Multi-Omics Approach to Elucidate Cerebrospinal Fluid Changes in Dogs with Intervertebral Disc Herniation
Abstract
1. Introduction
2. Results
2.1. Proteomic Analysis and Proteomic Data Validation
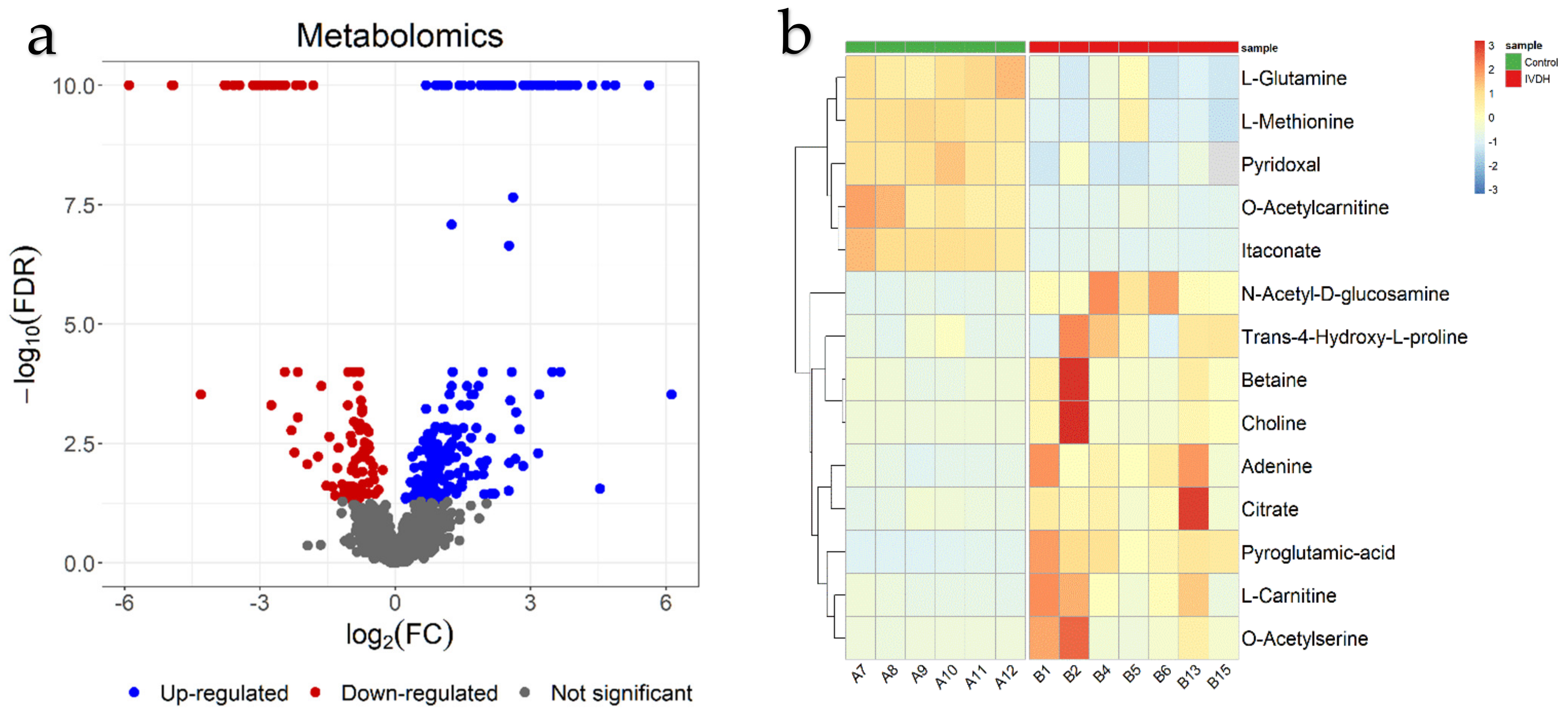
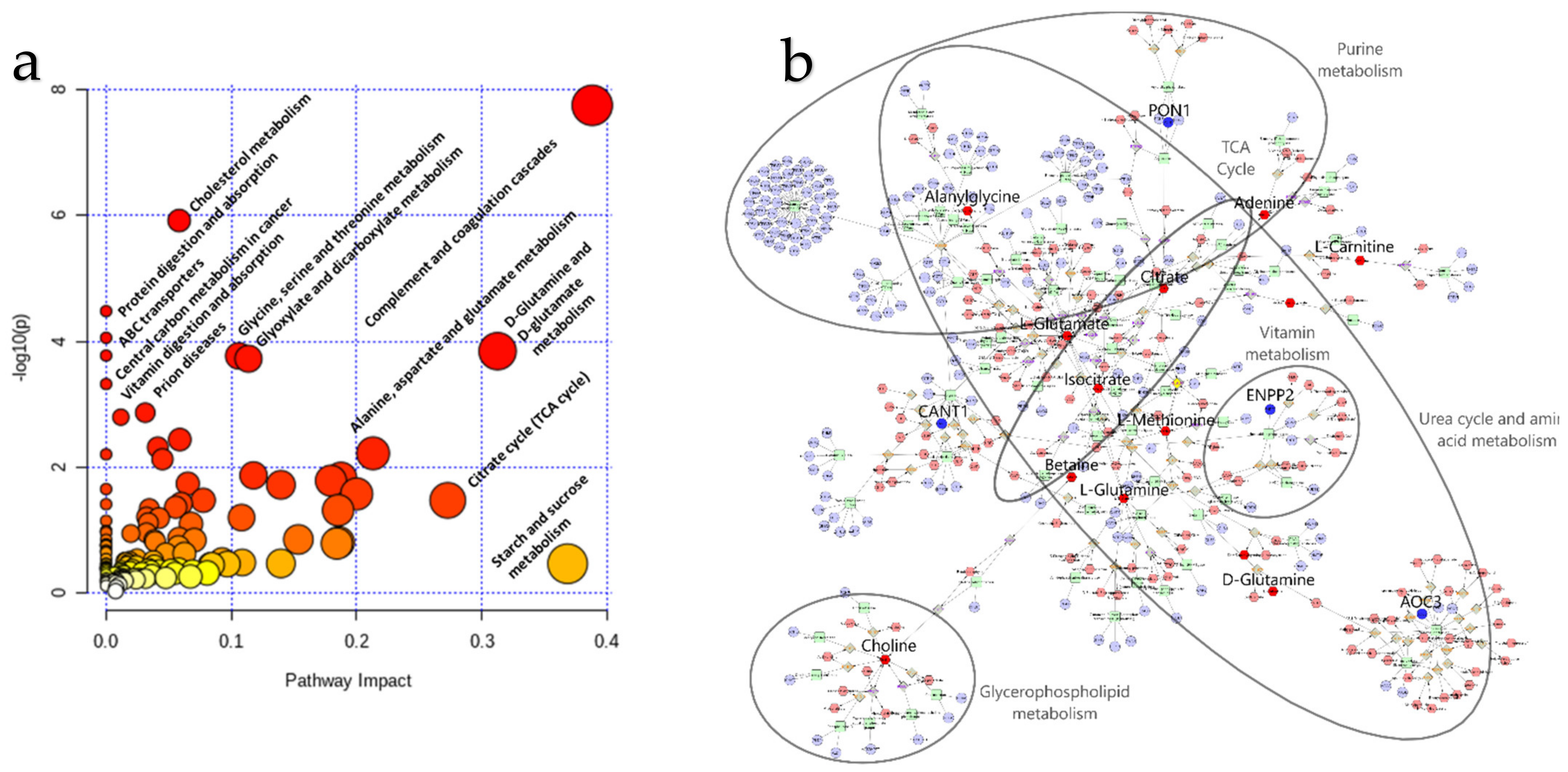
2.2. Metabolomics
2.3. Integration of Omics Data
3. Discussion
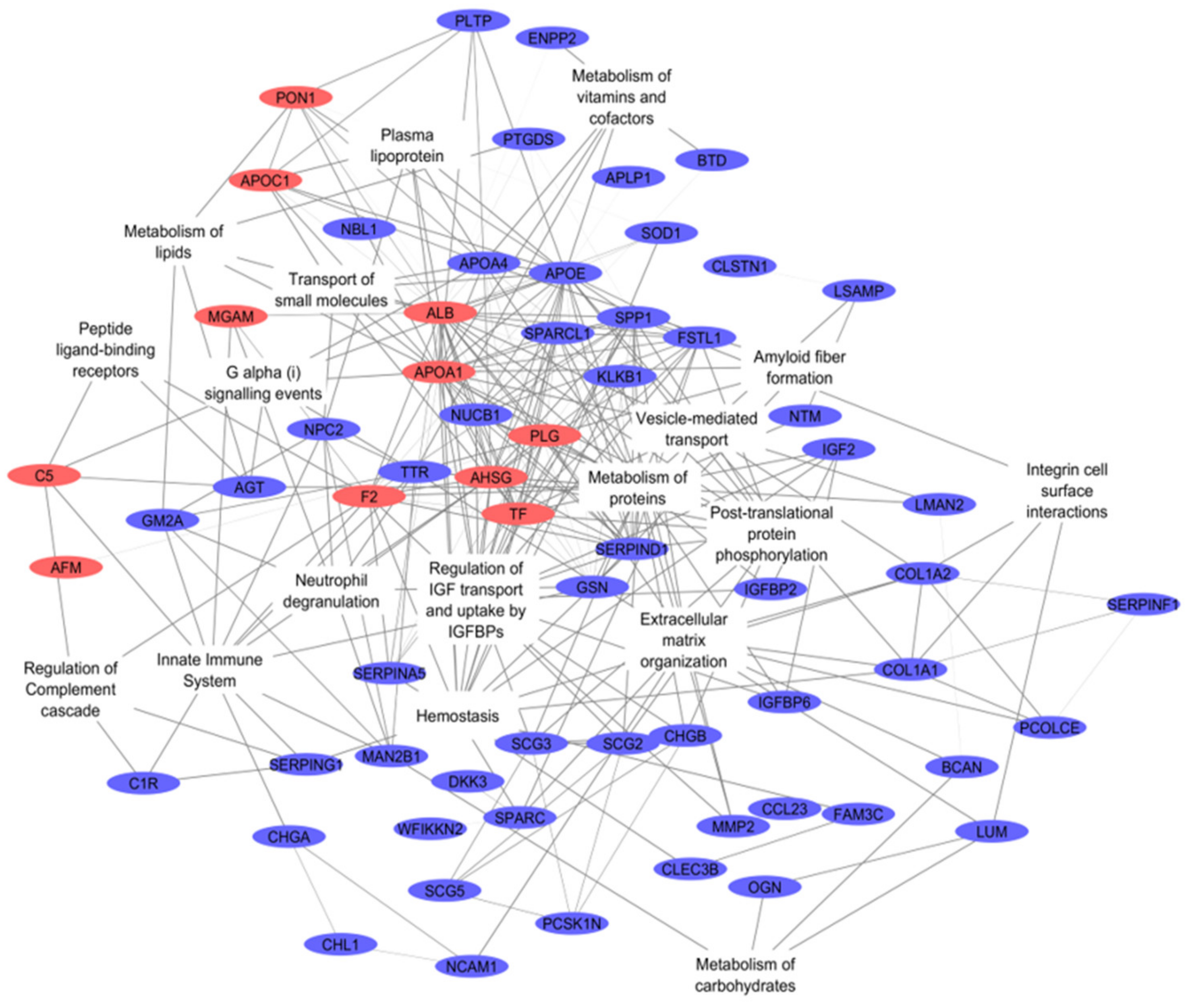
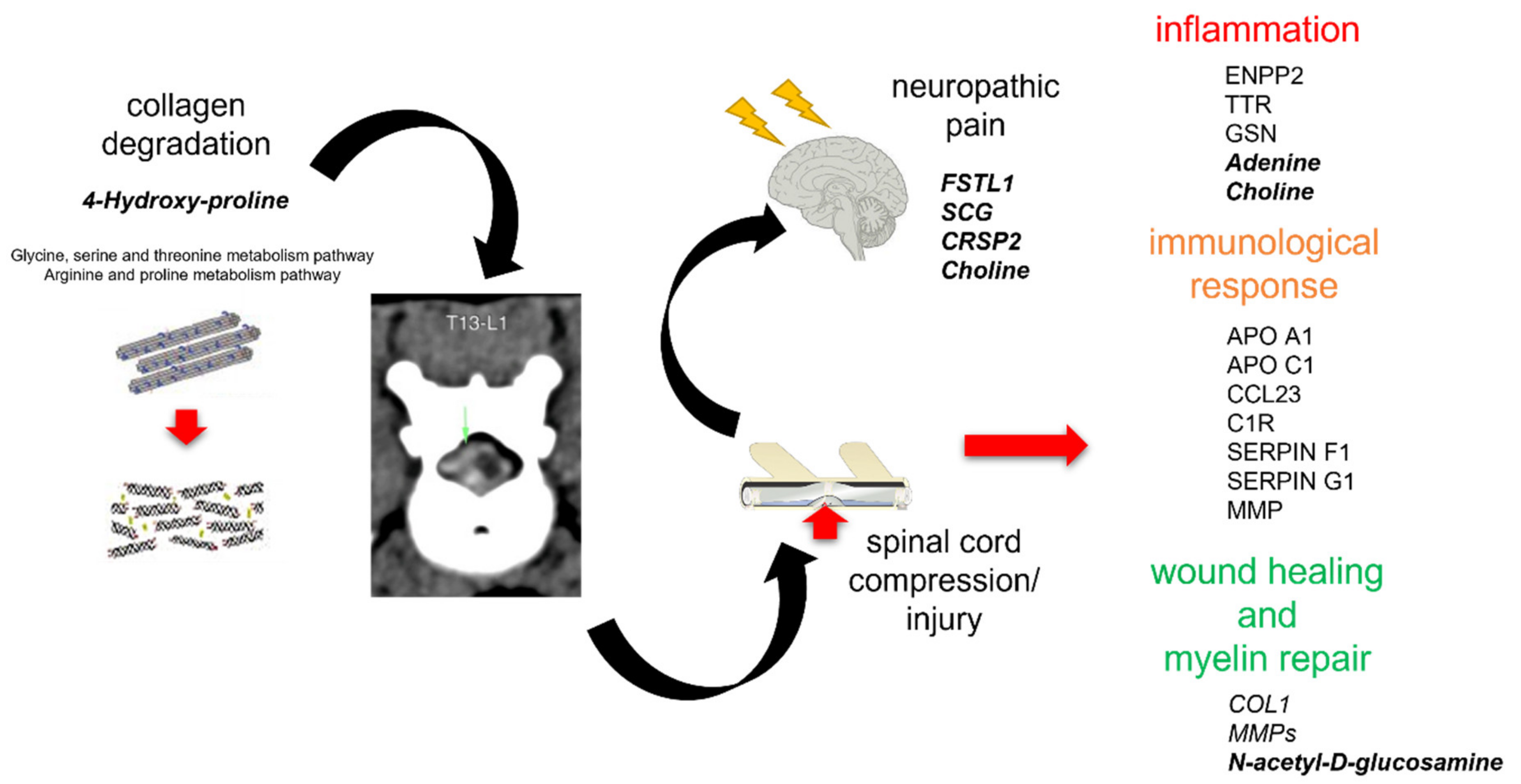
3.1. Integromics Model Reveals Key Molecules for IVDH
3.2. Integrative Omics Analysis Reveals Key Processes Referring to IVDH
4. Materials and Methods
4.1. Experimental Design
4.2. Sample Collection
4.3. Proteomic Analysis
4.4. Analytical Validation of Proteomic Results
4.5. Untargeted Metabolomic Analysis
4.6. Statistical and Bioinformatic Analyses
4.6.1. Proteomics
4.6.2. Proteomic Data Validation
4.6.3. Metabolomics
4.6.4. Statistical Omics Data Integration
4.6.5. Bioinformatic Omics Data Integration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bergknut, N.; Rutges, J.; Kranenburg, H.-J.; Smolders, L.; Hagman, R.; Smidt, H.-J.; Lagerstedt, A.-S.; Penning, L.; Voorhout, G.; Hazewinkel, H.; et al. The Dog as an Animal Model for Intervertebral Disc Degeneration? Spine 2011, 37, 351–358. [Google Scholar] [CrossRef]
- Grabovac, I.; Dorner, T.E. Association between low back pain and various everyday performances. Wien. Klin. Wochenschr. 2019, 131, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.; Moore, S.; Tang, S.; Wiet, M.; Purmessur, D. The chondrodystrophic dog: A clinically relevant intermediate-sized animal model for the study of intervertebral disc-associated spinal pain. JOR Spine 2018, 1, e1011. [Google Scholar] [CrossRef]
- Monchaux, M.; Forterre, S.; Spreng, D.; Karol, A.; Forterre, F.; Wuertz-Kozak, K. Inflammatory Processes Associated with Canine Intervertebral Disc Herniation. Front. Immunol. 2017, 8, 1681. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.A.; Roughley, P.J. What is intervertebral disc degeneration, and what causes it? Spine 2006, 31, 2151–2161. [Google Scholar] [CrossRef] [PubMed]
- Evans, H.E.; Christensen, G.C. Joints and ligaments. In Miller’s Anatomy of the Dog; WB Saunders Co.: Philadelphia, PA, USA, 1979; pp. 225–268. [Google Scholar]
- Liu, C.; Yang, M.; Liu, L.; Zhang, Y.; Zhu, Q.; Huang, C.; Wang, H.; Zhang, Y.; Li, H.; Li, C.; et al. Molecular basis of degenerative spinal disorders from a proteomic perspective (Review). Mol. Med. Rep. 2020, 21, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Risbud, M.V.; Shapiro, I.M. Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nat. Rev. Rheumatol. 2014, 10, 44–56. [Google Scholar] [CrossRef]
- Vergroesen, P.-P.A.; Kingma, I.; Emanuel, K.S.; Hoogendoorn, R.J.W.; Welting, T.J.; van Royen, B.J.; van Dieën, J.H.; Smit, T.H. Mechanics and biology in intervertebral disc degeneration: A vicious circle. Osteoarthr. Cartil. 2015, 23, 1057–1070. [Google Scholar] [CrossRef]
- Forterre, F.; Gorgas, D.; Dickomeit, M.; Jaggy, A.; Lang, J.; Spreng, D. Incidence of spinal compressive lesions in chondrodystrophic dogs with abnormal recovery after hemilaminectomy for treatment of thoracolumbar disc disease: A prospective magnetic resonance imaging study. Vet. Surg. 2010, 39, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.J.; Nerurkar, N.L.; Choi, K.-S.; Harfe, B.D.; Elliott, D.M. Degeneration and regeneration of the intervertebral disc: Lessons from development. Dis. Model. Mech. 2011, 4, 31–41. [Google Scholar] [CrossRef]
