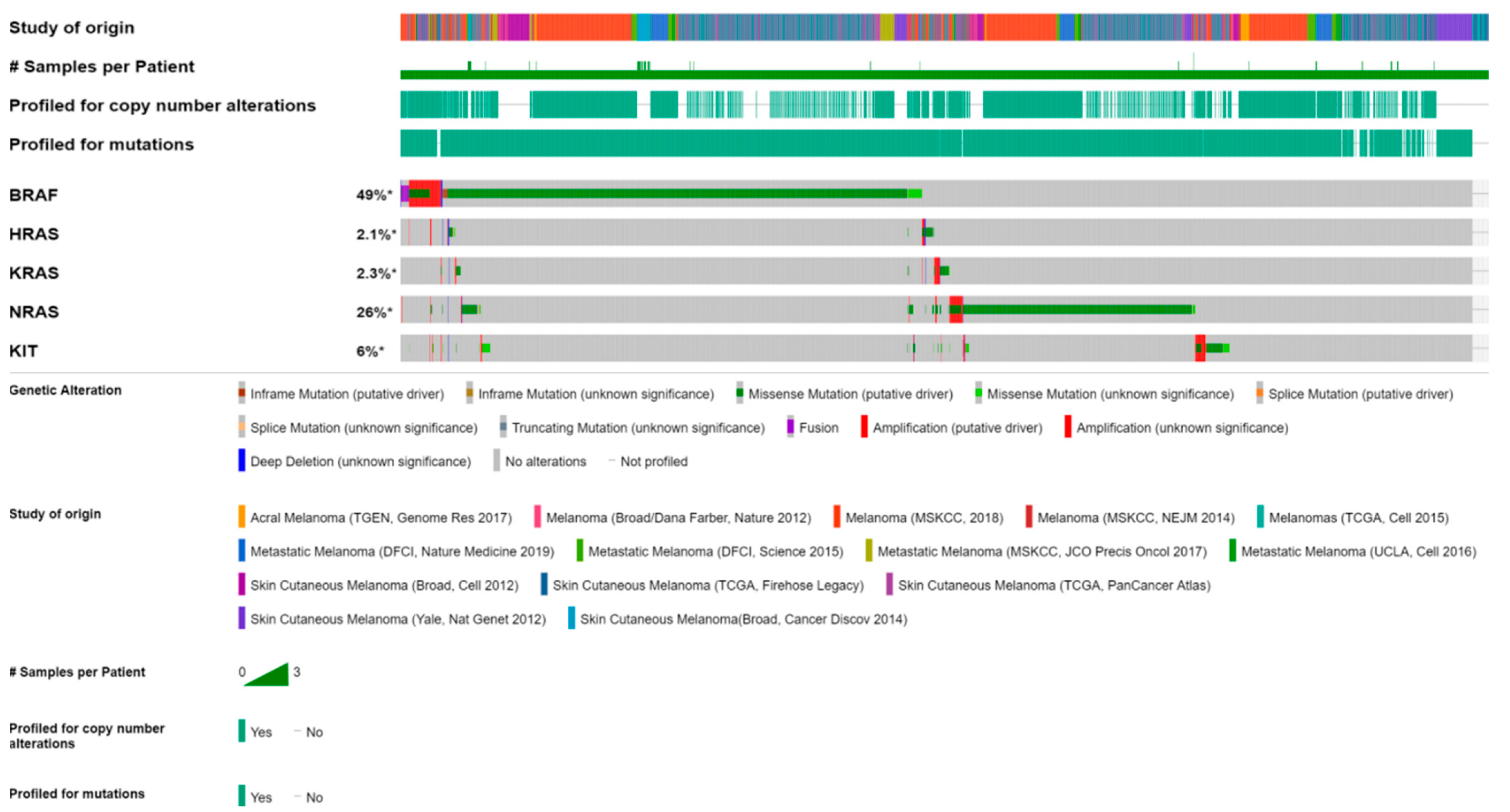
3.1. BRAF, KRAS, and KIT Mutational Landscape in Melanoma
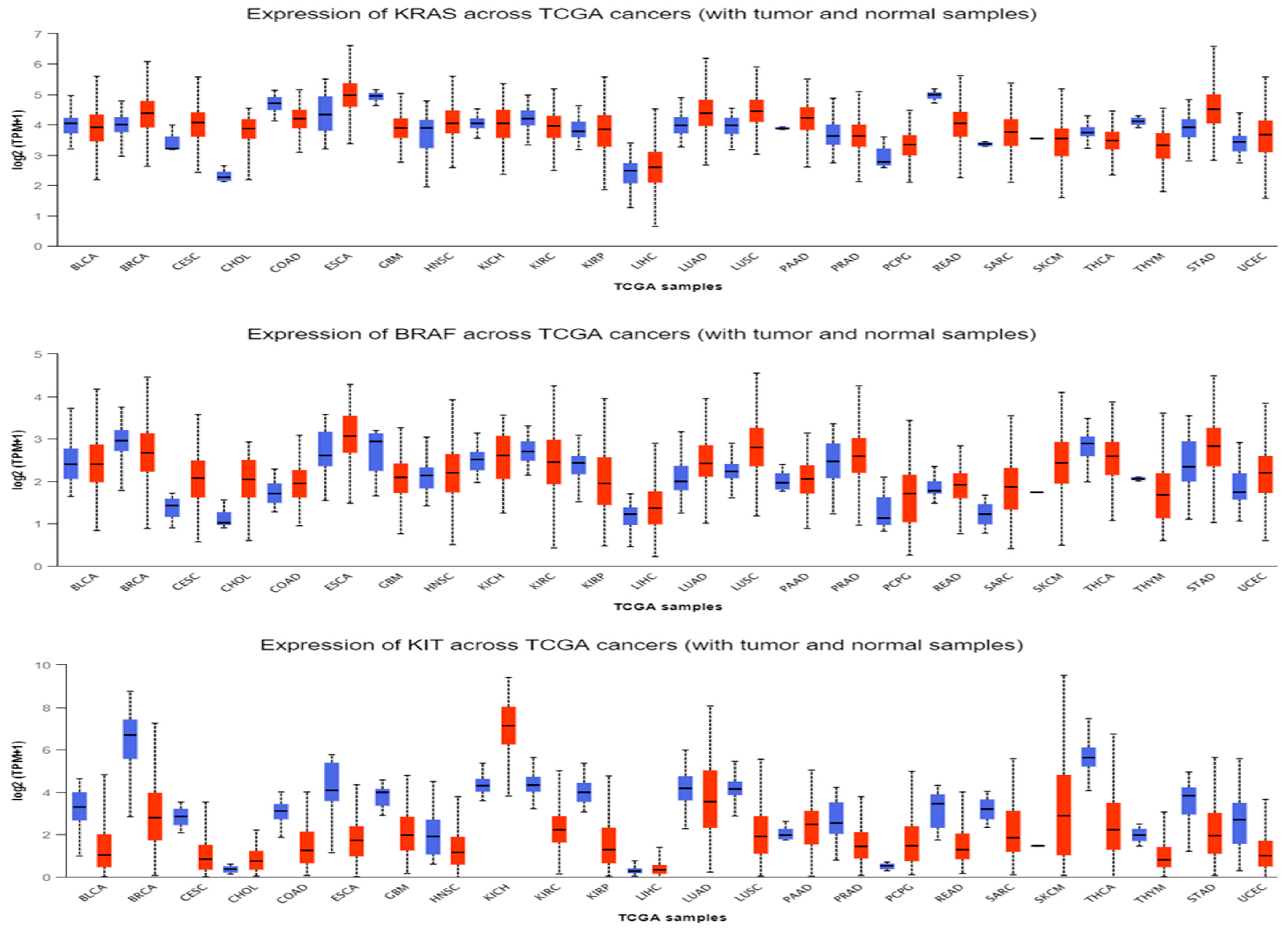
3.2. BRAF, KRAS, and KIT mRNA Expression Level in SKMs
3.3. Network Interaction
3.4. Protein Level Validation of the Selected Genes
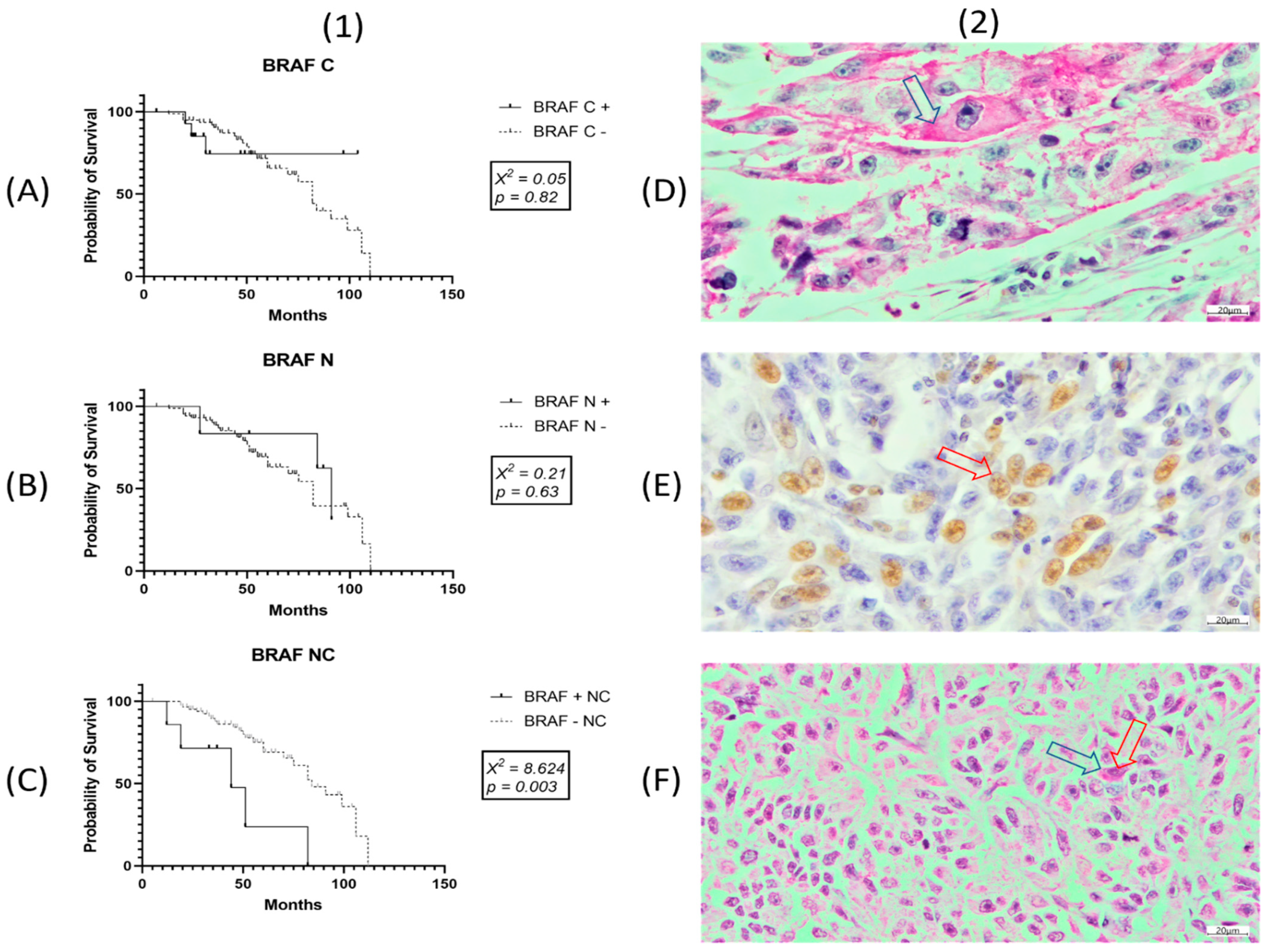
3.5. BRAF Subcellular Localization in SKM
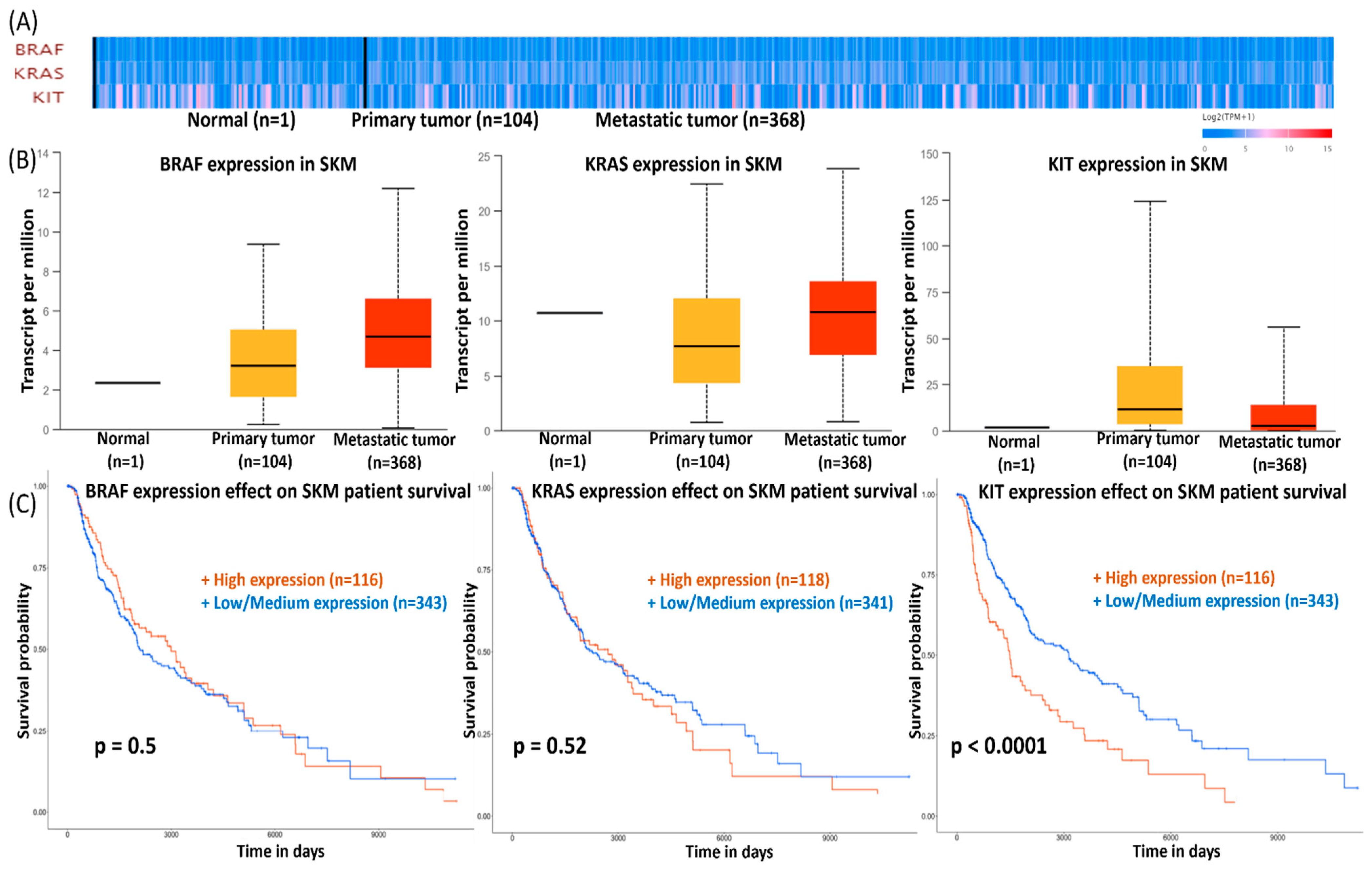
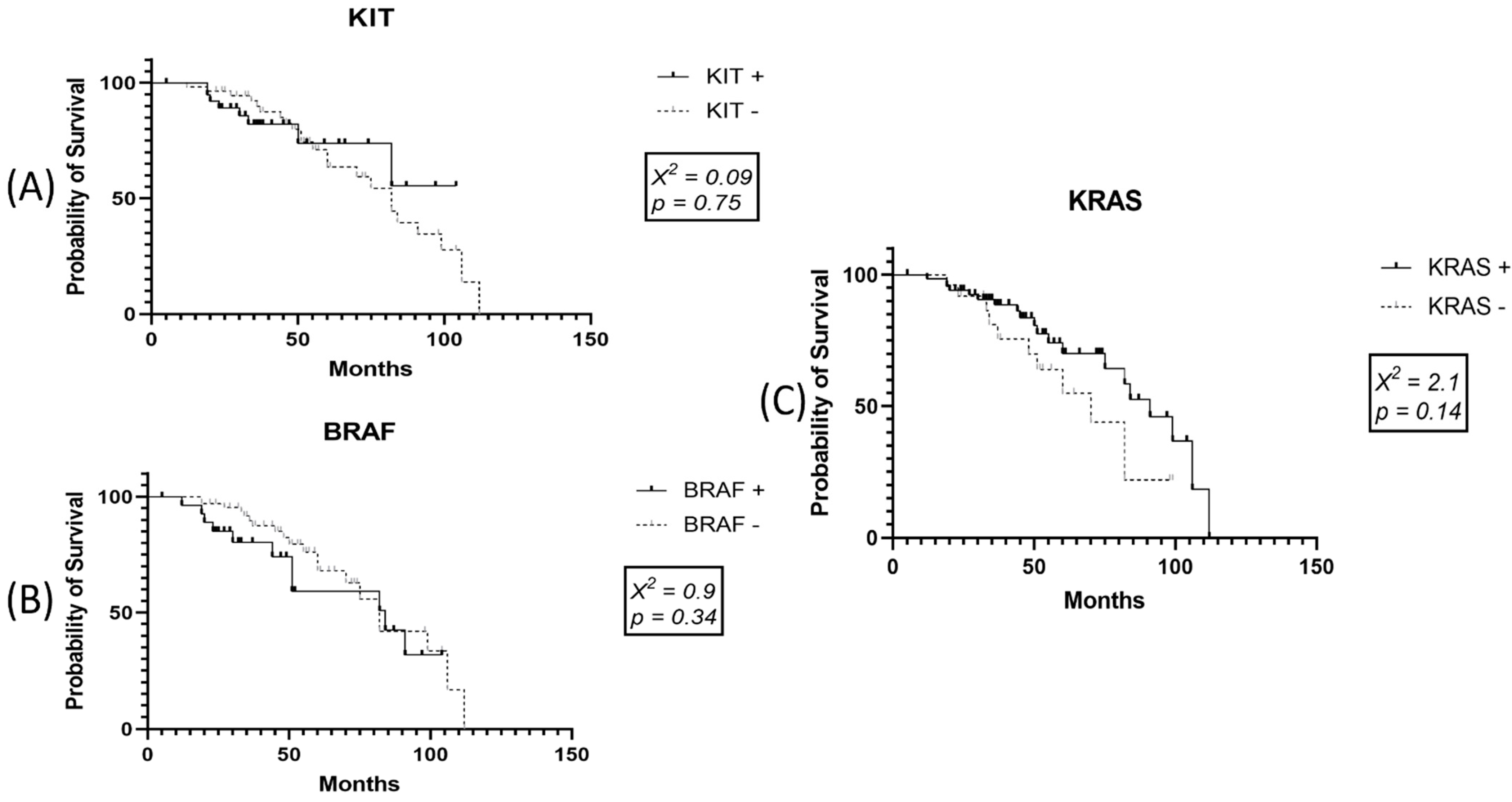
3.6. Survival Data for SKMs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Gurzu, S.; Beleaua, M.A.; Jung, I. The role of tumor microenvironment in development and progression of malignant mela-nomas—A systematic review. Rom. J. Morphol. Embryol. 2018, 59, 23–28. [Google Scholar]
- Akbani, R.; Akdemir, K.C.; Aksoy, B.A.; Albert, M.; Ally, A.; Amin, S.; Arachchi, H.; Arora, A.; Auman, J.T.; Ayala, B.; et al. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef] [PubMed]
- Bruford, E.A.; Braschi, B.; Denny, P.; Jones, T.E.M.; Seal, R.L.; Tweedie, S. Guidelines for human gene nomenclature. Nat. Genet. 2020, 52, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Braicu, C.; Buse, M.; Busuioc, C.; Drula, R.; Gulei, D.; Raduly, L.; Rusu, A.; Irimie, A.; Atanasov, A.G.; Slaby, O.; et al. A Comprehensive Review on MAPK: A Promising Therapeutic Target in Cancer. Cancers 2019, 11, 1618. [Google Scholar] [CrossRef] [PubMed]
- Babaei, M.A.; Kamalidehghan, B.; Saleem, M.; Huri, H.Z.; Ahmadipour, F. Receptor tyrosine kinase (c-Kit) inhibitors: A potential therapeutic target in cancer cells. Drug Des. Dev. Ther. 2016, 10, 2443–2459. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e10. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-Y.; Lee, J.-S. In Silico Analysis of Genomic Data for Construction of Nuclear Receptor Network. Methods Mol. Biol. 2014, 1204, 71–81. [Google Scholar] [CrossRef]
- Edfors, F.; Danielsson, F.; Hallström, B.M.; Käll, L.; Lundberg, E.; Pontén, F.; Forsström, B.; Uhlén, M. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol. Syst. Biol. 2016, 12, 883. [Google Scholar] [CrossRef]
- Ahrné, E.; Molzahn, L.; Glatter, T.; Schmidt, A. Critical assessment of proteome-wide label-free absolute abundance estimation strategies. Proteomics 2013, 13, 2567–2578. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Xia, J. miRNet—Functional Analysis and Visual Exploration of miRNA–Target Interactions in a Network Context. In Computational Cell Biology. Methods in Molecular Biology; von Stechow, L., Santos Delgado, A., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1819, pp. 215–233. