The Arabidopsis thaliana Kinesin-5 AtKRP125b Is a Processive, Microtubule-Sliding Motor Protein with Putative Plant-Specific Functions
Abstract
:1. Introduction
2. Results
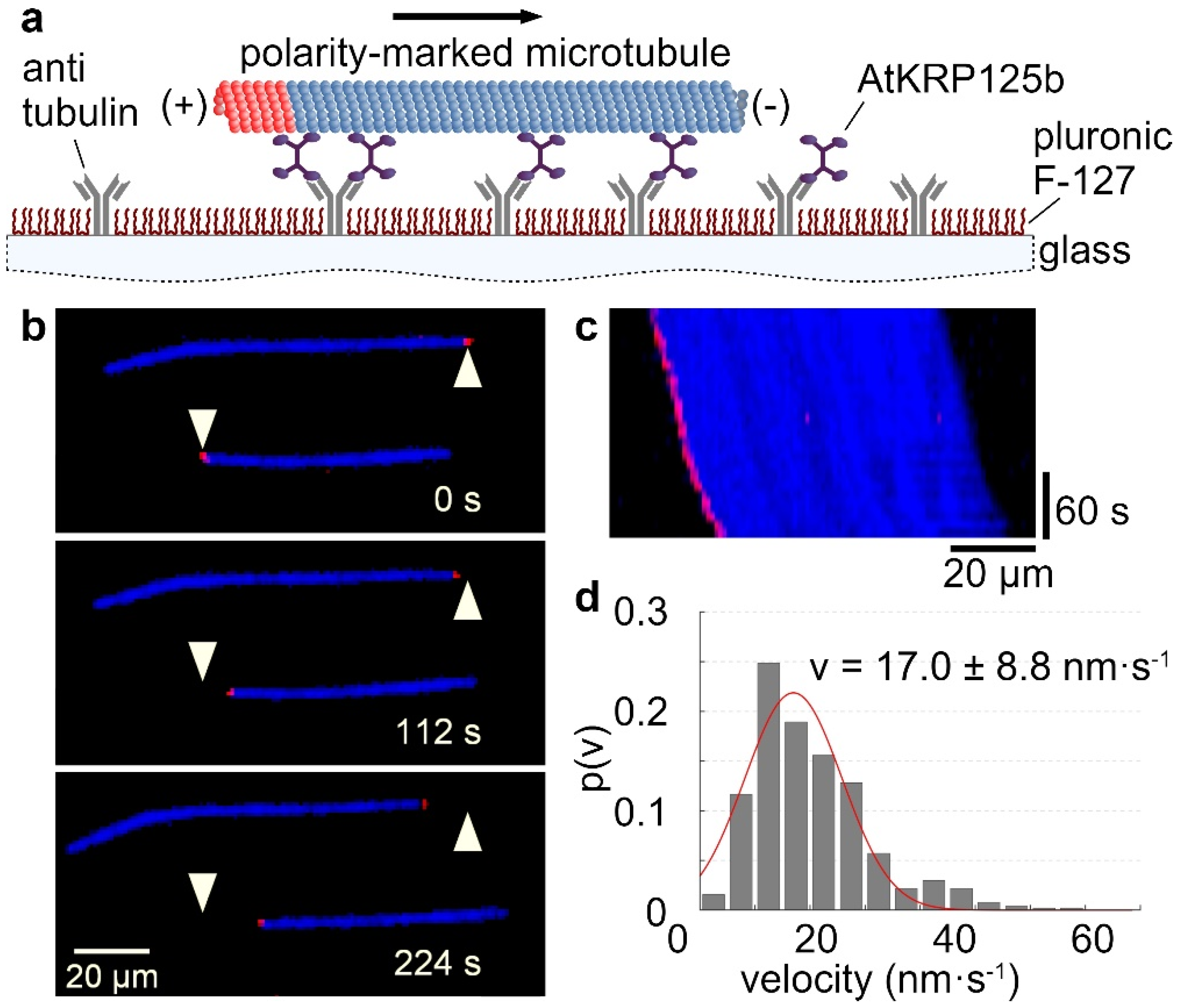
2.1. The Kinesin AtKRP125b Moves along Microtubules at a Slow Velocity
2.2. AtKRP125b Is a Processive Kinesin
2.3. AtKRP125b Moves along Microtubules toward Their plus End
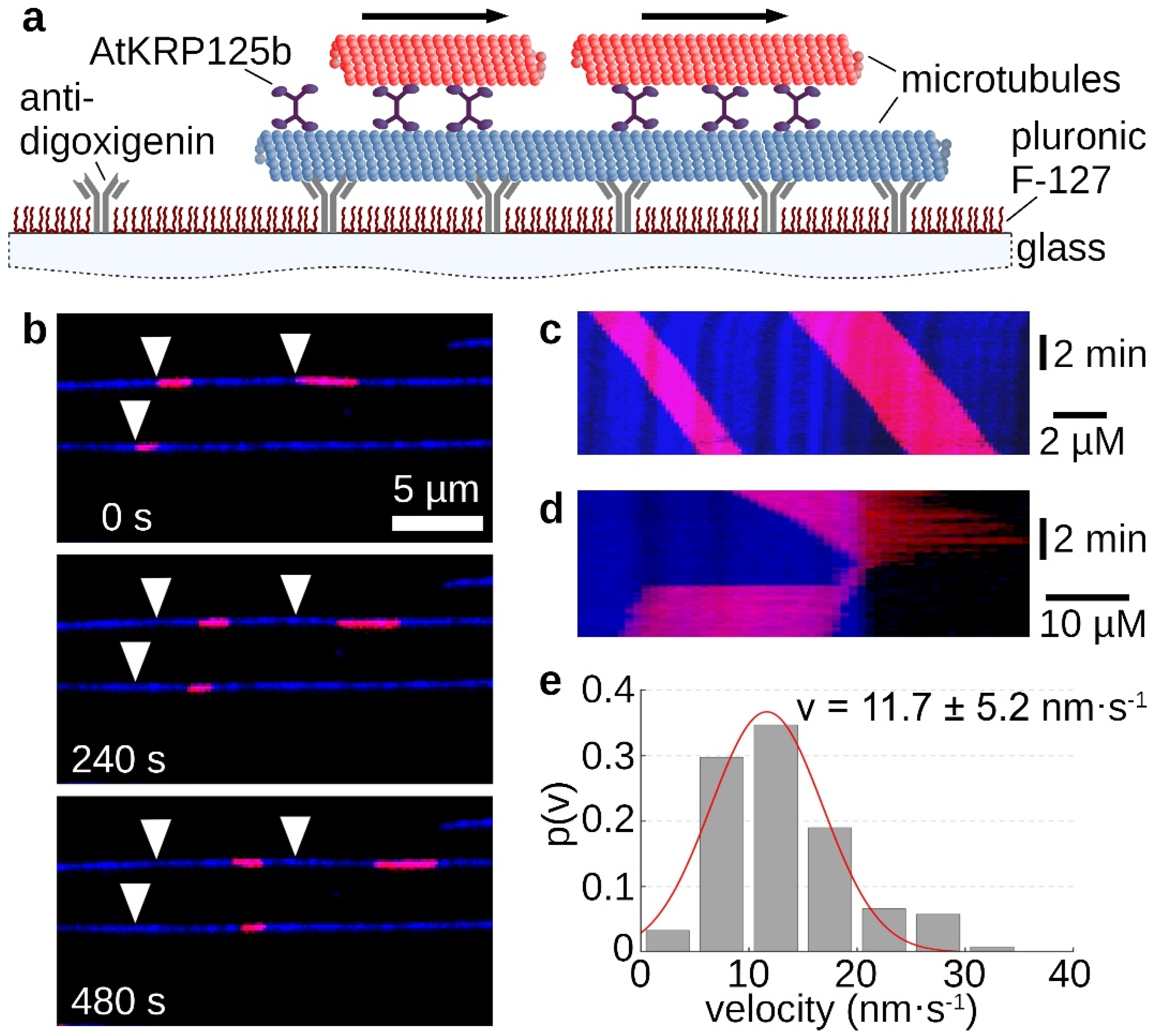
2.4. AtKRP125b Is Able to Cross-Link Microtubules and Slide Them Apart
2.5. The AtKRP125b Promoter Is Not Active in Mitotically Active Tissues
2.6. AtKRP125b Knockout Mutants Show No Developmental Abnormalities
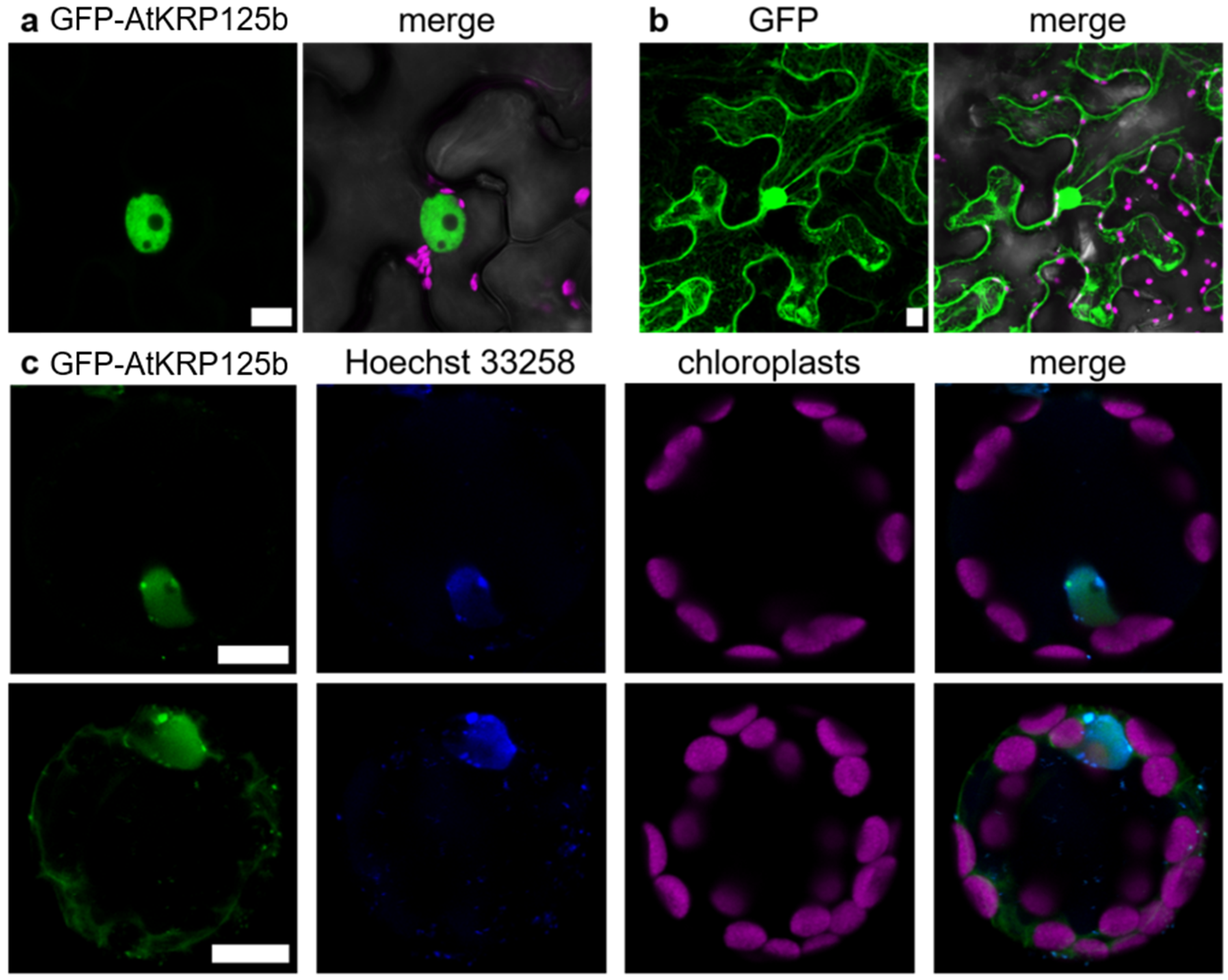
2.7. GFP-AtKRP125b Is Localized in the Nucleus and in the Cytoplasm
3. Discussion
4. Material and Methods
4.1. Protein Expression, Purification, and Labelling
4.2. In Vitro Motility Assays
4.3. Analysis of Promoter Activity
4.4. Plant Material and Growth Conditions
4.5. Subcellular Localization
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ogawa, T.; Nitta, R.; Okada, Y.; Hirokawa, N. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell 2004, 116, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Gigant, E.; Stefanutti, M.; Laband, K.; Gluszek-Kustusz, A.; Edwards, F.; Lacroix, B.; Maton, G.; Canman, J.C.; Welburn, J.P.I.; Dumont, J. Inhibition of ectopic microtubule assembly by the kinesin-13 KLP-7 prevents chromosome segregation and cytokinesis defects in oocytes. Development 2017, 144, 1674–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brady, S.T. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature 1985, 317, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Vale, R.D.; Reese, T.S.; Sheetz, M.P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 1985, 42, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Kapitein, L.C.; Peterman, E.J.G.; Kwok, B.H.; Kim, J.H.; Kapoor, T.M.; Schmidt, C.F. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 2005, 435, 114–118. [Google Scholar] [CrossRef]