- Feussner, I.; Polle, A. What the transcriptome does not tell—Proteomics and metabolomics are closer to the plants’ patho-phenotype. Curr. Opin. Plant Biol. 2015, 26, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Baka, R.; Eckersall, D.; Horvatic, A.; Gelemanovic, A.; Mrljak, V.; McLaughlin, M.; Athanasiou, L.V.; Papaioannou, N.; Stylianaki, I.; Hanh, H.Q.; et al. Quantitative proteomics of cerebrospinal fluid using tandem mass tags in dogs with recurrent epileptic seizures. J. Proteomics 2021, 231, 103997. [Google Scholar] [CrossRef] [PubMed]
- Willard, M.D.; Tvedten, T. Small Animal Clinical Diagnosis by Laboratory Methods, 5th ed; Elsevier Saunders: Saint Louis, MO, USA, 2004; pp. 322–331. ISBN 978-0-7216-8903-6. [Google Scholar]
- Khasawneh, A.H.; Garling, R.J.; Harris, C.A. Cerebrospinal fluid circulation: What do we know and how do we know it? Brain Circ. 2018, 4, 14–18. [Google Scholar] [CrossRef] [PubMed]
- McCabe, S.D.; Lin, D.-Y.; Love, M.I. Consistency and overfitting of multi-omics methods on experimental data. Brief. Bioinform. 2020, 21, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Vitrinel, B.; Koh, H.W.L.; Mujgan Kar, F.; Maity, S.; Rendleman, J.; Choi, H.; Vogel, C. Exploiting Interdata Relationships in Next-generation Proteomics Analysis. Mol. Cell. Proteomics 2019, 18, S5–S14. [Google Scholar] [CrossRef] [PubMed]
- Horvatić, A.; Guillemin, N.; Kaab, H.; McKeegan, D.; O’Reilly, E.; Bain, M.; Kuleš, J.; Eckersall, P.D. Quantitative proteomics using tandem mass tags in relation to the acute phase protein response in chicken challenged with Escherichia coli lipopolysaccharide endotoxin. J. Proteomics 2019, 192, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.N.; Kramer, J.S.; Stoker, A.M.; Bozynski, C.C.; Cook, C.R.; Stannard, J.T.; Choma, T.J.; Cook, J.L. Canine models of spine disorders. JOR SPINE 2020, 3, e1109. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.N.; Jacobsen, H.E.; Khan, J.; Filippi, C.G.; Levine, M.; Lehman, R.A., Jr.; Riew, K.D.; Lenke, L.G.; Chahine, N.O. Inflammatory biomarkers of low back pain and disc degeneration: A review. Ann. N. Y. Acad. Sci. 2017, 1410, 68–84. [Google Scholar] [CrossRef]
- Skouen, J.S.; Larsen, J.L.; Gjerde, I.O.; Hegrestad, S.E.; Vollset, S.E. Cerebrospinal fluid protein concentrations in patients with sciatica caused by lumbar disc herniation: An investigation of biochemical, neurologic, and radiologic predictors of long-term outcome. J. Spinal Disord. 1997, 10, 505–511. [Google Scholar] [CrossRef]
- Stolić, I.; Škrlin, B.; Bubić Špoljar, J.; Smolec, O.; Radišić, B.; Pećin, M.; Pirkić, B.; Lipar, M. Cerebrospinal fluid assessment in dogs with spinal tumors and intervertebral disk herniation. Vet. Stanica 2020, 51, 387–392. [Google Scholar] [CrossRef]
- Sarath Babu, N.; Krishnan, S.; Brahmendra Swamy, C.V.; Venkata Subbaiah, G.P.; Gurava Reddy, A.V.; Idris, M.M. Quantitative proteomic analysis of normal and degenerated human intervertebral disc. Spine J. 2016, 16, 989–1000. [Google Scholar] [CrossRef]
- Erwin, W.M.; DeSouza, L.; Funabashi, M.; Kawchuk, G.; Karim, M.Z.; Kim, S.; Mӓdler, S.; Matta, A.; Wang, X.; Mehrkens, K.A. The biological basis of degenerative disc disease: Proteomic and biomechanical analysis of the canine intervertebral disc. Arthritis Res. Ther. 2015, 17, 240. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-D.; Zeng, B.-F.; Xu, J.-G.; Zhu, H.-B.; Xia, Q.-C. Proteomic analysis of the cerebrospinal fluid of patients with lumbar disk herniation. Proteomics 2006, 6, 1019–1028. [Google Scholar] [CrossRef]