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Fan, Y.; Siklenka, K.; Arora, S.K.; Ribeiro, P.; Kimmins, S.; Xia, J. miRNet—Dissecting miRNA—Target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016, 44, W135–W141. [Google Scholar] [CrossRef]
- Elder, D.E.; Massi, D.; Scolyer, R.A.; Willemze, R. (Eds.) WHO Classification of Skin Tumors, 4th ed.; IARC, World Health Organization of Tumors: Lyon, France, 2018; Volume 11. [Google Scholar]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R. Melanoma of the Skin, In AJCC Cancer Staging Manual, 8th ed.; Amin, M.B., Edge, S., Greene, F., Eds.; Springer International Publishing: New York, NY, USA, 2017; pp. 563–585. [Google Scholar]
- Piepkorn, M.W.; Barnhill, R.L.; Elder, D.E.; Knezevich, S.R.; Carney, P.A.; Reisch, L.M.; Elmore, J.G. The MPATH-Dx reporting schema for melanocytic proliferations and melanoma. J. Am. Acad. Dermatol. 2014, 70, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Elmore, J.G.; Barnhill, R.L.; E Elder, D.; Longton, G.M.; Pepe, M.S.; Reisch, L.M.; Carney, P.A.; Titus, L.J.; Nelson, H.D.; Onega, T.; et al. Pathologists’ diagnosis of invasive melanoma and melanocytic proliferations: Observer accuracy and reproducibility study. BMJ 2017, 357, j2813. [Google Scholar] [CrossRef]
- Radick, A.C.; Reisch, L.M.; Shucard, H.L.; Piepkorn, M.W.; Kerr, K.F.; Elder, D.E.; Barnhill, R.L.; Knezevich, S.R.; Oster, N.; Elmore, J.G. Terminology for melanocytic skin lesions and the MPATH-Dx classification schema: A survey of dermatopathologists. J. Cutan. Pathol. 2021, 48, 733–738. [Google Scholar] [CrossRef]
- Gurzu, S.; Szentirmay, Z.; Popa, D.; Jung, I. Practical value of the new system for Maspin assessment, in colorectal cancer. Neoplasma 2013, 60, 373–383. [Google Scholar] [CrossRef][Green Version]
- Tarlock, K.; Alonzo, T.A.; Wang, Y.C.; Gerbing, R.B.; Ries, R.; Loken, M.R.; Pardo, L.; Hylkema, T.; Joaquin, J.; Sarukkai, L.; et al. Functional Properties of KIT Mutations Are Associated with Differential Clinical Outcomes and Response to Targeted Therapeutics in CBF Acute Myeloid Leukemia. Clin. Cancer Res. 2019, 25, 5038–5048. [Google Scholar] [CrossRef] [PubMed]
