Semi-Automated High-Throughput Substrate Screening Assay for Nucleoside Kinases
Abstract
1. Introduction
2. Results and Discussion
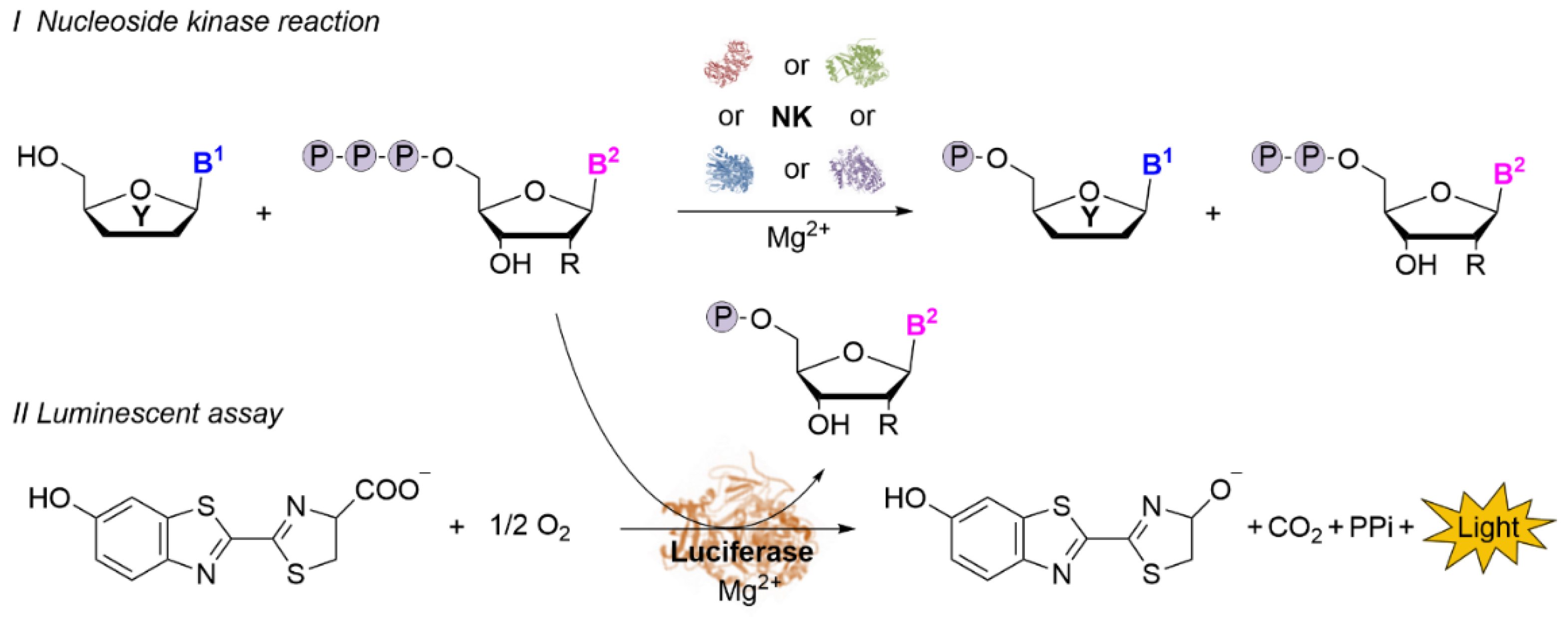
2.1. Establishment of the Luminescent Assay with (Deoxy)NTP Standards
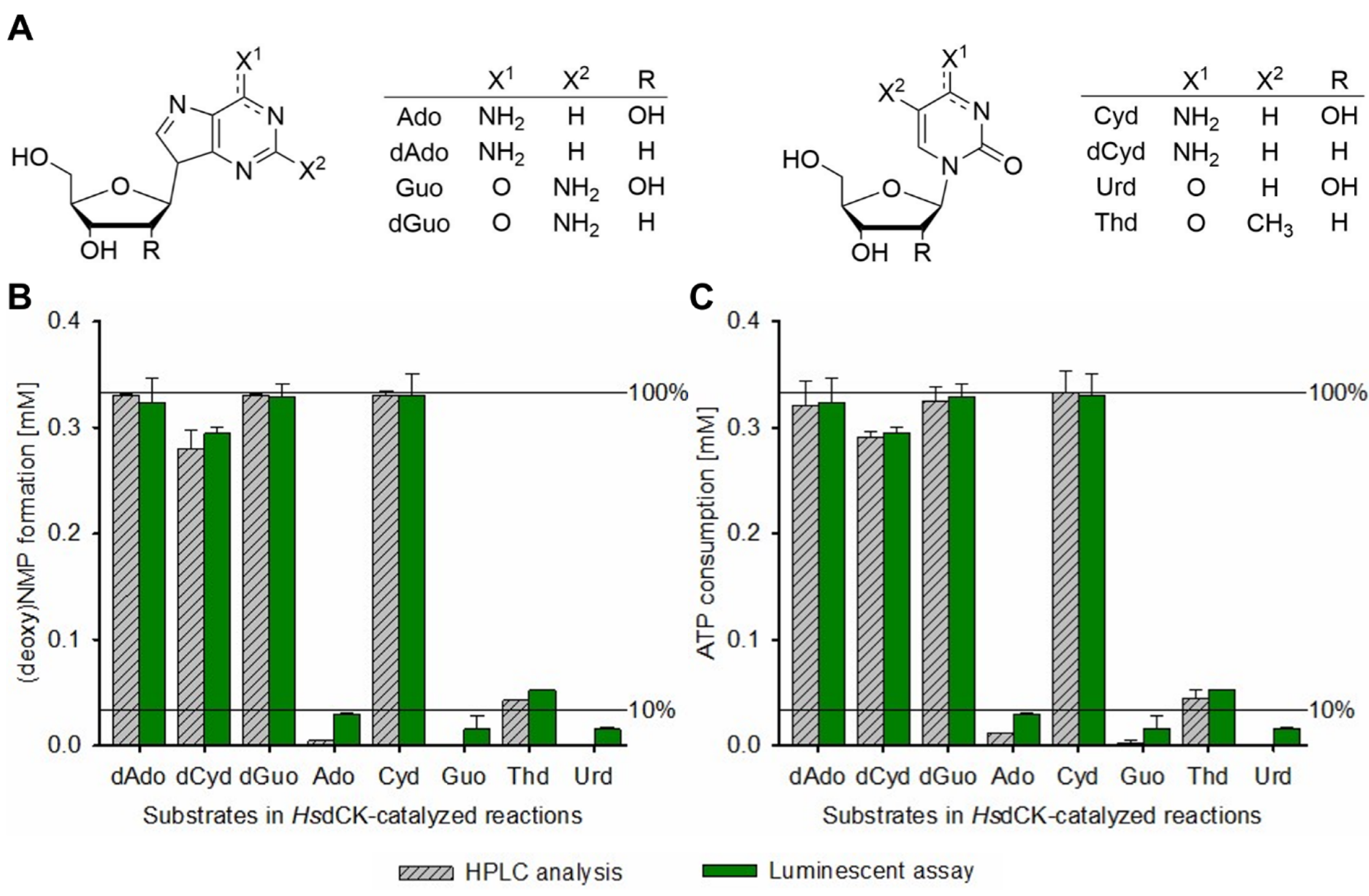
2.2. Establishment of the Luciferase-Based (d)NK Activity Assay Using Natural (Deoxy)Nucleosides as Substrates
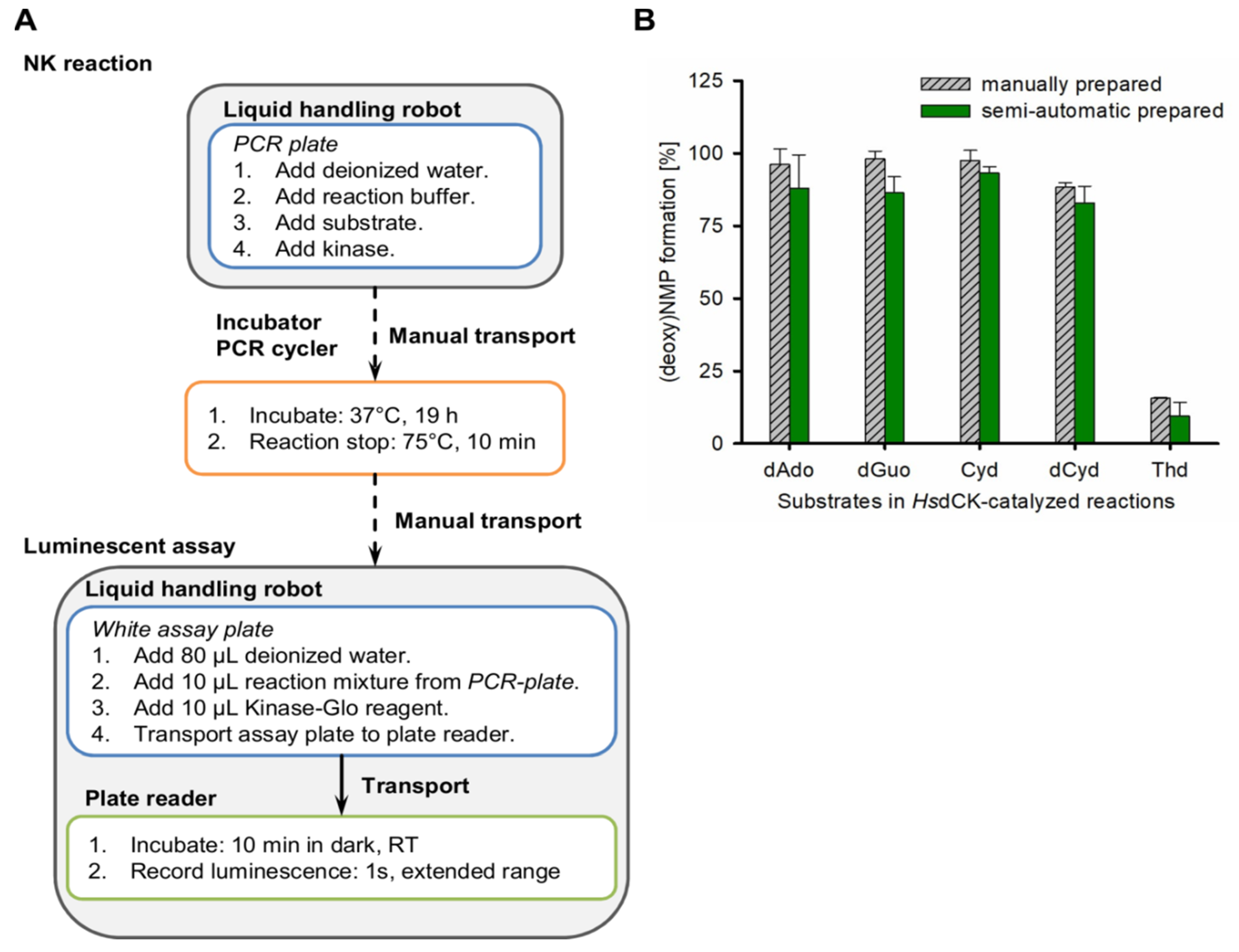
2.3. Development of a High-Throughput Semi-Automated Activity Assay for Nucleoside Kinases
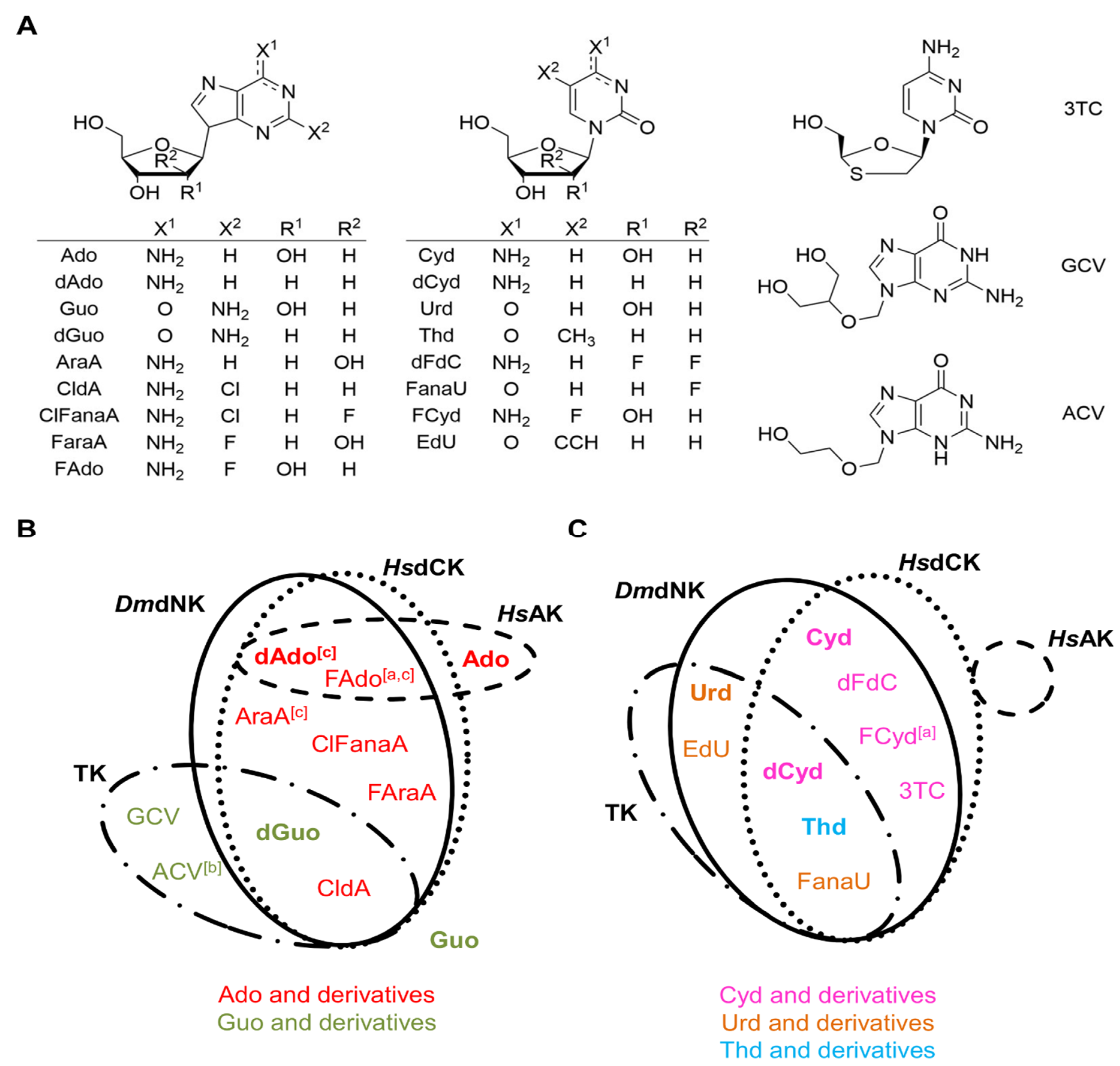
2.4. Application of the High-Throughput Semi-Automated Luminescent Assay to Compare the Substrate Spectra of Four NKs
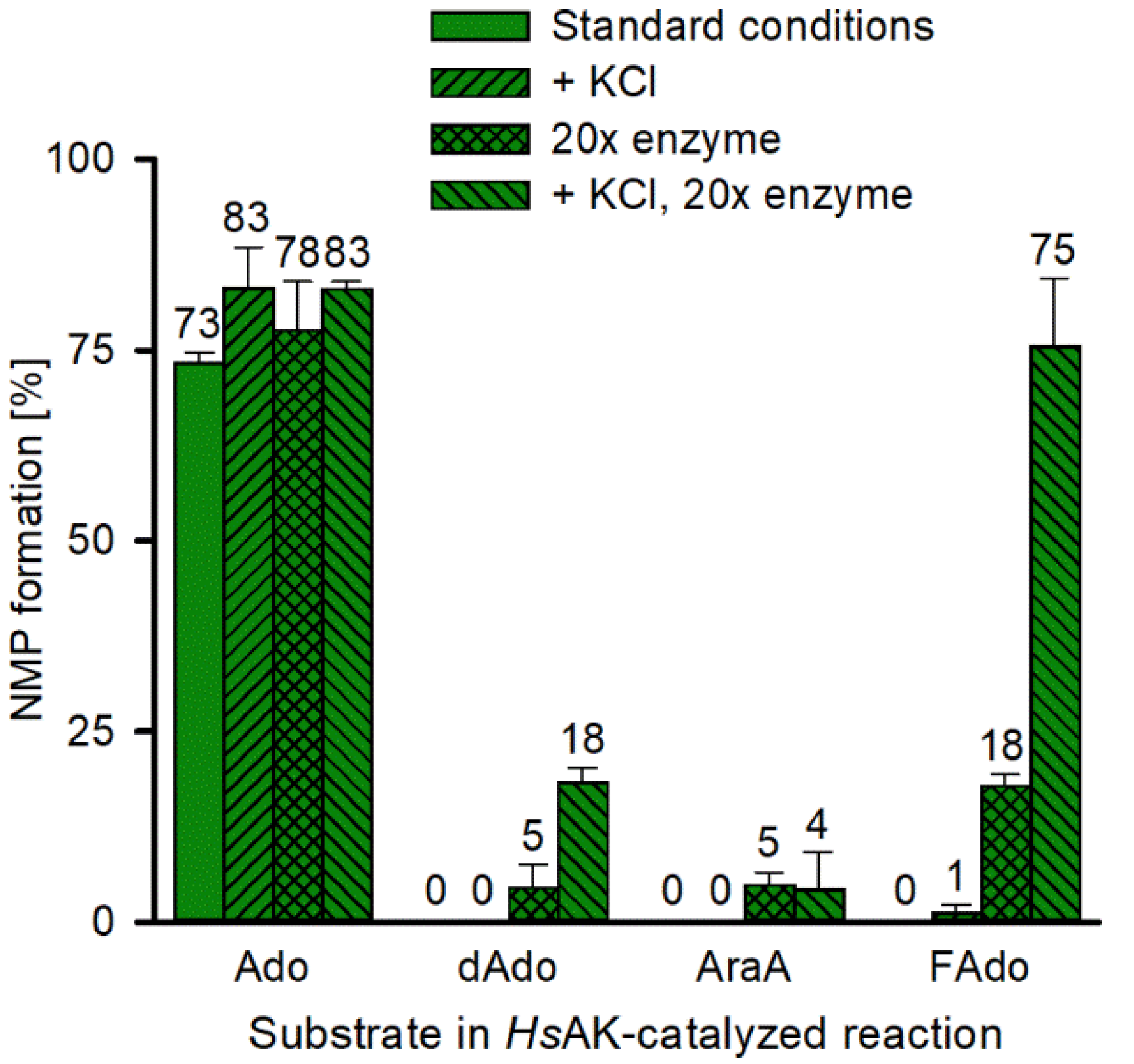
2.5. Application of the High-Throughput Semi-Automated Assay to Optimize Reaction Conditions for HsAK
3. Materials and Methods
3.1. General Remarks
3.2. Expression of DmdNK
3.3. The Luminescent Assay
3.4. Detection of (Deoxy)NTP and Mixed ATP Standards
3.5. Nucleoside Kinase Reactions
3.6. Semi-Automated Assay to Determine the Activity of Nucleoside Kinases
3.7. Accuracy of the Assay