- Simats, A.; Ramiro, L.; García-Berrocoso, T.; Briansó, F.; Gonzalo, R.; Martín, L.; Sabé, A.; Gill, N.; Penalba, A.; Colomé, N.; et al. A Mouse Brain-based Multi-omics Integrative Approach Reveals Potential Blood Biomarkers for Ischemic Stroke. Mol. Cell. Proteomics 2020, 19, 1921–1935. [Google Scholar] [CrossRef] [PubMed]
- Montaner, J.; Ramiro, L.; Simats, A.; Tiedt, S.; Makris, K.; Jickling, G.C.; Debette, S.; Sanchez, J.-C.; Bustamante, A. Multilevel omics for the discovery of biomarkers and therapeutic targets for stroke. Nat. Rev. Neurol. 2020, 16, 247–264. [Google Scholar] [CrossRef] [PubMed]
- Stoop, M.P.; Coulier, L.; Rosenling, T.; Shi, S.; Smolinska, A.M.; Buydens, L.; Ampt, K.; Stingl, C.; Dane, A.; Muilwijk, B.; et al. Quantitative Proteomics and Metabolomics Analysis of Normal Human Cerebrospinal Fluid Samples. Mol. Cell. Proteomics 2010, 9, 2063–2075. [Google Scholar] [CrossRef]
- French, C.D.; Willoughby, R.E.; Pan, A.; Wong, S.J.; Foley, J.F.; Wheat, L.J.; Fernandez, J.; Encarnacion, R.; Ondrush, J.M.; Fatteh, N.; et al. NMR metabolomics of cerebrospinal fluid differentiates inflammatory diseases of the central nervous system. PLoS Negl. Trop. Dis. 2018, 12, e0007045. [Google Scholar] [CrossRef]
- Sengupta, M.B.; Basu, M.; Iswarari, S.; Mukhopadhyay, K.K.; Sardar, K.P.; Acharyya, B.; Mohanty, P.K.; Mukhopadhyay, D. CSF Proteomics of Secondary Phase Spinal Cord Injury in Human Subjects: Perturbed Molecular Pathways Post Injury. PLoS ONE 2014, 9, e110885. [Google Scholar]
- Topsakal, C.; Kilic, N.; Ozveren, F.; Akdemir, I.; Kaplan, M.; Tiftikci, M.; Gursu, F. Effects of prostaglandin E1, melatonin, and oxytetracycline on lipid peroxidation, antioxidant defense system, paraoxonase (PON1) activities, and homocysteine levels in an animal model of spinal cord injury. Spine 2003, 28, 1643–1652. [Google Scholar] [CrossRef][Green Version]
- Willkommen, D.; Lucio, M.; Moritz, F.; Forcisi, S.; Kanawati, B.; Smirnov, K.S.; Schroeter, M.; Sigaroudi, A.; Schmitt-Kopplin, P.; Michalke, B. Metabolomic investigations in cerebrospinal fluid of Parkinson’s disease. PLoS ONE 2018, 13, e0208752. [Google Scholar] [CrossRef]
- Murgia, F.; Lorefice, L.; Poddighe, S.; Fenu, G.; Secci, M.A.; Marrosu, M.G.; Cocco, E.; Atzori, L. Multi-Platform Characterization of Cerebrospinal Fluid and Serum Metabolome of Patients Affected by Relapsing–Remitting and Primary Progressive Multiple Sclerosis. J. Clin. Med. 2020, 9, 863. [Google Scholar] [CrossRef] [PubMed]
- Sperringer, J.E.; Addington, A.; Hutson, S.M. Branched-Chain Amino Acids and Brain Metabolism. Neurochem. Res. 2017, 42, 1697–1709. [Google Scholar] [CrossRef]
- Liu, Y.; Hyde, A.S.; Simpson, M.A.; Barycki, J.J. Emerging Regulatory Paradigms in Glutathione Metabolism. Adv. Cancer Res. 2014, 122, 69–101. [Google Scholar] [PubMed]
- Fenves, A.Z.; Kirkpatrick, H.M.; Patel, V.V.; Sweetman, L.; Emmett, M. Increased Anion Gap Metabolic Acidosis as a Result of 5-Oxoproline (Pyroglutamic Acid): A Role for Acetaminophen. Clin. J. Am. Soc. Nephrol. 2006, 1, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Shin, Y.; Kim, T.H.; Kim, D.-H.; Lee, A. Plasma metabolites as possible biomarkers for diagnosis of breast cancer. PLoS ONE 2019, 14, e0225129. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Bruno, D.; Nierenberg, J.; Marmar, C.R.; Zetterberg, H.; Blennow, K.; Pomara, N. Abnormality in glutamine–glutamate cycle in the cerebrospinal fluid of cognitively intact elderly individuals with major depressive disorder: A 3-year follow-up study. Transl. Psychiatry 2016, 6, e744. [Google Scholar] [CrossRef] [PubMed]