- Scheffler, M.; Ihle, M.A.; Hein, R.; Merkelbach-Bruse, S.; Scheel, A.H.; Siemanowski, J.; Brägelmann, J.; Kron, A.; Abedpour, N.; Roth, R.; et al. K-ras Mutation Subtypes in NSCLC and Associated Co-occuring Mutations in Other Oncogenic Pathways. J. Thorac. Oncol. 2019, 14, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-N.; Zhao, L.; Yu, L.-F.; Wei, M.-J. BRAF and KRAS mutations in metastatic colorectal cancer: Future perspectives for personalized therapy. Gastroenterol. Rep. 2020, 8, 192–205. [Google Scholar] [CrossRef]
- Ng, J.Y.-S.; Lu, C.T.; Lam, A.K.-Y. BRAF mutation: Current and future clinical pathological applications in colorectal carcinoma. Histol. Histopathol. 2019, 34, 469–477. [Google Scholar] [PubMed]
- Inamura, K. Clinicopathological Characteristics and Mutations Driving Development of Early Lung Adenocarcinoma: Tumor Initiation and Progression. Int. J. Mol. Sci. 2018, 19, 1259. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, F.; Xu, D.; Hou, K.; Fang, W.; Li, Y. The Function of RAS Mutation in Cancer and Advances in its Drug Research. Curr. Pharm. Des. 2019, 25, 1105–1114. [Google Scholar] [CrossRef]
- Ritterhouse, L.L.; Barletta, J.A. BRAF V600E mutation-specific antibody: A review. Semin. Diagn. Pathol. 2015, 32, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.-J.; Shi, X.-H.; Chen, W.-J.; Pu, X.-M.; Sun, Z.-Z.; Halifu, Y.; Wu, X.-J.; Yu, S.-R.; Liu, W.-X.; Liang, J.-Q.; et al. Analysis of KIT mutations and c-KIT expression in Chinese Uyghur and Han patients with melanoma. Clin. Exp. Dermatol. 2016, 41, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Pham, D.D.M.; Guhan, S.; Tsao, H. KIT and Melanoma: Biological Insights and Clinical Implications. Yonsei Med. J. 2020, 61, 562–571. [Google Scholar] [CrossRef]
- Nassar, K.; Tan, A.C. The mutational landscape of mucosal melanoma. Semin. Cancer Biol. 2020, 61, 139–148. [Google Scholar] [CrossRef]
- Doma, V.; Barbai, T.; Beleaua, M.-A.; Kovalszky, I.; Rásó, E.; Tímár, J. KIT Mutation Incidence and Pattern of Melanoma in Central Europe. Pathol. Oncol. Res. 2020, 26, 17–22. [Google Scholar] [CrossRef]
- Oyama, S.; Funasaka, Y.; Watanabe, A.; Takizawa, T.; Kawana, S.; Saeki, H. BRAF, KIT and NRAS mutations and expression of c-KIT, phosphorylated extracellular signal-regulated kinase and phosphorylated AKT in Japanese melanoma patients. J. Dermatol. 2015, 42, 477–484. [Google Scholar] [CrossRef]
- Radu, A.; Bejenaru, C.; Ţolea, I.; Maranduca, M.A.; Brănişteanu, D.C.; Bejenaru, L.E.; Petrariu, F.D.; Stoleriu, G.; Brănişteanu, D.E. Immunohistochemical study of CD117 in various cutaneous melanocytic lesions. Exp. Ther. Med. 2020, 21, 78. [Google Scholar] [CrossRef]