- Miki, T.; Naito, H.; Nishina, M.; Goshima, G. Endogenous localizome identifies 43 mitotic kinesins in a plant cell. Proc. Natl. Acad. Sci. USA 2014, 111, E1053–E1061. [Google Scholar] [CrossRef] [Green Version]
- Peters, N.T.; Miller, A.C.; Kropf, D.L. Localization and function of Kinesin-5-like proteins during assembly and maintenance of mitotic spindles in Silvetia compressa. BMC Res. Notes 2009, 2, 106. [Google Scholar] [CrossRef] [Green Version]
- Kashina, A.S.; Baskin, R.J.; Cole, D.G.; Wedaman, K.P.; Saxton, W.M.; Scholey, J.M. A bipolar kinesin. Nature 1996, 379, 270–272. [Google Scholar] [CrossRef]
- Mann, B.J.; Wadsworth, P. Kinesin-5 Regulation and Function in Mitosis. Trends Cell Biol. 2019, 29, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Asada, T.; Kuriyama, R.; Shibaoka, H. TKRP125, a kinesin-related protein involved in the centrosome-independent organization of the cytokinetic apparatus in tobacco BY-2 cells. J. Cell Sci. 1997, 110, 179–189. [Google Scholar] [CrossRef]
- Bannigan, A.; Scheible, W.-R.; Lukowitz, W.; Fagerstrom, C.; Wadsworth, P.; Somerville, C.; Baskin, T.I. A conserved role for kinesin-5 in plant mitosis. J. Cell Sci. 2007, 120, 2819–2827. [Google Scholar] [CrossRef] [Green Version]
- Richardson, D.N.; Simmons, M.P.; Reddy, A.S.N. Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes. BMC Genom. 2006, 7, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, A.S.; Day, I.S. Kinesins in the Arabidopsis genome: A comparative analysis among eukaryotes. BMC Genom. 2001, 2, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Dixit, R. Functions of the Arabidopsis kinesin superfamily of microtubule-based motor proteins. Protoplasma 2012, 249, 887–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishihama, R.; Soyano, T.; Ishikawa, M.; Araki, S.; Tanaka, H.; Asada, T.; Irie, K.; Ito, M.; Terada, M.; Banno, H.; et al. Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 2002, 109, 87–99. [Google Scholar] [CrossRef] [Green Version]
- Itoh, R.; Fujiwara, M.; Yoshida, S. Kinesin-related proteins with a mitochondrial targeting signal. Plant Physiol. 2001, 127, 724–726. [Google Scholar] [CrossRef]