- Belov Kirdajova, D.; Kriska, J.; Tureckova, J.; Anderova, M. Ischemia-Triggered Glutamate Excitotoxicity From the Perspective of Glial Cells. Front. Cell. Neurosci. 2020, 14, 51. [Google Scholar] [CrossRef]
- Chen, Q.; Meng, Z.; Su, R. WERFE: A Gene Selection Algorithm Based on Recursive Feature Elimination and Ensemble Strategy. Front. Bioeng. Biotechnol. 2020, 8, 496. [Google Scholar] [CrossRef]
- EL-Manzalawy, Y.; Hsieh, T.-Y.; Shivakumar, M.; Kim, D.; Honavar, V. Min-redundancy and max-relevance multi-view feature selection for predicting ovarian cancer survival using multi-omics data. BMC Med. Genom. 2018, 11, 71. [Google Scholar] [CrossRef] [PubMed]
- Johannes, M.; Brase, J.C.; Fröhlich, H.; Gade, S.; Gehrmann, M.; Fälth, M.; Sültmann, H.; Beißbarth, T. Integration of pathway knowledge into a reweighted recursive feature elimination approach for risk stratification of cancer patients. Bioinformatics 2010, 26, 2136–2144. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.-B.; Rajapakse, J.C.; Wang, H.; Azuaje, F. Multiple SVM-RFE for gene selection in cancer classification with expression data. IEEE Trans. Nanobioscience 2005, 4, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Li, K.-C.; Wang, F.; Zhong, Y.-Q.; Lu, Y.-J.; Wang, Q.; Zhang, F.-X.; Xiao, H.-S.; Bao, L.; Zhang, X. Reduction of follistatin-like 1 in primary afferent neurons contributes to neuropathic pain hypersensitivity. Cell Res. 2011, 21, 697–699. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tulke, S.; Williams, P.; Hellysaz, A.; Ilegems, E.; Wendel, M.; Broberger, C. Nucleobindin 1 (NUCB1) is a Golgi-resident marker of neurons. Neuroscience 2016, 314, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Sy, M.; Brandt, A.U.; Lee, S.-U.; Newton, B.L.; Pawling, J.; Golzar, A.; Rahman, A.A.; Yu, Z.; Cooper, G.; Scheel, M.; et al. N-Acetylglucosamine drives myelination by triggering oligodendrocyte precursor cell differentiation. J. Biol. Chem. 2020, 295, 17413–17424. [Google Scholar] [CrossRef]
- Yang, X.; Chen, S.; Shao, Z.; Li, Y.; Wu, H.; Li, X.; Mao, L.; Zhou, Z.; Bai, L.; Mei, X.; et al. Apolipoprotein E Deficiency Exacerbates Spinal Cord Injury in Mice: Inflammatory Response and Oxidative Stress Mediated by NF-κB Signaling Pathway. Front. Cell. Neurosci. 2018, 12, 142. [Google Scholar] [CrossRef]
- Lind, A.-L.; Just, D.; Mikus, M.; Fredolini, C.; Ioannou, M.; Gerdle, B.; Ghafouri, B.; Bäckryd, E.; Tanum, L.; Gordh, T.; et al. CSF levels of apolipoprotein C1 and autotaxin found to associate with neuropathic pain and fibromyalgia. J. Pain Res. 2019, 12, 2875–2889. [Google Scholar] [CrossRef]
- Salsas-Escat, R.; Stultz, C.M. The Molecular Mechanics of Collagen Degradation: Implications for Human Disease. Exp. Mech. 2009, 49, 65–77. [Google Scholar] [CrossRef]