- Uprety, D.; Adjei, A.A. KRAS: From undruggable to a druggable Cancer Target. Cancer Treat. Rev. 2020, 89, 102070. [Google Scholar] [CrossRef] [PubMed]
- Cicenas, J.; Tamosaitis, L.; Kvederaviciute, K.; Tarvydas, R.; Staniute, G.; Kalyan, K.; Meskinyte-Kausiliene, E.; Stankevicius, V.; Valius, M. K-RAS, N-RAS and BRAF mutations in colorectal cancer and melanoma. Med. Oncol. 2017, 34, 26. [Google Scholar] [CrossRef]
- Yu, X.; Ambrosini, G.; Roszik, J.; Eterovic, A.K.; Stempke-Hale, K.; Seftor, E.A.; Chattopadhyay, C.; Grimm, E.; Carvajal, R.D.; Hendrix, M.J.C.; et al. Genetic analysis of the ‘uveal melanoma’ C918 cell line reveals atypical BRAF and common KRAS mutations and single tandem repeat profile identical to the cutaneous melanoma C8161 cell line. Pigment. Cell Melanoma Res. 2015, 28, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Lokhandwala, P.M.; Tseng, L.-H.; Rodriguez, E.; Zheng, G.; Pallavajjalla, A.; Gocke, C.D.; Eshleman, J.R.; Lin, M.-T. Clinical mutational profiling and categorization of BRAF mutations in melanomas using next generation sequencing. BMC Cancer 2019, 19, 665. [Google Scholar] [CrossRef] [PubMed]
- Tsuyama, S.; Kohsaka, S.; Hayashi, T.; Suehara, Y.; Hashimoto, T.; Kajiyama, Y.; Tsurumaru, M.; Ueno, T.; Mano, H.; Yao, T.; et al. Comprehensive clinicopathological and molecular analysis of primary malignant melanoma of the oesophagus. Histopathology 2021, 78, 240–251. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, L.; Gilmour, J.; Bonifer, C. The Role of the Ubiquitously Expressed Transcription Factor Sp1 in Tissue-specific Transcriptional Regulation and in Disease. Yale J. Boil. Med. 2016, 89, 513–525. [Google Scholar]
- Bang, W.; Jeon, Y.-J.; Cho, J.H.; Lee, R.H.; Park, S.-M.; Shin, J.-C.; Choi, N.-J.; Choi, Y.H.; Cho, J.-J.; Seo, J.-M.; et al. β-lapachone suppresses the proliferation of human malignant melanoma cells by targeting specificity protein 1. Oncol. Rep. 2016, 35, 1109–1116. [Google Scholar] [CrossRef][Green Version]
- Carroll, P.A.; Freie, B.W.; Mathsyaraja, H.; Eisenman, R.N. The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 2018, 12, 412–425. [Google Scholar] [CrossRef]
- Pilloni, L.; Bianco, P.; DiFelice, E.; Cabras, S.; Castellanos, M.E.; Atzori, L.; Ferreli, C.; Mulas, P.; Nemolato, S.; Faa, G. The usefulness of c-Kit in the immunohistochemical assessment of melanocytic lesions. Eur. J. Histochem. 2011, 55, e20. [Google Scholar] [CrossRef]