3.8. High-Performance Liquid Chromatography (HPLC)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zaccolo, M.; Williams, D.M.; Brown, D.M.; Gherardi, E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J. Mol. Biol. 1996, 255, 589–603. [Google Scholar] [CrossRef]
- Silvia, F.; Joana, B.; Pedro, M.; Céu, F.; Jesper, W.; Filipe, A.N. Mismatch discrimination in fluorescent in situ hybridization using different types of nucleic acids. Appl. Microbiol. Biotechnol. 2015, 99, 3961–3969. [Google Scholar] [CrossRef]
- Bumgarner, R. Overview of DNA Microarrays: Types, Applications, and Their Future. Curr. Protoc. Mol. Biol. 2013, 101, 22.1.1–22.1.11. [Google Scholar]
- Gragoudas, E.S.; Adamis, A.P.; Cunningham, E.T.; Feinsod, M.; Guyer, D.R. Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med. 2004, 351, 2805–2816. [Google Scholar] [CrossRef]
- Kalota, A.; Karabon, L.; Swider, C.R.; Viazovkina, E.; Elzagheid, M.; Damha, M.J.; Gewirtz, A.M. 2’-deoxy-2’-fluoro-beta-D-arabinonucleic acid (2’F-ANA) modified oligonucleotides (ON) effect highly efficient, and persistent, gene silencing. Nucleic Acids Res. 2006, 34, 451–461. [Google Scholar] [CrossRef]