- Radek, M.; Pacholczyk-Sienicka, B.; Jankowski, S.; Albrecht, Ł.; Grodzka, M.; Depta, A.; Radek, A. Assessing the correlation between the degree of disc degeneration on the Pfirrmann scale and the metabolites identified in HR-MAS NMR spectroscopy. Magn. Reson. Imaging 2016, 34, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.A.; Pappan, K.; Thompson, P.A.; Want, E.J.; Siskos, A.P.; Keun, H.C.; Wulff, J.; Hu, C.; Lang, J.E.; Chow, H.-H.S. Plasma Metabolomic Profiles of Breast Cancer Patients after Short-term Limonene Intervention. Cancer Prev. Res. 2015, 8, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Pinto-Sanchez, M.I.; Bercik, P.; Britz-McKibbin, P. Metabolomics reveals elevated urinary excretion of collagen degradation and epithelial cell turnover products in irritable bowel syndrome patients. Metabolomics 2019, 15, 82. [Google Scholar] [CrossRef]
- Ågren, M.S.; Jorgensen, I.N.; Andersen, M.; Viljanto, J.; Gottrup, P. Matrix metalloproteinase 9 level predicts optimal collagen deposition during early wound repair in humans. Brit. J. Surgery 1998, 85, 68–71. [Google Scholar] [CrossRef]
- Cao, X.; Feng, S.; Fu, C.; Gao, K.; Guo, J.; Guo, X.; He, X.; Huang, Z.; Li, Z.; Liu, L.; et al. Repair, protection and regeneration of spinal cord injury. Neural Regen. Res. 2015, 10, 1953. [Google Scholar] [CrossRef] [PubMed]
- Albayar, A.A.; Roche, A.; Swiatkowski, P.; Antar, S.; Ouda, N.; Emara, E.; Smith, D.H.; Ozturk, A.K.; Awad, B.I. Biomarkers in Spinal Cord Injury: Prognostic Insights and Future Potentials. Front. Neurol. 2019, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Streijger, F.; Wang, Y.; Lin, G.; Christie, S.; Mac-Thiong, J.-M.; Parent, S.; Bailey, C.S.; Paquette, S.; Boyd, M.C.; et al. Parallel Metabolomic Profiling of Cerebrospinal Fluid and Serum for Identifying Biomarkers of Injury Severity after Acute Human Spinal Cord Injury. Sci. Rep. 2016, 6, 38718. [Google Scholar] [CrossRef] [PubMed]
- Mangaraj, M.; Nanda, R.; Panda, S. Apolipoprotein A-I: A Molecule of Diverse Function. Indian J. Clin. Biochem. 2016, 31, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Lepennetier, G.; Hracsko, Z.; Unger, M.; van Griensven, M.; Grummel, V.; Krumbholz, M.; Berthele, A.; Hemmer, B.; Kowarik, M.C. Cytokine and immune cell profiling in the cerebrospinal fluid of patients with neuro-inflammatory diseases. J. Neuroinflamm. 2019, 16, 219. [Google Scholar] [CrossRef]
- Simats, A.; García-Berrocoso, T.; Penalba, A.; Giralt, D.; Llovera, G.; Jiang, Y.; Ramiro, L.; Bustamante, A.; Martinez-Saez, E.; Canals, F.; et al. CCL 23: A new CC chemokine involved in human brain damage. J. Intern. Med. 2018, 283, 461–475. [Google Scholar] [CrossRef]
- Rabin, R.L. CC, C, and CX3C Chemokines, Encyclopedia of Hormones; Academic Press: Cambridge, MA, USA, 2003; pp. 255–263. [Google Scholar] [CrossRef]
- Könnecke, H.; Bechmann, I. The Role of Microglia and Matrix Metalloproteinases Involvement in Neuroinflammation and Gliomas. Clin. Dev. Immunol. 2013, 2013, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mlekusch, R.; Humpel, C. Matrix metalloproteinases-2 and -3 are reduced in cerebrospinal fluid with low beta-amyloid1–42 levels. Neurosci. Lett. 2009, 466, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef] [PubMed]
- Magdalon, J.; Mansur, F.; Teles e Silva, A.L.; de Goes, V.A.; Reiner, O.; Sertié, A.L. Complement System in Brain Architecture and Neurodevelopmental Disorders. Front. Neurosci. 2020, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Gompels, M.M.; Lock, R.J.; Abinun, M.; Bethune, C.A.; Davies, G.; Grattan, C.; Fay, A.C.; Longhurst, H.J.; Morrison, L.; Price, A.; et al. C1 inhibitor deficiency: Consensus document. Clin. Exp. Immunol. 2005, 139, 379–394. [Google Scholar] [CrossRef]