- Shen, S.S.; Zhang, P.S.; Eton, O.; Prieto, V.G. Analysis of protein tyrosine kinase expression in melanocytic lesions by tissue array. J. Cutan. Pathol. 2003, 30, 539–547. [Google Scholar] [CrossRef]
- Kiuru, M.; Tartar, D.M.; Qi, L.; Chen, D.; Yu, L.; Konia, T.; McPherson, J.; Murphy, W.J.; Fung, M.A. Improving classification of melanocytic nevi: Association of BRAF V600E expression with distinct histomorphologic features. J. Am. Acad. Dermatol. 2018, 79, 221–229. [Google Scholar] [CrossRef] [PubMed]
- da Silva, V.M.; Martinez-Barrios, E.; Tell-Marti, G.; Dabad, M.; Carrera, C.; Aguilera, P.; Brualla, D.; Esteve-Codina, A.; Vicente, A.; Puig, S.; et al. Genetic Abnormalities in Large to Giant Congenital Nevi: Beyond NRAS Mutations. J. Investig. Dermatol. 2019, 139, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Xu, W.-H.; Wang, J.-X.; Hou, J.-M.; Zhang, H.-L.; Zhao, X.-Y.; Shen, G.-L. Identification, Validation, and Functional Annotations of Genome-Wide Profile Variation between Melanocytic Nevus and Malignant Melanoma. BioMed Res. Int. 2020, 2020, 1840415. [Google Scholar] [CrossRef]
- Li, X.; Yan, X.; Wang, F.; Yang, Q.; Luo, X.; Kong, J.; Ju, S. Down-regulated lncRNA SLC25A5-AS1 facilitates cell growth and inhibits apoptosis via miR-19a-3p/PTEN/PI3K/AKT signalling pathway in gastric cancer. J. Cell. Mol. Med. 2019, 23, 2920–2932. [Google Scholar] [CrossRef]
- Wa, Q.; Li, L.; Lin, H.; Peng, X.; Ren, N.; Huang, Y.; He, P.; Huang, S. Downregulation of miR-19a-3p promotes invasion, migration and bone metastasis via activating TGF-β signaling in prostate cancer. Oncol. Rep. 2018, 39, 81–90. [Google Scholar] [CrossRef]
- Sun, H.-X.; Yang, Z.-F.; Tang, W.-G.; Ke, A.-W.; Liu, W.-R.; Li, Y.; Gao, C.; Hu, B.; Fu, P.-Y.; Yu, M.-C.; et al. MicroRNA-19a-3p regulates cell growth through modulation of the PIK3IP1-AKT pathway in hepatocellular carcinoma. J. Cancer 2020, 11, 2476–2484. [Google Scholar] [CrossRef]
- He, X.; Zhu, Z.; Johnson, C.; Stoops, J.; Eaker, A.E.; Bowen, W.; DeFrances, M.C. PIK3IP1, a negative regulator of PI3K, suppresses the development of hepatocellular carcinoma. Cancer Res. 2008, 68, 5591–5598. [Google Scholar] [CrossRef]