- Watts, J.K.; Corey, D.R. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012, 226, 365–379. [Google Scholar] [CrossRef]
- Granados-Riveron, J.T.; Aquino-Jarquin, G. Engineering of the current nucleoside-modified mRNA-LNP vaccines against SARS-CoV-2. Biomed. Pharm. 2021, 142, 111953. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef]
- Thomson, J.M.; Lamont, I.L. Nucleoside Analogues as Antibacterial Agents. Front. Microbiol. 2019, 10, 952. [Google Scholar] [CrossRef] [PubMed]
- Deville-Bonne, D.; El Amri, C.; Meyer, P.; Chen, Y.; Agrofoglio, L.A.; Janin, J. Human and viral nucleoside/nucleotide kinases involved in antiviral drug activation: Structural and catalytic properties. Antiviral Res. 2010, 86, 101–120. [Google Scholar] [CrossRef]
- Fehlau, M.; Kaspar, F.; Hellendahl, K.F.; Schollmeyer, J.; Neubauer, P.; Wagner, A. Modular Enzymatic Cascade Synthesis of Nucleotides Using a (d)ATP Regeneration System. Front. Bioeng. Biotechnol. 2020, 8, 854. [Google Scholar] [CrossRef]
- Kore, A.; Srinivasan, B. Recent Advances in the Syntheses of Nucleoside Triphosphates. Curr. Org. Synth. 2014, 10, 903–934. [Google Scholar] [CrossRef]