- Hiwatashi, Y.; Obara, M.; Sato, Y.; Fujita, T.; Murata, T.; Hasebe, M. Kinesins are indispensable for interdigitation of phragmoplast microtubules in the moss Physcomitrella patens. Plant Cell 2008, 20, 3094–3106. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.R.; Liu, B. Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr. Biol. 2000, 10, 797–800. [Google Scholar] [CrossRef] [Green Version]
- Lipka, E.; Gadeyne, A.; Stöckle, D.; Zimmermann, S.; de Jaeger, G.; Ehrhardt, D.W.; Kirik, V.; van Damme, D.; Müller, S. The Phragmoplast-Orienting Kinesin-12 Class Proteins Translate the Positional Information of the Preprophase Band to Establish the Cortical Division Zone in Arabidopsis thaliana. Plant Cell 2014, 26, 2617–2632. [Google Scholar] [CrossRef] [Green Version]
- Shimamoto, Y.; Forth, S.; Kapoor, T.M. Measuring Pushing and Braking Forces Generated by Ensembles of Kinesin-5 Crosslinking Two Microtubules. Dev. Cell 2015, 34, 669–681. [Google Scholar] [CrossRef] [Green Version]
- Endow, S.A.; Waligora, K.W. Determinants of kinesin motor polarity. Science 1998, 281, 1200–1202. [Google Scholar] [CrossRef] [Green Version]
- Fridman, V.; Gerson-Gurwitz, A.; Shapira, O.; Movshovich, N.; Lakämper, S.; Schmidt, C.F.; Gheber, L. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J. Cell Sci. 2013, 126, 4147–4159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerson-Gurwitz, A.; Thiede, C.; Movshovich, N.; Fridman, V.; Podolskaya, M.; Danieli, T.; Lakämper, S.; Klopfenstein, D.R.; Schmidt, C.F.; Gheber, L. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 2011, 30, 4942–4954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roostalu, J.; Hentrich, C.; Bieling, P.; Telley, I.A.; Schiebel, E.; Surrey, T. Directional switching of the kinesin Cin8 through motor coupling. Science 2011, 332, 94–99. [Google Scholar] [CrossRef] [Green Version]
- Vanstraelen, M.; Inzé, D.; Geelen, D. Mitosis-specific kinesins in Arabidopsis. Trends Plant Sci. 2006, 11, 167–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, M.; Lansky, Z.; Szuba, A.; Schwarz, F.W.; Mitra, A.; Gao, M.; Lüdecke, A.; ten Wolde, P.R.; Diez, S. Changes in microtubule overlap length regulate kinesin-14-driven microtubule sliding. Nat. Chem. Biol. 2017, 13, 1245–1252. [Google Scholar] [CrossRef]