- Jinnin, M. Recent progress in studies of miRNA and skin diseases. J. Dermatol. 2015, 42, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, L.; Huang, S.; Heynen, G.J.J.E.; Prahallad, A.; Robert, C.; Haanen, J.; Blank, C.; Wesseling, J.; Willems, S.M.; et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014, 508, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Beleaua, M.-A.; Jung, I.; Braicu, C.; Milutin, D.; Gurzu, S. SOX11, SOX10 and MITF Gene Interaction: A Possible Diagnostic Tool in Malignant Melanoma. Life 2021, 11, 281. [Google Scholar] [CrossRef]
- Cloonan, N.; Brown, M.K.; Steptoe, A.L.; Wani, S.; Chan, W.L.; Forrest, A.R.; Kolle, G.; Gabrielli, B.; Grimmond, S.M. The miR-17–5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008, 9, R127. [Google Scholar] [CrossRef]
- Greenberg, E.; Hershkovitz, L.; Itzhaki, O.; Hajdu, S.; Nemlich, Y.; Ortenberg, R.; Gefen, N.; Edry, L.; Modai, S.; Keisari, Y.; et al. Regulation of Cancer Aggressive Features in Melanoma Cells by MicroRNAs. PLoS ONE 2011, 6, e18936. [Google Scholar] [CrossRef]
- Audrito, V.; Serra, S.; Stingi, A.; Orso, F.; Gaudino, F.; Bologna, C.; Neri, F.; Garaffo, G.; Nassini, R.; Baroni, G.; et al. PD-L1 up-regulation in melanoma increases disease aggressiveness and is mediated through miR-17-5p. Oncotarget 2017, 8, 15894–15911. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, S.; Lai, Q.; Fang, Y.; Wu, C.; Liu, Y.; Li, Q.; Wang, X.; Gu, C.; Chen, J.; et al. Cancer-associated fibroblasts-derived exosomal miR-17-5p promotes colorectal cancer aggressive phenotype by initiating a RUNX3/MYC/TGF-β1 positive feedback loop. Cancer Lett. 2020, 491, 22–35. [Google Scholar] [CrossRef]
- Gruszka, R.; Zakrzewski, K.; Liberski, P.P.; Zakrzewska, M. microRNA interaction with MAPK and AKT pathways in paediatric brain tumours—Preliminary results and review of the literature. Folia Neuropathol. 2020, 58, 123–132. [Google Scholar] [CrossRef]
- Babapoor, S.; Wu, R.; Kozubek, J.; Auidi, D.; Grant-Kels, J.M.; Dadras, S.S. Identification of microRNAs associated with invasive and aggressive phenotype in cutaneous melanoma by next-generation sequencing. Lab. Investig. 2017, 97, 636–648. [Google Scholar] [CrossRef][Green Version]