- Gerdle, B.; Ghafouri, B. Proteomic studies of common chronic pain conditions—A systematic review and associated network analyses. Expert Rev. Proteom. 2020, 17, 483–505. [Google Scholar] [CrossRef]
- Teckchandani, S.; Nagana Gowda, G.A.; Raftery, D.; Curatolo, M. Metabolomics in chronic pain research. Eur. J. Pain 2021, 25, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Pfyffer, D.; Wyss, P.O.; Huber, E.; Curt, A.; Henning, A.; Freund, P. Metabolites of neuroinflammation relate to neuropathic pain after spinal cord injury. Neurology 2020, 95, e805–e814. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Ichesco, E.; Ratai, E.-M.; Gonzalez, R.G.; Burdo, T.; Loggia, M.L.; Harris, R.E.; Napadow, V. Magnetic resonance imaging of neuroinflammation in chronic pain: A role for astrogliosis? Pain 2020, 161, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Simpson, K.L.; Lancashire, L.J.; Walker, M.J.; Dawson, M.J.; Unwin, R.D.; Rembielak, A.; Price, P.; West, C.; Dive, C.; et al. Statistical Considerations of Optimal Study Design for Human Plasma Proteomics and Biomarker Discovery. J. Proteome Res. 2012, 11, 2103–2113. [Google Scholar] [CrossRef]
- Bharucha, T.; Gangadharan, B.; Kumar, A.; de Lamballerie, X.; Newton, P.N.; Winterberg, M.; Dubot-Pérès, A.; Zitzmann, N. Mass spectrometry-based proteomic techniques to identify cerebrospinal fluid biomarkers for diagnosing suspected central nervous system infections. A systematic review. J. Infect. 2019, 79, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Bednarski, R.; Grimm, K.; Harvey, R.; Lukasik, V.M.; Penn, W.S.; Sargent, B.; Spelts, K. American Animal Hospital Association. AAHA anesthesia guidelines for dogs and cats. J. Am. Anim. Hosp. Assoc. 2011, 7, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Tvarijonaviciute, A.; Tecles, F.; Caldin, M.; Tasca, S.; Cerón, J. Validation of spectrophotometric assays for serum paraoxonase type-1 measurement in dogs. Am. J. Vet. Res. 2012, 73, 34–41. [Google Scholar] [CrossRef]
- Weidmeyer, C.E.; Solter, P.F. Validation of human haptoglobin immunoturbidimetric assay for detection of haptoglobin in equine and canine serum and plasma. Vet. Clin. Pathol. 1996, 25, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.C.; Mudaliar, M.; Tassi, R.; McNeilly, T.N.; Burchmore, R.; Burgess, K.; Herzyk, P.; Zadoks, R.N.; Eckersall, P.D. Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 3. Untargeted metabolomics. Mol. Biosyst. 2016, 12, 2762–2769. [Google Scholar] [CrossRef]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2019, 48, D440–D444. [Google Scholar] [CrossRef] [PubMed]
- Gloaguen, Y.; Morton, F.; Daly, R.; Gurden, R.; Rogers, S.; Wandy, J.; Wilson, D.; Barrett, M.; Burgess, K. PiMP my metabolome: An integrated, web-based tool for LC-MS metabolomics data. Bioinformatics 2017, 33, 4007–4009. [Google Scholar] [CrossRef] [PubMed]
- Holman, J.D.; Tabb, D.L.; Mallick, P. Employing ProteoWizard to Convert Raw Mass Spectrometry Data. Curr. Protoc. Bioinforma. 2014, 46, 13–24. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Komsta, L. Outliers: Tests for outliers. In R Package Version 0.14; R Core Team: Vienna, Austria, 2011. [Google Scholar]
- Wickham, H. About the ggplot2 Package. J. Appl. Comput. Math. 2016, 5, 4. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Kuhn, M. Classification and Regression Training. R Package Version 6.0-86. 2020. Available online: https://cran.r-project.org/web/packages/caret/index.html (accessed on 14 May 2021).
