PPR647 Protein Is Required for Chloroplast RNA Editing, Splicing and Chloroplast Development in Maize
Abstract
:1. Introduction
2. Results
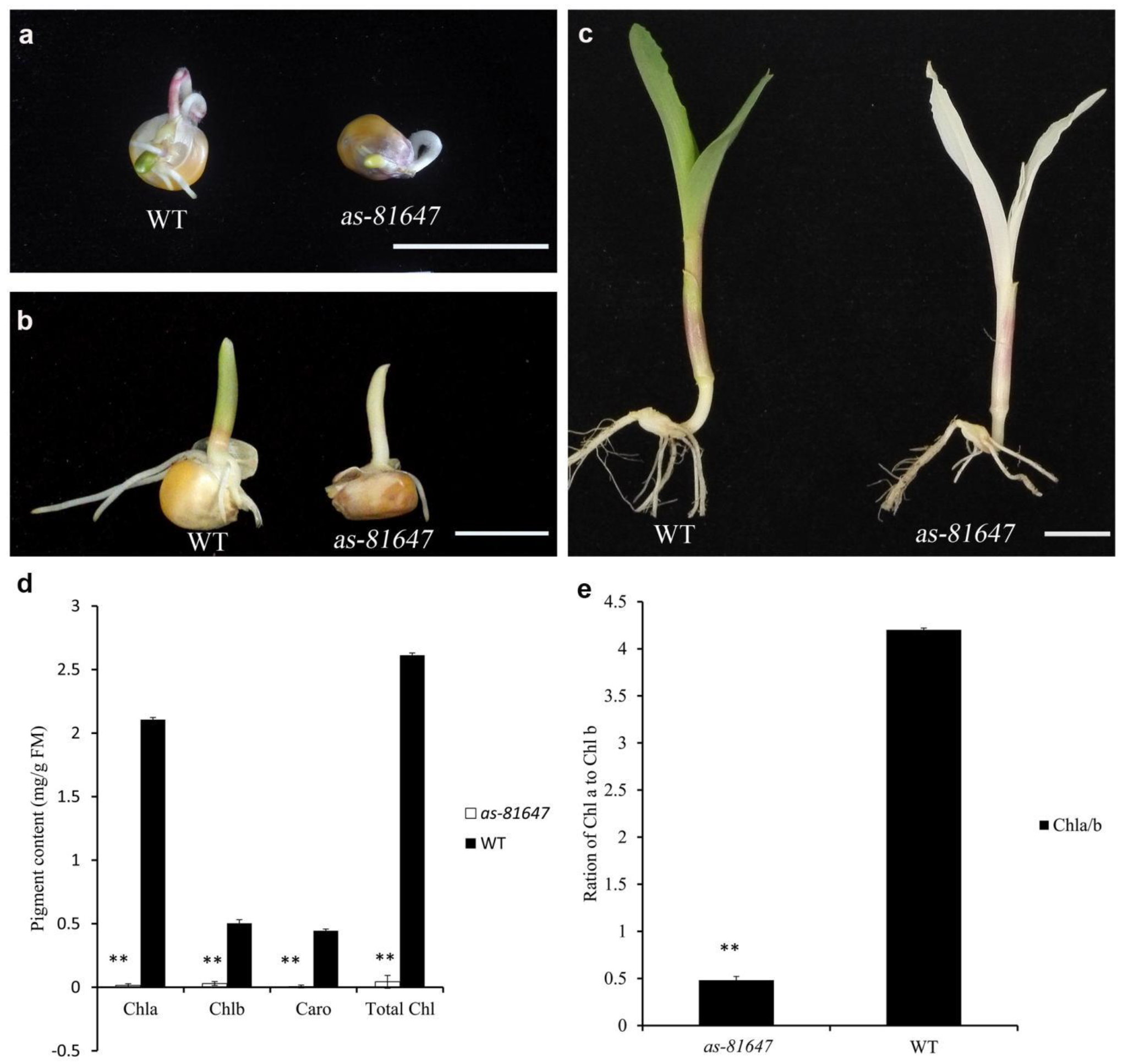
2.1. Phenotypic Characterization of as-81647
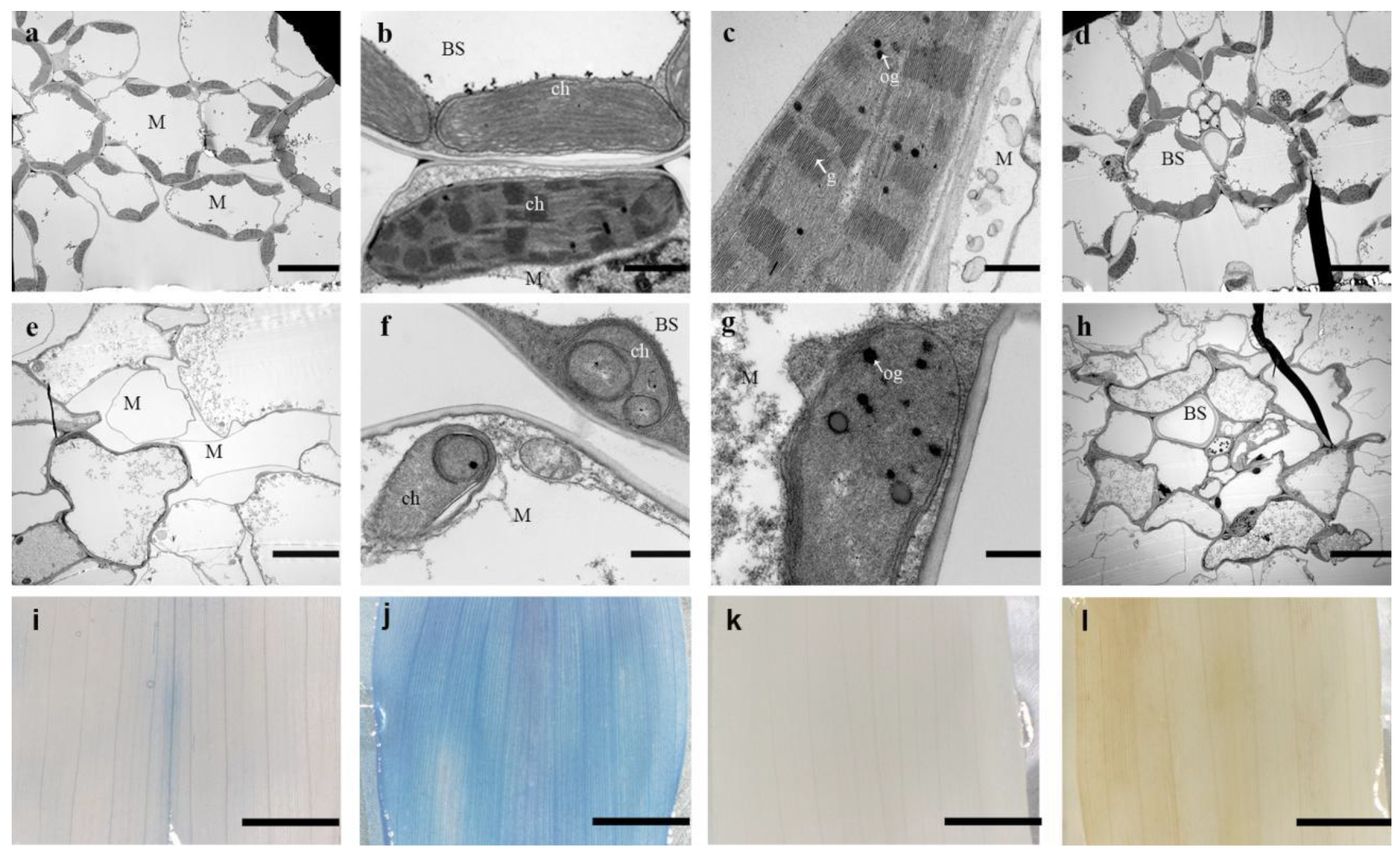
2.2. Abnormal Chloroplast Morphology in as-81647
2.3. Increased Reactive Oxygen Species (ROS) Levels in as-81647
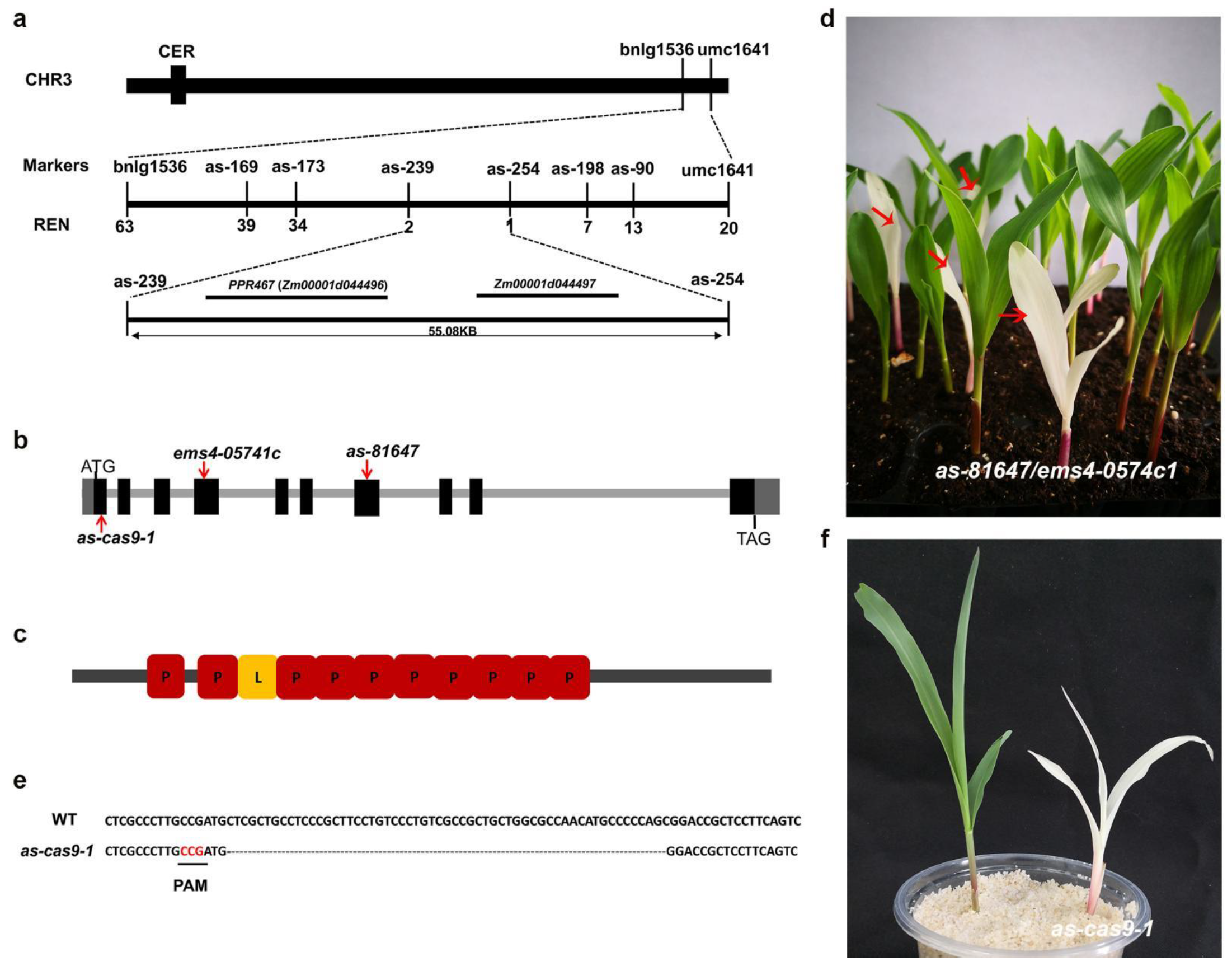
2.4. PLS-Type Pentatricopeptide Repeat (PPR) Protein PPR647, Is Responsible for the as-81647 Albino Phenotype
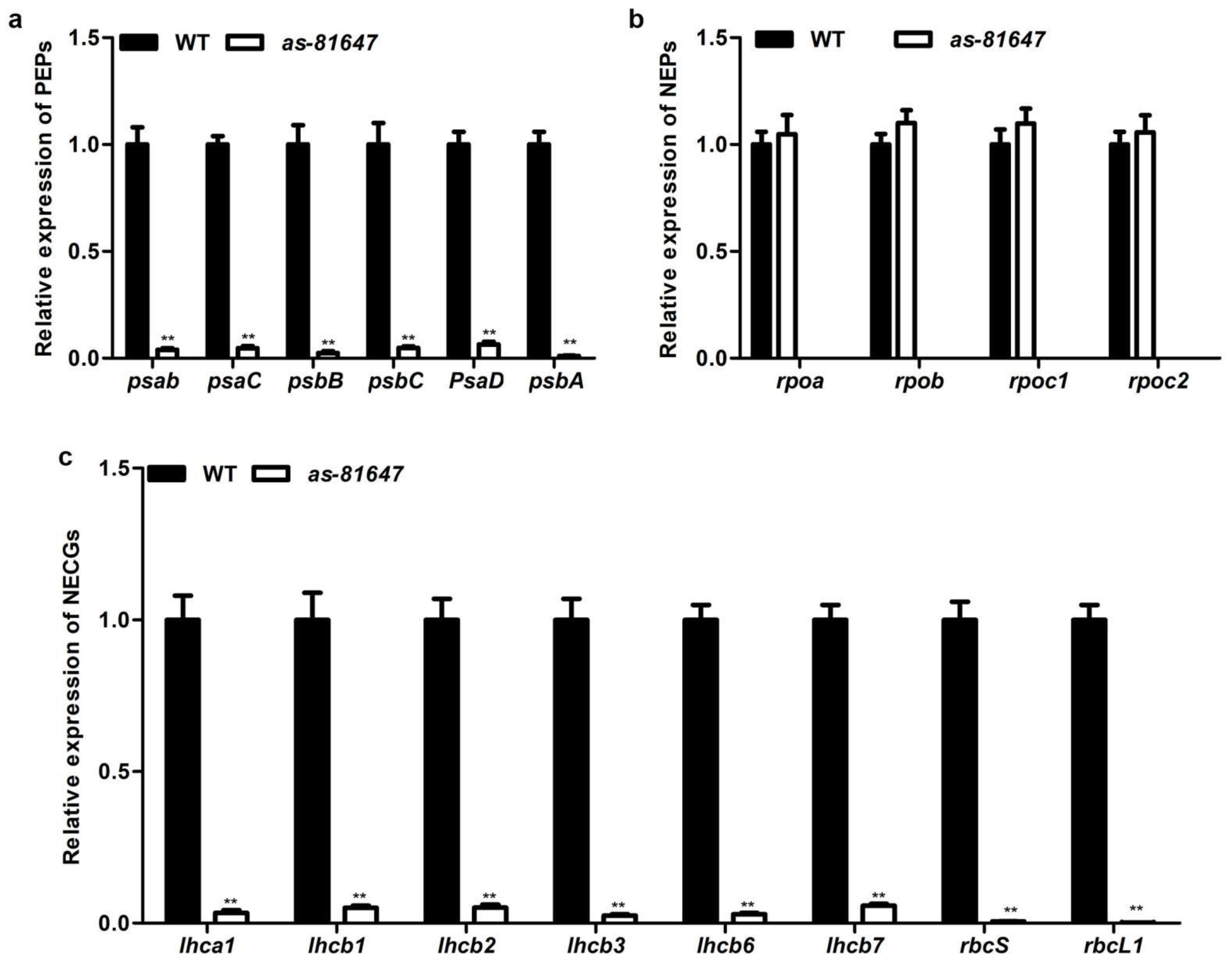
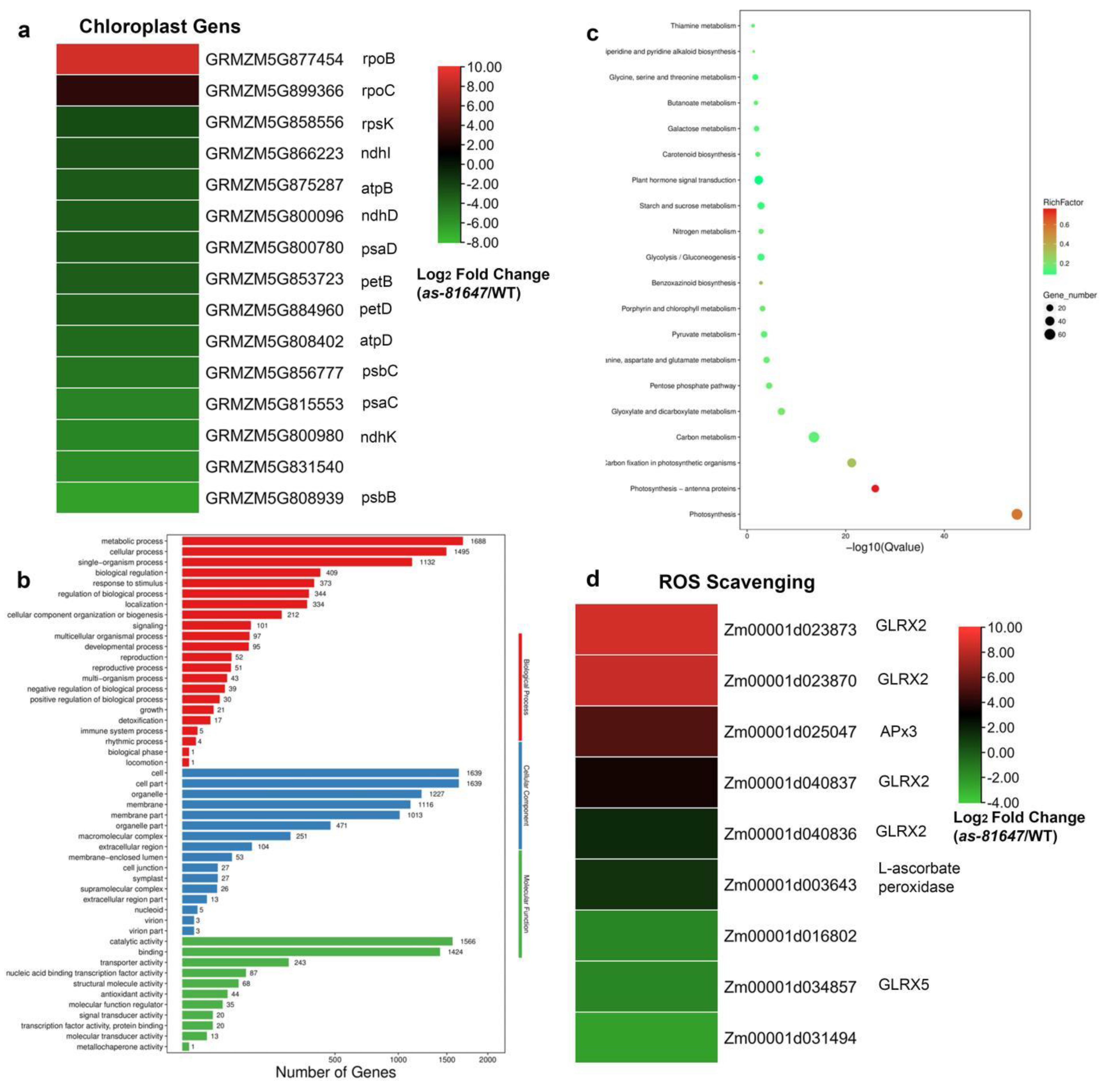
2.5. PPR647 Mutation Affects Chloroplast-Associated Gene Expression during Leaves Development
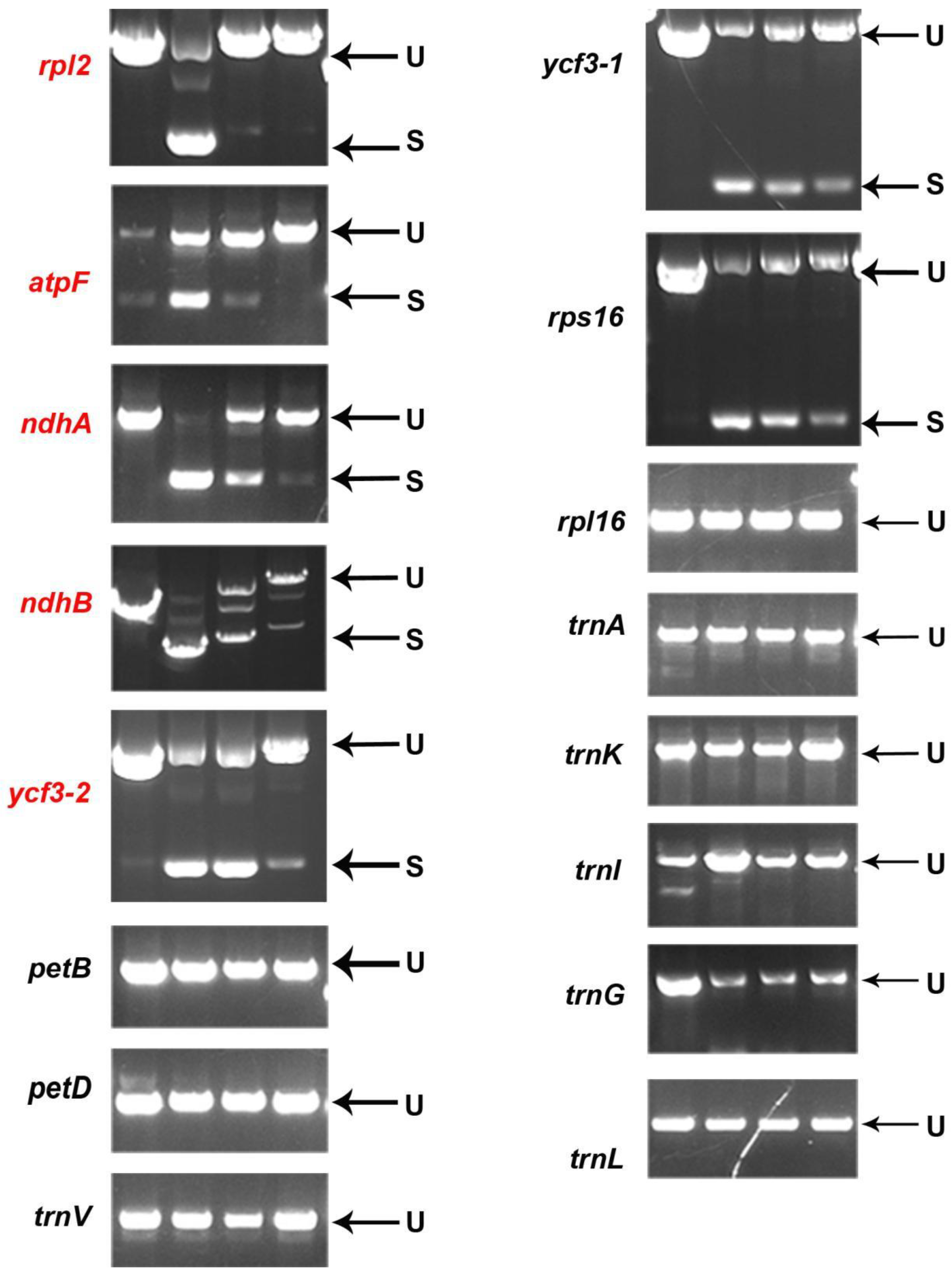
2.6. PPR647 Is Required for C-to-U RNA Editing of Multiple Chloroplast Transcripts
2.7. PPR647 Affects the Splicing of rpl2 Transcripts in Chloroplasts
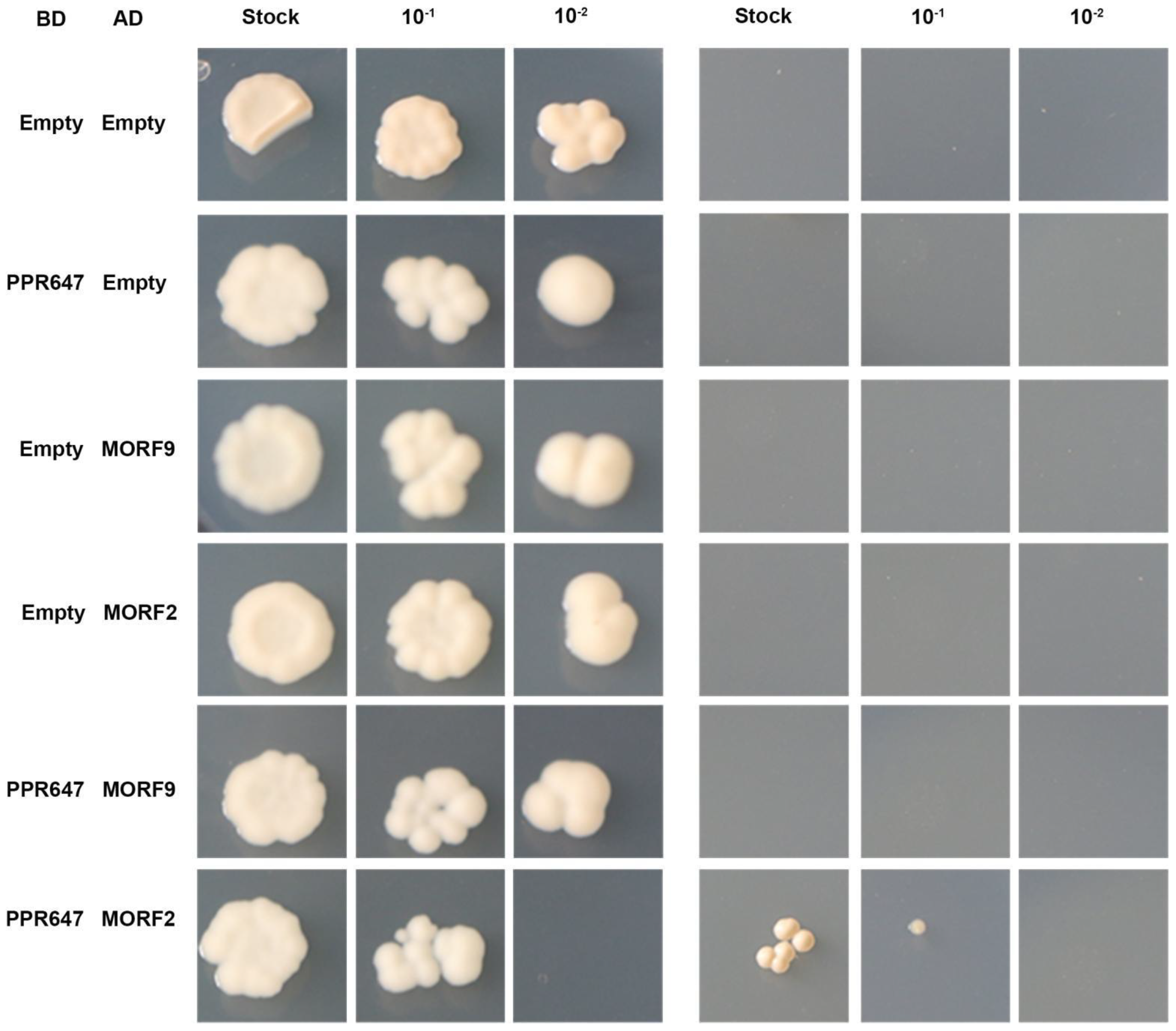
2.8. PPR647 Might Affect RNA Editing by Interacting with ZmMORF2
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Photosynthetic Pigment Content Analysis
4.3. Transmission Electron Microscopy (TEM)
4.4. Histochemical Analysis
4.5. Map-Based Cloning and Allelism Test
4.6. Generation of CRISPR-Cas9-Edited Mutant Alleles
4.7. RNA Sequencing (RNA-seq) and Data Analysis
4.8. RT-PCR and qRT- PCR Analysis
4.9. Analysis of RNA Editing and Splicing of Chloroplast Genes
4.10. Phylogenetic Analysis
4.11. Y2H Assays
4.12. Subcellular Localization of PPR647
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kleffmann, T.; Russenberger, D.; von Zychlinski, A.; Christopher, W.; Sjolander, K.; Gruissem, W.; Baginsky, S. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 2004, 14, 354–362. [Google Scholar] [CrossRef] [Green Version]