- Ding, Y.; Li, M.; Tayier, T.; Zhang, M.; Chen, L.; Feng, S. Bioinformatics analysis of lncRNA-associated ceRNA network in melanoma. J. Cancer 2021, 12, 2921–2932. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hao, C.; Zhai, R.; Wang, D.; Zhang, J.; Bao, L.; Li, Y.; Yao, W. Downregulation of exosomal let-7a-5p in dust exposed- workers contributes to lung cancer development. Respir. Res. 2018, 19, 235. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pang, Z.; Li, G.; Gu, T. Bioinformatics analysis of differentially expressed miRNAs in non-small cell lung cancer. J. Clin. Lab. Anal. 2021, 35, e23588. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-P.; Huang, C.-C.; Yeh, K.-T.; Ke, T.-W.; Wei, P.-L.; Yang, J.-R.; Cheng, Y.-W. Down-regulation of let-7a-5p predicts lymph node metastasis and prognosis in colorectal cancer: Implications for chemotherapy. Surg. Oncol. 2016, 25, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tang, Q.; Dong, W.; Wang, Y. CircBACH1/let-7a-5p axis enhances the proliferation and metastasis of colorectal cancer by upregulating CREB5 expression. J. Gastrointest. Oncol. 2020, 11, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Tai, Y.; Ma, J. LncRNA NEAT1/let-7a-5p axis regulates the cisplatin resistance in nasopharyngeal carcinoma by targeting Rsf-1 and modulating the Ras-MAPK pathway. Cancer Biol. Ther. 2018, 19, 534–542. [Google Scholar] [CrossRef] [PubMed]
| Parameters | n
(%) 96 | BRAF (28 +) | KRAS (69 +) | KIT (40 +) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| + | − | r | p | + | − | r | p | + | − | r | p | ||
| Gender | |||||||||||||
| Male | 46 (47.91%) | 10 | 36 | −0.16 | 0.12 | 30 | 16 | −0.14 | 0.16 | 16 | 30 | −0.13 | 0.19 |
| Female | 50 (52.09%) | 18 | 32 | 39 | 11 | 24 | 26 | ||||||
| Age (years) | |||||||||||||
| ≤60 | 34 (35.42%) | 8 | 26 | 0.11 | 0.28 | 25 | 9 | −0.13 | 0.19 | 17 | 17 | −0.17 | 0.09 |
| >60 | 62 (64.58%) | 20 | 42 | 44 | 18 | 23 | 39 | ||||||
| Histologic type | |||||||||||||
| Nodular | 71 (73.95%) | 24 | 47 | −0.17 | 0.09 | 50 | 21 | 0.03 | 0.75 | 23 | 48 | 0.34 | 0.0007 |
| Superficial | 17 (17.7%) | 3 | 14 | 12 | 5 | 14 | 3 | ||||||
| Lentiginous | 8 (8.35%) | 1 | 7 | 7 | 1 | 3 | 5 | ||||||
| Breslow thickness | |||||||||||||
| ≤1 mm | 17 (17.7%) | 4 | 13 | 0.17 | 0.09 | 15 | 2 | −0.19 | 0.05 | 13 | 4 | −0.21 | 0.03 |
| >1 to ≤2 mm | 11 (11.45%) | 2 | 9 | 9 | 2 | 7 | 4 | ||||||
| >2 to ≤4 mm | 14 (14.58%) | 2 | 12 | 9 | 5 | 3 | 11 | ||||||
| >4 mm | 54 (56.27%) | 20 | 34 | 36 | 18 | 17 | 37 | ||||||
| Ulceration | |||||||||||||
| Present | 71 (73.95%) | 22 | 49 | 0.07 | 0.49 | 47 | 24 | −0.2 | 0.04 | 22 | 49 | −0.36 | 0.0003 |
| Absent | 25 (26.05%) | 6 | 19 | 22 | 3 | 18 | 7 | ||||||
| Microsatellites | |||||||||||||