- Winkler, C.K.; Schrittwieser, J.H.; Kroutil, W. Power of Biocatalysis for Organic Synthesis. ACS Cent. Sci. 2021, 7, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Acker, M.G.; Auld, D.S. Considerations for the design and reporting of enzyme assays in high-throughput screening applications. Perspect. Sci. 2014, 1, 56–73. [Google Scholar] [CrossRef]
- Baki, A.; Bielik, A.; Molnár, L.; Szendrei, G.; Keserü, G.M. A high throughput luminescent assay for glycogen synthase kinase-3β inhibitors. Assay Drug Dev. Technol. 2007, 5, 75–83. [Google Scholar] [CrossRef]
- Ma, H.; Deacon, S.; Horiuchi, K. The challenge of selecting protein kinase assays for lead discovery optimization. Expert Opin. Drug Discov. 2008, 3, 607–621. [Google Scholar] [CrossRef]
- Feng, J.; Chen, Y.; Pu, J.; Yang, X.; Zhang, C.; Zhu, S.; Zhao, Y.; Yuan, Y.; Yuan, H.; Liao, F. An improved malachite green assay of phosphate: Mechanism and application. Anal. Biochem. 2011, 409, 144–149. [Google Scholar] [CrossRef]
- Hellendahl, K.F.; Kamel, S.; Wetterwald, A.; Neubauer, P.; Wagner, A. Human Deoxycytidine Kinase Is a Valuable Biocatalyst for the Synthesis of Nucleotide Analogues. Catalysts 2019, 9, 997. [Google Scholar] [CrossRef]