- Hentrich, C.; Surrey, T. Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. J. Cell Biol. 2010, 189, 465–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hepperla, A.J.; Willey, P.T.; Coombes, C.E.; Schuster, B.M.; Gerami-Nejad, M.; McClellan, M.; Mukherjee, S.; Fox, J.; Winey, M.; Odde, D.J.; et al. Minus-end-directed Kinesin-14 motors align antiparallel microtubules to control metaphase spindle length. Dev. Cell 2014, 31, 61–72. [Google Scholar] [CrossRef] [Green Version]
- Mitra, A.; Meißner, L.; Gandhimathi, R.; Renger, R.; Ruhnow, F.; Diez, S. Kinesin-14 motors drive a right-handed helical motion of antiparallel microtubules around each other. Nat. Commun. 2020, 11, 2565. [Google Scholar] [CrossRef]
- Sawin, K.E.; Mitchison, T.J. Poleward microtubule flux mitotic spindles assembled in vitro. J. Cell Biol. 1991, 112, 941–954. [Google Scholar] [CrossRef] [Green Version]
- Bodrug, T.; Wilson-Kubalek, E.M.; Nithianantham, S.; Thompson, A.F.; Alfieri, A.; Gaska, I.; Major, J.; Debs, G.; Inagaki, S.; Gutierrez, P.; et al. The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding. eLife 2020, 9, e51131. [Google Scholar] [CrossRef] [PubMed]
- Barroso, C.; Chan, J.; Allan, V.; Doonan, J.; Hussey, P.; Lloyd, C. Two kinesin-related proteins associated with the cold-stable cytoskeleton of carrot cells: Characterization of a novel kinesin, DcKRP120-2. Plant J. 2000, 24, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Ari, C.; Borysov, S.I.; Wu, J.; Padmanabhan, J.; Potter, H. Alzheimer amyloid beta inhibition of Eg5/kinesin 5 reduces neurotrophin and/or transmitter receptor function. Neurobiol. Aging 2014, 35, 1839–1849. [Google Scholar] [CrossRef] [Green Version]
- Falnikar, A.; Tole, S.; Baas, P.W. Kinesin-5, a mitotic microtubule-associated motor protein, modulates neuronal migration. Mol. Biol. Cell 2011, 22, 1561–1574. [Google Scholar] [CrossRef]