- Hedtke, B.; Borner, T.; Weihe, A. Mitochondrial and Chloroplast Phage-Type RNA Polymerases in Arabidopsis. Science 1997, 227, 809–811. [Google Scholar] [CrossRef]
- Pogson, B.J.; Albrecht, V. Genetic dissection of chloroplast biogenesis and development: An overview. Plant Physiol. 2011, 155, 1545–1551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Q.B.; Huang, C.; Yang, Z.N. Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front. Plant Sci. 2014, 5, 316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- del Campo, E.M. Post-Transcriptional Control of Chloroplast Gene Expression. Gene Regul. Syst. Biol. 2009, 3, 31–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusumi, K.; Yara, A.; Mitsui, N.; Tozawa, Y.; Iba, K. Characterization of a Rice Nuclear-Encoded Plastid RNA Polymerase Gene OsRpoTp. Plant Cell Physiol. 2004, 1194–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beick, S.; Schmitz-Linneweber, C.; Williams-Carrier, R.; Jensen, B.; Barkan, A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell Biol. 2008, 28, 5337–5347. [Google Scholar] [CrossRef] [Green Version]
- Schmitz-Linneweber, C.; Williams-Carrier, R.E.; Williams-Voelker, P.M.; Kroeger, T.S.; Vichas, A.; Barkan, A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 2006, 18, 2650–2663. [Google Scholar] [CrossRef] [Green Version]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–420. [Google Scholar] [CrossRef]
- Cheng, S.; Gutmann, B.; Zhong, X.; Ye, Y.; Fisher, M.F.; Bai, F.; Castleden, I.; Song, Y.; Song, B.; Huang, J.; et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016, 85, 532–547. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.H.; Liu, N.Y.; Tang, Z.S.; Liu, J.; Yang, W.C. Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 2006, 18, 815–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammani, K.; Gobert, A.; Hleibieh, K.; Choulier, L.; Small, I.; Giege, P. An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 2011, 23, 730–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colcombet, J.; Lopez-Obando, M.; Heurtevin, L.; Bernard, C.; Martin, K.; Berthome, R.; Lurin, C. Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol. 2013, 10, 1557–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haili, N.; Planchard, N.; Arnal, N.; Quadrado, M.; Vrielynck, N.; Dahan, J.; des Francs-Small, C.C.; Mireau, H. The MTL1 Pentatricopeptide Repeat Protein Is Required for Both Translation and Splicing of the Mitochondrial NADH DEHYDROGENASE SUBUNIT7 mRNA in Arabidopsis. Plant Physiol. 2016, 170, 354–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Zhong, M.; Shuai, B.; Song, J.; Zhang, J.; Han, L.; Ling, H.; Tang, Y.; Wang, G.; Song, R. E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol. 2017, 214, 1563–1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, R.C.; Wang, L.L.; Zhang, L.; Zhao, Y.J.; Wu, J.W.; Wei, Y.M.; Zhang, X.S.; Zhao, X.Y. DEK43 is a P-type pentatricopeptide repeat (PPR) protein responsible for the Cis-splicing of nad4 in maize mitochondria. J. Integr. Plant Biol. 2020, 62, 299–313. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Z.; Zhang, Y.; Zhou, W.; Zhang, A.; Lu, C. Pentatricopeptide repeat protein PHOTOSYSTEM I BIOGENESIS FACTOR2 is required for splicing of ycf3. J. Integr. Plant Biol. 2020, 62, 1741–1761. [Google Scholar] [CrossRef]
- Ren, R.C.; Lu, X.; Zhao, Y.J.; Wei, Y.M.; Wang, L.L.; Zhang, L.; Zhang, W.T.; Zhang, C.; Zhang, X.S.; Zhao, X.Y. Pentatricopeptide repeat protein DEK40 is required for mitochondrial function and kernel development in maize. J. Exp. Bot. 2019, 70, 6163–6179. [Google Scholar] [CrossRef]