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [PubMed]
- De Jay, N.; Papillon-Cavanagh, S.; Olsen, C.; El-Hachem, N.; Bontempi, G.; Haibe-Kains, B. mRMRe: An R package for parallelized mRMR ensemble feature selection. Bioinformatics 2013, 29, 2365–2368. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
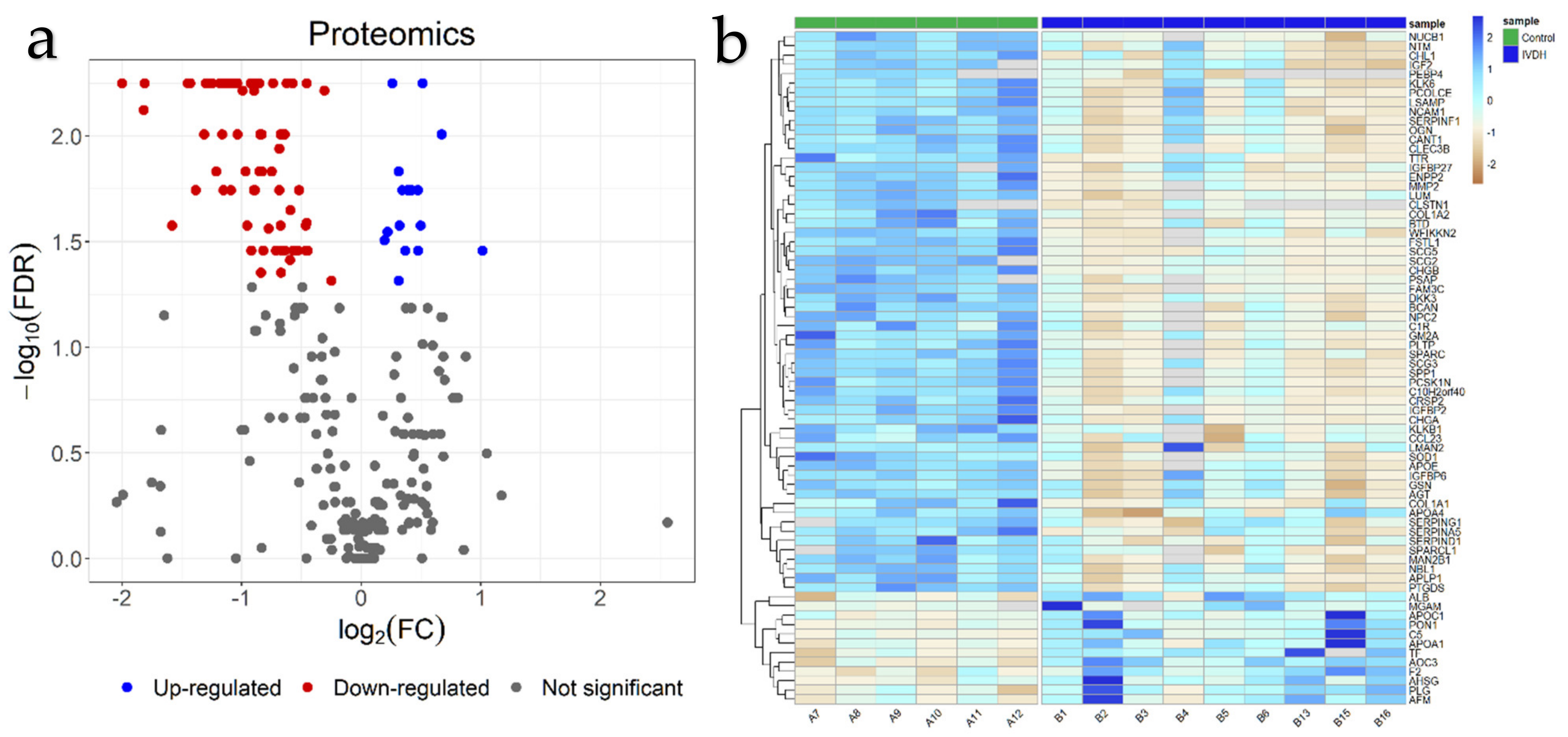

| Higher in Abundance in IVDH Group (Upregulated) | Higher in Abundance in Control Group (Downregulated) | ||||||
|---|---|---|---|---|---|---|---|
| Accession No. (NCBI) | Adjusted p-Value | log2FC | Gene Symbol | Accession No. (NCBI) | Adjusted p-Value | log2FC | Gene Symbol |
| 359321961 | 0.0147 | 0.31 | F2 | 345789648 | 0.0056 | −2.00 | CHGB |
| 73975389 | 0.0349 | 0.37 | AFM | 1418342647 | 0.0075 | −1.82 | SCG2 |
| 1239925760 | 0.0181 | 0.39 | C5 | 345795204 | 0.0056 | −1.81 | SCG5 |
| 18139619 | 0.0181 | 0.41 | PLG | 3046976 | 0.0266 | −1.58 | COL1A1 |
| 1418515495 | 0.0181 | 0.43 | AHSG | 1239931864 | 0.0056 | −1.43 | ENPP2 |
| 2147092 | 0.0181 | 0.48 | ALB | 73970011 | 0.0181 | −1.38 | C10H2orf40 |
| 3915607 | 0.0266 | 0.50 | APOA1 | 1418314244 | 0.0098 | −1.31 | TTR |
| 1239918269 | 0.0056 | 0.52 | AOC3 | 1239976444 | 0.0056 | −1.30 | FSTL1 |
| 73975797 | 0.0098 | 0.68 | PON1 | 1418222978 | 0.0056 | −1.30 | CRSP2 |
| 1418304654 | 0.0349 | 1.01 | APOC1 | 197085524 | 0.0056 | −1.30 | CRSP4 |
| REACTOME Pathway Name | ID | # Genes | # Background Genes | Adjusted p-Value |
|---|---|---|---|---|
| Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) | HSA-381426 | 19 | 123 | 5.86 × 10−23 |
| Post-translational protein phosphorylation | HSA-8957275 | 13 | 106 | 2.79 × 10−14 |
| Platelet activation, signaling and aggregation | HSA-76002 | 16 | 256 | 1.04 × 10−13 |
| Hemostasis | HSA-109582 | 19 | 601 | 1.14 × 10−11 |
| Extracellular matrix organization | HSA-1474244 | 12 | 298 | 2.69 × 10−8 |
| Metabolism of proteins | HSA-392499 | 26 | 1948 | 3.46 × 10−8 |
| Binding and uptake of ligands by scavenger receptors | HSA-2173782 | 6 | 40 | 3.17 × 10−7 |
| Intrinsic pathway of fibrin clot formation | HSA-140837 | 5 | 22 | 7.83 × 10−7 |
| ECM proteoglycans | HSA-3000178 | 6 | 75 | 7.48 × 10−6 |
| Degradation of the extracellular matrix | HSA-1474228 | 7 | 139 | 1.30 × 10−5 |
| Post-translational protein modification | HSA-597592 | 17 | 1366 | 6.26 × 10−5 |
| Plasma lipoprotein remodeling | HSA-8963899 | 4 | 30 | 7.28 × 10−5 |
| Chylomicron remodeling | HSA-8963901 | 3 | 10 | 1.20 × 10−4 |
| Innate immune system | HSA-168249 | 14 | 1012 | 1.20 × 10−4 |
| Regulation of complement cascade | HSA-977606 | 4 | 47 | 2.70 × 10−4 |
| Neutrophil degranulation | HSA-6798695 | 9 | 471 | 3.70 × 10−4 |
| Crosslinking of collagen fibrils | HSA-2243919 | 3 | 18 | 3.90 × 10−4 |
| Metabolism of vitamins and cofactors | HSA-196854 | 6 | 185 | 4.20 × 10−4 |
| Amyloid fiber formation | HSA-977225 | 4 | 78 | 1.30 × 10−3 |
| Integrin cell surface interactions | HSA-216083 | 4 | 83 | 1.60 × 10−3 |
| Activation of matrix metalloproteinases | HSA-1592389 | 3 | 33 | 1.60 × 10−3 |
| Diseases associated with glycosaminoglycan metabolism | HSA-3560782 | 3 | 40 | 2.50 × 10−3 |
| Collagen degradation | HSA-1442490 | 3 | 64 | 7.20 × 10−3 |
| Glycosphingolipid metabolism | HSA-1660662 | 2 | 44 | 3.68 × 10−2 |
| Glycosaminoglycan metabolism | HSA-1630316 | 3 | 122 | 3.22 × 10−2 |
| Higher in Abundance in IVDH Group (Upregulated) | Higher in Abundance in Control Group (Downregulated) | ||||||
|---|---|---|---|---|---|---|---|
| Peak ID | Metabolite | log2FC | Adjusted p-Value | Peak ID | Metabolite | log2FC | Adjusted p-Value |
| 693 | Hydroxyproline | 0.76 | 3.00 × 10−2 | 2325 | Itaconate | −3.78 | 5.1 × 10−10 |
| 23 | Betaine | 1.08 | 4.16 × 10−2 | 858 | O-Acetylcarnitine | −2.73 | 4.6 × 10−6 |
| 1789 | Citrate | 1.19 | 1.44 × 10−2 | 847 | Pyridoxal | −2.30 | 0.0017 |
| 1410 | N-Acetyl-D-glucosamine | 1.46 | 5.00 × 10−4 | 2717 | L-Methionine | −2.16 | 0.0009 |