- Wakana, Y.; Villeneuve, J.; van Galen, J.; Cruz-Garcia, D.; Tagaya, M.; Malhotra, V. Kinesin-5/Eg5 is important for transport of CARTS from the trans-Golgi network to the cell surface. J. Cell Biol. 2013, 202, 241–250. [Google Scholar] [CrossRef] [Green Version]
- Ganguly, A.; Zhu, C.; Chen, W.; Dixit, R. FRA1 Kinesin Modulates the Lateral Stability of Cortical Microtubules through Cellulose Synthase-Microtubule Uncoupling Proteins. Plant Cell 2020, 32, 2508–2524. [Google Scholar] [CrossRef]
- Bryksin, A.V.; Matsumura, I. Overlap extension PCR cloning: A simple and reliable way to create recombinant plasmids. Biotechniques 2010, 48, 463–465. [Google Scholar] [CrossRef] [PubMed]
- Castoldi, M.; Popov, A.V. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 2003, 32, 83–88. [Google Scholar] [CrossRef]
- Hyman, A.; Drechsel, D.; Kellogg, D.; Salser, S.; Sawin, K.; Steffen, P.; Wordeman, L.; Mitchison, T. Preparation of modified tubulins. Methods Enzymol. 1991, 196, 478–485. [Google Scholar] [CrossRef]
- Howard, J.; Hyman, A.A. Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence microscopy. Methods Cell Biol. 1993, 39, 105–113. [Google Scholar] [CrossRef]
- Fink, G.; Hajdo, L.; Skowronek, K.J.; Reuther, C.; Kasprzak, A.A.; Diez, S. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat. Cell Biol. 2009, 11, 717–723. [Google Scholar] [CrossRef]
- Walter, W.J.; Beránek, V.; Fischermeier, E.; Diez, S. Tubulin acetylation alone does not affect kinesin-1 velocity and run length in vitro. PLoS ONE 2012, 7, e42218. [Google Scholar] [CrossRef] [PubMed]
- Walter, W.J.; Koonce, M.P.; Brenner, B.; Steffen, W. Two independent switches regulate cytoplasmic dynein’s processivity and directionality. Proc. Natl. Acad. Sci. USA 2012, 109, 5289–5293. [Google Scholar] [CrossRef] [Green Version]
- Griffin, B.A.; Adams, S.R.; Tsien, R.Y. Specific covalent labeling of recombinant protein molecules inside live cells. Science 1998, 281, 269–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Widengren, J.; Chmyrov, A.; Eggeling, C.; Löfdahl, P.