- Xu, C.; Song, S.; Yang, Y.Z.; Lu, F.; Zhang, M.D.; Sun, F.; Jia, R.; Song, R.; Tan, B.C. DEK46 performs C-to-U editing of a specific site in mitochondrial nad7 introns that is critical for intron splicing and seed development in maize. Plant J. 2020, 103, 1767–1782. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Huang, W.L.; Yang, H.H.; Jiang, R.C.; Sun, F.; Wang, H.C.; Zhao, J.; Xu, C.H.; Tan, B.C. EMP18 functions in mitochondrial atp6 and cox2 transcript editing and is essential to seed development in maize. New Phytol. 2019, 221, 896–907. [Google Scholar] [CrossRef] [Green Version]
- Khrouchtchova, A.; Monde, R.A.; Barkan, A. A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA 2012, 18, 1197–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Ren, Y.; Zhou, K.; Liu, L.; Wang, J.; Xu, Y.; Zhang, H.; Zhang, L.; Feng, Z.; Wang, L.; et al. WHITE STRIPE LEAF4 Encodes a Novel P-Type PPR Protein Required for Chloroplast Biogenesis during Early Leaf Development. Front. Plant Sci. 2017, 8, 1116. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Park, S.J.; Colas des Francs-Small, C.; Whitby, M.; Small, I.; Kang, H. The coordinated action of PPR4 and EMB2654 on each intron half mediates trans-splicing of rps12 transcripts in plant chloroplasts. Plant J. 2019, 100, 1193–1207. [Google Scholar] [CrossRef]
- Liu, X.Y.; Jiang, R.C.; Wang, Y.; Tang, J.J.; Sun, F.; Yang, Y.Z.; Tan, B.C. ZmPPR26, a DYW-type pentatricopeptide repeat protein, is required for C-to-U RNA editing at atpA-1148 in maize chloroplasts. J. Exp. Bot. 2021, 72, 4809–4821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Guo, Y.; Fang, Q.; Zhu, Y.; Zhang, Y.; Liu, X.; Lin, Y.; Barkan, A.; Zhou, F. The PPR-SMR Protein ATP4 Is Required for Editing the Chloroplast rps8 mRNA in Rice and Maize. Plant Physiol. 2020, 184, 2011–2021. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhang, J.; Qu, S.; Zhao, Y.; Su, B.; Lv, X.; Li, R.; Wan, Y.; Xiao, J. The Pentratricopeptide Repeat Protein Pigment-Defective Mutant2 is Involved in the Regulation of Chloroplast Development and Chloroplast Gene Expression in Arabidopsis. Plant Cell Physiol. 2017, 58, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Hammani, K.; Takenaka, M.; Miranda, R.; Barkan, A. A PPR protein in the PLS subfamily stabilizes the 5′-end of processed rpl16 mRNAs in maize chloroplasts. Nucleic Acids Res. 2016, 44, 4278–4288. [Google Scholar] [CrossRef] [Green Version]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Kugelmann, M.; Hartel, B.; Brennicke, A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5104–5109. [Google Scholar] [CrossRef] [Green Version]
- Igarashi, K.; Kazama, T.; Toriyama, K. A Gene Encoding Pentatricopeptide Repeat Protein Partially Restores Fertility in RT98-Type Cytoplasmic Male-Sterile Rice. Plant Cell Physiol. 2016, 57, 2187–2193. [Google Scholar] [CrossRef] [Green Version]
- Thyssen, G.N.; Fang, D.D.; Zeng, L.; Song, X.; Delhom, C.D.; Condon, T.L.; Li, P.; Kim, H.J. The Immature Fiber Mutant Phenotype of Cotton (Gossypium hirsutum) Is Linked to a 22-bp Frame-Shift Deletion in a Mitochondria Targeted Pentatricopeptide Repeat Gene. G3 Genes Genomes Genet. 2016, 6, 1627–1633. [Google Scholar] [CrossRef] [Green Version]
- Schallenberg-Rudinger, M.; Oldenkott, B.; Hiss, M.; Trinh, P.L.; Knoop, V.; Rensing, S.A. A Single-Target Mitochondrial RNA Editing Factor of Funaria hygrometrica Can Fully Reconstitute RNA Editing at Two Sites in Physcomitrella patens. Plant Cell Physiol. 2017, 58, 496–507. [Google Scholar] [CrossRef]
- Liu, C.; Zhu, H.; Xing, Y.; Tan, J.; Chen, X.; Zhang, J.; Peng, H.; Xie, Q.; Zhang, Z. Albino Leaf 2 is involved in the splicing of chloroplast group I and II introns in rice. J. Exp. Bot. 2016, 67, 5339–5347. [Google Scholar] [CrossRef]