- Singh, M.; Tiwari, P.; Arora, G.; Agarwal, S.; Kidwai, S.; Singh, R. Establishing Virulence Associated Polyphosphate Kinase 2 as a drug target for Mycobacterium tuberculosis. Sci. Rep. 2016, 6, 26900. [Google Scholar] [CrossRef][Green Version]
- Helenius, M.; Jalkanen, S.; Yegutkin, G.G. Enzyme-coupled assays for simultaneous detection of nanomolar ATP, ADP, AMP, adenosine, inosine and pyrophosphate concentrations in extracellular fluids. Biochim. Biophys. Acta-Mol. Cell Res. 2012, 1823, 1967–1975. [Google Scholar] [CrossRef]
- Karamohamed, S.; Nordstrom, T.; Nyren, P. Real-time bioluminometric method for detection of nucleoside diphosphate kinase activity. Biotechniques 1999, 26, 728–734. [Google Scholar] [CrossRef]
- Hu, C.M.; Chang, Z.F. A bioluminescent method for measuring thymidylate kinase activity suitable for high-throughput screening of inhibitor. Anal. Biochem. 2010, 398, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, S.; Nikkhah, M.; Nazari, M.; Hosseinkhani, S. Design of a Coupled Bioluminescent Assay for a Recombinant Pyruvate Kinase from a Thermophilic Geobacillus. Photochem. Photobiol. 