| Present | 19 (19.79%) | 7 | 12 | 0.09 | 0.33 | 10 | 9 | −0.18 | 0.07 | 7 | 12 | −0.03 | 0.76 |
| Absent | 77 (80.21%) | 21 | 56 | 59 | 18 | 33 | 44 | ||||||
| Mitotic rate (mm2) | |||||||||||||
| <10 | 64 (66.67%) | 18 | 46 | 0.15 | 0.12 | 46 | 18 | −0.001 | 0.99 | 31 | 33 | −0.3 | 0.002 |
| ≥10 | 32 (33.33%) | 10 | 22 | 23 | 9 | 9 | 23 | ||||||
| TILs | |||||||||||||
| Present | 69 (71.88%) | 19 | 50 | −0.01 | 0.89 | 46 | 18 | −0.18 | 0.08 | 27 | 42 | −0.003 | 0.002 |
| Absent | 27 (28.12%) | 9 | 18 | 23 | 9 | 13 | 14 | ||||||
| Lymphovascular invasion | |||||||||||||
| Present | 21 (21.88%) | 10 | 11 | 0.21 | 0.03 | 17 | 4 | 0.09 | 0.33 | 6 | 15 | −0.14 | 0.15 |
| Absent | 75 (78.12%) | 18 | 57 | 52 | 23 | 34 | 41 | ||||||
| Neurotropism | |||||||||||||
| Present | 9 (9.37%) | 5 | 4 | 0.18 | 0.07 | 4 | 5 | −0.2 | 0.04 | 2 | 7 | −0.13 | 0.2 |
| Absent | 87 (90.63%) | 23 | 64 | 65 | 22 | 38 | 49 | ||||||
| Tumor regression | |||||||||||||
| Present | 31 (32.29%) | 7 | 24 | −0.1 | 0.31 | 24 | 7 | 0.07 | 0.47 | 15 | 16 | 0.08 | 0.39 |
| Absent | 65 (67.71%) | 21 | 44 | 45 | 20 | 25 | 40 | ||||||
| TNM stage | |||||||||||||
| ≤pT2 | 29 (30.21%) | 6 | 23 | 0.16 | 0.11 | 7 | 22 | −0.16 | 0.1 | 20 | 9 | −0.3 | 0.002 |
| ≥pT3 | 67 (69.79%) | 22 | 45 | 62 | 5 | 20 | 47 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beleaua, M.-A.; Jung, I.; Braicu, C.; Milutin, D.; Gurzu, S. Relevance of BRAF Subcellular Localization and Its Interaction with KRAS and KIT Mutations in Skin Melanoma. Int. J. Mol. Sci. 2021, 22, 11918. https://doi.org/10.3390/ijms222111918
Beleaua M-A, Jung I, Braicu C, Milutin D, Gurzu S. Relevance of BRAF Subcellular Localization and Its Interaction with KRAS and KIT Mutations in Skin Melanoma. International Journal of Molecular Sciences. 2021; 22(21):11918. https://doi.org/10.3390/ijms222111918
Chicago/Turabian StyleBeleaua, Marius-Alexandru, Ioan Jung, Cornelia Braicu, Doina Milutin, and Simona Gurzu. 2021. "Relevance of BRAF Subcellular Localization and Its Interaction with KRAS and KIT Mutations in Skin Melanoma" International Journal of Molecular Sciences 22, no. 21: 11918. https://doi.org/10.3390/ijms222111918
APA StyleBeleaua, M.-A., Jung, I., Braicu, C., Milutin, D., & Gurzu, S. (2021). Relevance of BRAF Subcellular Localization and Its Interaction with KRAS and KIT Mutations in Skin Melanoma. International Journal of Molecular Sciences, 22(21), 11918. https://doi.org/10.3390/ijms222111918