- Zhang, Z.; Tan, J.; Shi, Z.; Xie, Q.; Xing, Y.; Liu, C.; Chen, Q.; Zhu, H.; Wang, J.; Zhang, J.; et al. Albino Leaf1 That Encodes the Sole Octotricopeptide Repeat Protein Is Responsible for Chloroplast Development. Plant Physiol. 2016, 171, 1182–1191. [Google Scholar]
- Kobayashi, K.; Fujii, S.; Sasaki, D.; Baba, S.; Ohta, H.; Masuda, T.; Wada, H. Transcriptional regulation of thylakoid galactolipid biosynthesis coordinated with chlorophyll biosynthesis during the development of chloroplasts in Arabidopsis. Front. Plant Sci. 2014, 5, 272. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; An, Y.; Li, Y.; Xiao, J. A PPR Protein ACM1 Is Involved in Chloroplast Gene Expression and Early Plastid Development in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 2512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xiao, J.; Li, Y.; Su, B.; Xu, H.; Shan, X.; Song, C.; Xie, J.; Li, R. PDM3, a pentatricopeptide repeat-containing protein, affects chloroplast development. J. Exp. Bot. 2017, 68, 5615–5627. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Tan, Z.; Wu, F.; Sheng, P.; Heng, Y.; Wang, X.; Ren, Y.; Wang, J.; Guo, X.; Zhang, X.; et al. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant 2014, 7, 1329–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowotny, N.; Nierhaus, K.H. Initiator proteins for the assembly of the 50S subunit from Escherichia coli ribosomes. Proc. Natl. Acad. Sci. USA 1982, 79, 7238–7242. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, C.; Wang, Y.; Niu, M.; Ren, Y.; Zhou, K.; Zhang, H.; Lin, Q.; Wu, F.; Cheng, Z.; et al. WSL3, a component of the plastid-encoded plastid RNA polymerase, is essential for early chloroplast development in rice. Plant Mol. Biol. 2016, 92, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Shao, G.; Qiu, J.; Jiao, G.; Sheng, Z.; Xie, L.; Wu, Y.; Tang, S.; Wei, X.; Hu, P. White Leaf and Panicle 2, encoding a PEP-associated protein, is required for chloroplast biogenesis under heat stress in rice. J. Exp. Bot. 2017, 68, 5147–5160. [Google Scholar] [CrossRef] [Green Version]
- Arsova, B.; Hoja, U.; Wimmelbacher, M.; Greiner, E.; Ustun, S.; Melzer, M.; Petersen, K.; Lein, W.; Bornke, F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: Evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 2010, 22, 1498–1515. [Google Scholar] [CrossRef] [Green Version]
- Gilkerson, J.; Perez-Ruiz, J.M.; Chory, J.; Callis, J. The plastid-localized pfkB-type carbohydrate kinases FRUCTOKINASE-LIKE 1 and 2 are essential for growth and development of Arabidopsis thaliana. BMC Plant Biol. 2012, 12, 102–117. [Google Scholar] [CrossRef] [Green Version]
- Pfalz, J.; Holtzegel, U.; Barkan, A.; Weisheit, W.; Mittag, M.; Pfannschmidt, T. ZmpTAC12 binds single-stranded nucleic acids and is essential for accumulation of the plastid-encoded polymerase complex in maize. New Phytol. 2015, 206, 1024–1037. [Google Scholar] [CrossRef]
- Chi, W.; Mao, J.; Li, Q.; Ji, D.; Zou, M.; Lu, C.; Zhang, L. Interaction of the pentatricopeptide-repeat protein DELAYED GREENING 1 with sigma factor SIG6 in the regulation of chloroplast gene expression in Arabidopsis cotyledons. Plant J. 2010, 64, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. polyphenoloxidase in beta vulgaris. Plant Physiol. 1949, 24, 549–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, A.L.; Wang, M.L.; Zhou, R.H.; Kong, X.Y.; Huo, N.X.; Wang, W.S.; Jia, J.Z. Comparative analysis of early H2O2 accumulation in compatible and incompatible wheat-powdery mildew interactions. Plant Pathol. 2005, 54, 308–316. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Qi, W.; Li, X.; Yang, Q.; Yan, S.; Ling, H.; Wang, G.; Wang, G.; Song, R. Maize opaque 10 Encodes a Cereal-Specific Protein That Is Essential for the Proper Distribution of Zeins in Endosperm Protein Bodies. PLoS Genet. 2016, 12, e1006270. [Google Scholar] [CrossRef]


| Number | Total | Normal Plants | Mutant Plants | Ratio of Segregation | χ20.05 (1) |
|---|---|---|---|---|---|
| 1 | 410 | 309 | 101 | 3/1 | 0.029 |
| 2 | 426 | 316 | 110 | 3/1 | 0.153 |
| 3 | 392 | 296 | 96 | 3/1 | 0.05 |
| 4 | 370 | 283 | 87 | 3/1 | 0.436 |
| 5 | 398 | 295 | 103 | 3/1 | 0.164 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Xu, W.; Zhang, Y.; Sun, S.; Wang, L.; Zhong, S.; Zhao, X.; Liu, B. PPR647 Protein Is Required for Chloroplast RNA Editing, Splicing and Chloroplast Development in Maize. Int. J. Mol. Sci. 2021, 22, 11162. https://doi.org/10.3390/ijms222011162
Zhao Y, Xu W, Zhang Y, Sun S, Wang L, Zhong S, Zhao X, Liu B. PPR647 Protein Is Required for Chloroplast RNA Editing, Splicing and Chloroplast Development in Maize. International Journal of Molecular Sciences. 2021; 22(20):11162. https://doi.org/10.3390/ijms222011162
Chicago/Turabian StyleZhao, Yan, Wei Xu, Yongzhong Zhang, Shilei Sun, Lijing Wang, Shiyi Zhong, Xiangyu Zhao, and Baoshen Liu. 2021. "PPR647 Protein Is Required for Chloroplast RNA Editing, Splicing and Chloroplast Development in Maize" International Journal of Molecular Sciences 22, no. 20: 11162. https://doi.org/10.3390/ijms222011162
APA StyleZhao, Y., Xu, W., Zhang, Y., Sun, S., Wang, L., Zhong, S., Zhao, X., & Liu, B. (2021). PPR647 Protein Is Required for Chloroplast RNA Editing, Splicing and Chloroplast Development in Maize. International Journal of Molecular Sciences, 22(20), 11162. https://doi.org/10.3390/ijms222011162