2011, 87, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Goto, H.; Ogasawara, N. Adenosine kinase from human liver. BBA-Enzymol. 1981, 660, 36–43. [Google Scholar] [CrossRef]
- Munch-Petersen, B.; Piskur, J.; Søndergaard, L. Four deoxynucleoside kinase activities from Drosophila melanogaster are contained within a single monomeric enzyme, a new multifunctional deoxynucleoside kinase. J. Biol. Chem. 1998, 273, 3926–3931. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, M.F.; Shaw, R.W.; Moses, J.D.; Kim, H.J.; Kim, M.J.; Kim, M.S.; Hoshika, S.; Karalkar, N.; Benner, S.A. Assays to Detect the Formation of Triphosphates of Unnatural Nucleotides: Application to Escherichia coli Nucleoside Diphosphate Kinase. ACS Synth. Biol. 2016, 5, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Leitão, J.M.M.; Esteves da Silva, J.C.G. Firefly luciferase inhibition. J. Photochem. Photobiol. B Biol. 2010, 101, 1–8. [Google Scholar] [CrossRef]
- Sabini, E.; Ort, S.; Monnerjahn, C.; Konrad, M.; Lavie, A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nat. Struct. Mol. Biol. 2003, 10, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Datta, N.S.; Shewach, D.S.; Hurley, M.C.; Mitchell, B.S.; Fox, I.H. Human T-lymphoblast deoxycytidine kinase: Purification and properties. Biochemistry 1989, 28, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Hazra, S.; Ort, S.; Konrad, M.; Lavie, A. Structural and kinetic characterization of human deoxycytidine kinase variants able to phosphorylate 5-substituted deoxycytidine and thymidine analogues. Biochemistry 2010, 49, 6784–6790. [Google Scholar] [CrossRef]
- Iyidogan, P.; Lutz, S. Systematic Exploration of Active Site Mutations on Human Deoxycytidine Kinase Substrate Specificity. Biochemistry 2008, 47, 4711–4720. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mani, R.S.; Usova, E.V.; Eriksson, S.; Cass, C.E. Fluorescence studies of substrate binding to human recombinant deoxycytidine kinase. Nucleosides Nucleotides Nucleic Acids 2004, 23, 1343–1346. [Google Scholar] [CrossRef] [PubMed]
- Hellendahl, K.F.; Fehlau, M. Supplementary Information for “Semi-automated high-throughput substrate screening assay for nucleoside kinases”. Zenodo 2021. Available online: https://doi.org/10.5281/zenodo.5363310 (accessed on 29 September 2021).