-A.; Seidel, C.A.M. Strategies to improve photostabilities in ultrasensitive fluorescence spectroscopy. J. Phys. Chem. A 2007, 111, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Ruhnow, F.; Zwicker, D.; Diez, S. Tracking single particles and elongated filaments with nanometer precision. Biophys. J. 2011, 100, 2820–2828. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Henriques, R.; Lin, S.-S.; Niu, Q.-W.; Chua, N.-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Harrison, S.J.; Mott, E.K.; Parsley, K.; Aspinall, S.; Gray, J.C.; Cottage, A. A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2006, 2, 19. [Google Scholar] [CrossRef] [Green Version]
- Tong, M.; Kotur, T.; Liang, W.; Vogelmann, K.; Kleine, T.; Leister, D.; Brieske, C.; Yang, S.; Lüdke, D.; Wiermer, M.; et al. E3 ligase SAUL1 serves as a positive regulator of PAMP-triggered immunity and its homeostasis is monitored by immune receptor SOC3. New Phytol. 2017, 215, 1516–1532. [Google Scholar] [CrossRef] [Green Version]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strauß, T.; Schattner, S.; Hoth, S.; Walter, W.J. The Arabidopsis thaliana Kinesin-5 AtKRP125b Is a Processive, Microtubule-Sliding Motor Protein with Putative Plant-Specific Functions. Int. J. Mol. Sci. 2021, 22, 11361. https://doi.org/10.3390/ijms222111361
Strauß T, Schattner S, Hoth S, Walter WJ. The Arabidopsis thaliana Kinesin-5 AtKRP125b Is a Processive, Microtubule-Sliding Motor Protein with Putative Plant-Specific Functions. International Journal of Molecular Sciences. 2021; 22(21):11361. https://doi.org/10.3390/ijms222111361
Chicago/Turabian StyleStrauß, Tobias, Saskia Schattner, Stefan Hoth, and Wilhelm J. Walter. 2021. "The Arabidopsis thaliana Kinesin-5 AtKRP125b Is a Processive, Microtubule-Sliding Motor Protein with Putative Plant-Specific Functions" International Journal of Molecular Sciences 22, no. 21: 11361. https://doi.org/10.3390/ijms222111361
APA StyleStrauß, T., Schattner, S., Hoth, S., & Walter, W. J. (2021). The Arabidopsis thaliana Kinesin-5 AtKRP125b Is a Processive, Microtubule-Sliding Motor Protein with Putative Plant-Specific Functions. International Journal of Molecular Sciences, 22(21), 11361. https://doi.org/10.3390/ijms222111361