| 688 | Adenine | 1.51 | 1.50 × 10−2 | 2070 | L-Glutamine | −0.83 | 0.0002 |
| 217 | L-Carnitine | 1.65 | 1.51 × 10−2 | ||||
| 24 | Choline | 1.79 | 1.46 × 10−2 | ||||
| 466 | L-Glutamate | 2.51 | 3.10 × 10−2 | ||||
| 245 | Pyroglutamic acid | 2.52 | 2.30 × 10−7 | ||||
| KEGG Pathway Name | Pathway ID | Assigned Formulas | Total Pathway Formulas | Pathway Coverage (%) |
|---|---|---|---|---|
| C5-Branched dibasic acid metabolism | 99 | 7 | 21 | 33.33 |
| D-Glutamine and D-glutamate metabolism | 55 | 4 | 7 | 57.14 |
| Glyoxylate and dicarboxylate metabolism | 93 | 11 | 48 | 22.92 |
| Ascorbate and aldarate metabolism | 7 | 12 | 25 | 48.00 |
| Arginine and proline metabolism | 35 | 25 | 79 | 31.65 |
| GABAergic synapse | 218 | 3 | 9 | 33.33 |
| Histidine metabolism | 37 | 21 | 41 | 51.22 |
| Butanoate metabolism | 98 | 6 | 30 | 20.00 |
| Pentose and glucuronate interconversions | 4 | 8 | 23 | 34.78 |
| Chlorocyclohexane and chlorobenzene degradation | 41 | 3 | 57 | 5.26 |
| Citrate cycle (TCA cycle) | 2 | 4 | 15 | 26.67 |
| Phosphotransferase system (PTS) | 155 | 3 | 25 | 12.00 |
| Cysteine and methionine metabolism | 28 | 11 | 52 | 21.15 |
| Glutathione metabolism | 58 | 5 | 29 | 17.24 |
| Proximal tubule bicarbonate reclamation | 243 | 5 | 16 | 31.25 |
| Glutamatergic synapse | 215 | 2 | 7 | 28.57 |
| Nitrogen metabolism | 124 | 2 | 17 | 11.76 |
| Vitamin B6 metabolism | 106 | 4 | 29 | 13.79 |
| Glycerolipid metabolism | 73 | 4 | 14 | 28.57 |
| ABC transporters | 152 | 32 | 80 | 40.00 |
| Alanine, aspartate and glutamate metabolism | 24 | 9 | 23 | 39.13 |
| Purine metabolism | 20 | 10 | 78 | 12.82 |
| Glycerophospholipid metabolism | 76 | 3 | 23 | 13.04 |
| HIF-1 signaling pathway | 167 | 3 | 12 | 25.00 |
| Zeatin biosynthesis | 122 | 4 | 30 | 13.33 |
| Bile secretion | 251 | 10 | 89 | 11.24 |
| Pyrimidine metabolism | 23 | 13 | 56 | 23.21 |
| Galactose metabolism | 6 | 5 | 21 | 23.81 |
| Mineral absorption | 253 | 13 | 26 | 50.00 |
| KEGG Pathway Name | Total Features in Pathway | p/FDR * | Impact * |
|---|---|---|---|
| Complement and coagulation cascades | 80 | 5.83 × 10−2 | 0.3881 |
| Cholesterol metabolism | 60 | 1.99 × 10−4 | 0.0583 |
| D-Glutamine and D-glutamate metabolism | 18 | 7.86 × 10−3 | 0.3125 |
| Glycine, serine and threonine metabolism | 90 | 7.86 × 10−3 | 0.1059 |
| Glyoxylate and dicarboxylate metabolism | 92 | 7.86 × 10−3 | 0.1136 |
| Prion diseases | 38 | 4.54 × 10−2 | 0.0313 |
| Mineral absorption | 87 | 4.87 × 10−2 | 0.0118 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horvatić, A.; Gelemanović, A.; Pirkić, B.; Smolec, O.; Beer Ljubić, B.; Rubić, I.; Eckersall, P.D.; Mrljak, V.; McLaughlin, M.; Samardžija, M.; et al. Multi-Omics Approach to Elucidate Cerebrospinal Fluid Changes in Dogs with Intervertebral Disc Herniation. Int. J. Mol. Sci. 2021, 22, 11678. https://doi.org/10.3390/ijms222111678
Horvatić A, Gelemanović A, Pirkić B, Smolec O, Beer Ljubić B, Rubić I, Eckersall PD, Mrljak V, McLaughlin M, Samardžija M, et al. Multi-Omics Approach to Elucidate Cerebrospinal Fluid Changes in Dogs with Intervertebral Disc Herniation. International Journal of Molecular Sciences. 2021; 22(21):11678. https://doi.org/10.3390/ijms222111678
Chicago/Turabian StyleHorvatić, Anita, Andrea Gelemanović, Boris Pirkić, Ozren Smolec, Blanka Beer Ljubić, Ivana Rubić, Peter David Eckersall, Vladimir Mrljak, Mark McLaughlin, Marko Samardžija, and et al. 2021. "Multi-Omics Approach to Elucidate Cerebrospinal Fluid Changes in Dogs with Intervertebral Disc Herniation" International Journal of Molecular Sciences 22, no. 21: 11678. https://doi.org/10.3390/ijms222111678
APA StyleHorvatić, A., Gelemanović, A., Pirkić, B., Smolec, O., Beer Ljubić, B., Rubić, I., Eckersall, P. D., Mrljak, V., McLaughlin, M., Samardžija, M., & Lipar, M. (2021). Multi-Omics Approach to Elucidate Cerebrospinal Fluid Changes in Dogs with Intervertebral Disc Herniation. International Journal of Molecular Sciences, 22(21), 11678. https://doi.org/10.3390/ijms222111678