- Serra, I.; Conti, S.; Piškur, J.; Clausen, A.R.; Munch-Petersen, B.; Terreni, M.; Ubiali, D. Immobilized Drosophila melanogaster Deoxyribonucleoside Kinase (DmdNK) as a High Performing Biocatalyst for the Synthesis of Purine Arabinonucleotides. Adv. Synth. Catal. 2014, 356, 563–570. [Google Scholar] [CrossRef]
- Munch-Petersen, B.; Knecht, W.; Lenz, C.; Søndergaard, L.; Piskur, J. Functional expression of a multisubstrate deoxyribonucleoside kinase from Drosophila melanogaster and its C-terminal deletion mutants. J. Biol. Chem. 2000, 275, 6673–6679. [Google Scholar] [CrossRef]
- Eriksson, S.; Munch-Petersen, B.; Johansson, K.; Ecklund, H. Structure and function of cellular deoxyribonucleoside kinases. Cell. Mol. Life Sci. 2002, 59, 1327–1346. [Google Scholar] [CrossRef]
- Kierdaszuk, B.; Krawiec, K.; Kazimierczuk, Z.; Jacobsson, U.; Johansson, N.G.; Munch-Petersen, B.; Eriksson, S.; Shugar, D. Substrate/Inhibitor Properties of Human Deoxycytidine Kinase (dCK) and Thymidine Kinases (Tk1 And Tk2) Towards the Sugar Moiety of Nucleosides, Including O′-Alkyl Analogues. Nucleosides Nucleotides 1999, 18, 1883–1903. [Google Scholar] [CrossRef]
- Zhang, Y.; Secrist, J.A.; Ealick, S.E. The structure of human deoxycytidine kinase in complex with clofarabine reveals key interactions for prodrug activation. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, S.; Kierdaszuk, B.; Munch-Petersen, B.; Oberg, B.; Gunnar Johansson, N. Comparison of the substrate specificities of human thymidine kinase 1 and 2 and deoxycytidine kinase toward antiviral and cytostatic nucleoside analogs. Biochem. Biophys. Res. Commun. 1991, 176, 586–592. [Google Scholar] [CrossRef]
- Nagai, S.; Takenaka, K.; Nachagari, D.; Rose, C.; Domoney, K.; Sun, D.; Sparreboom, A.; Schuetz, J.D. Deoxycytidine kinase modulates the impact of the ABC transporter ABCG2 on clofarabine cytotoxicity. Cancer Res. 2011, 71, 1781–1791. [Google Scholar] [CrossRef] [PubMed]
- Knecht, W.; Mikkelsen, N.E.; Clausen, A.R.; Willer, M.; Eklund, H.; Gojković, Z.; Piškur, J. Drosophila melanogaster deoxyribonucleoside kinase activates gemcitabine. Biochem. Biophys. Res. Commun. 2009, 382, 430–433. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hellendahl, K.F.; Fehlau, M.; Hans, S.; Neubauer, P.; Kurreck, A. Semi-Automated High-Throughput Substrate Screening Assay for Nucleoside Kinases. Int. J. Mol. Sci. 2021, 22, 11558. https://doi.org/10.3390/ijms222111558
Hellendahl KF, Fehlau M, Hans S, Neubauer P, Kurreck A. Semi-Automated High-Throughput Substrate Screening Assay for Nucleoside Kinases. International Journal of Molecular Sciences. 2021; 22(21):11558. https://doi.org/10.3390/ijms222111558
Chicago/Turabian StyleHellendahl, Katja F., Maryke Fehlau, Sebastian Hans, Peter Neubauer, and Anke Kurreck. 2021. "Semi-Automated High-Throughput Substrate Screening Assay for Nucleoside Kinases" International Journal of Molecular Sciences 22, no. 21: 11558. https://doi.org/10.3390/ijms222111558
APA StyleHellendahl, K. F., Fehlau, M., Hans, S., Neubauer, P., & Kurreck, A. (2021). Semi-Automated High-Throughput Substrate Screening Assay for Nucleoside Kinases. International Journal of Molecular Sciences, 22(21), 11558. https://doi.org/10.3390/ijms222111558






