Isolation and Characterization of Compounds from Glycyrrhiza uralensis as Therapeutic Agents for the Muscle Disorders
Abstract
1. Introduction
2. Results
2.1. The CWE of G. uralensis Promoted Myoblast Proliferation and Differentiation
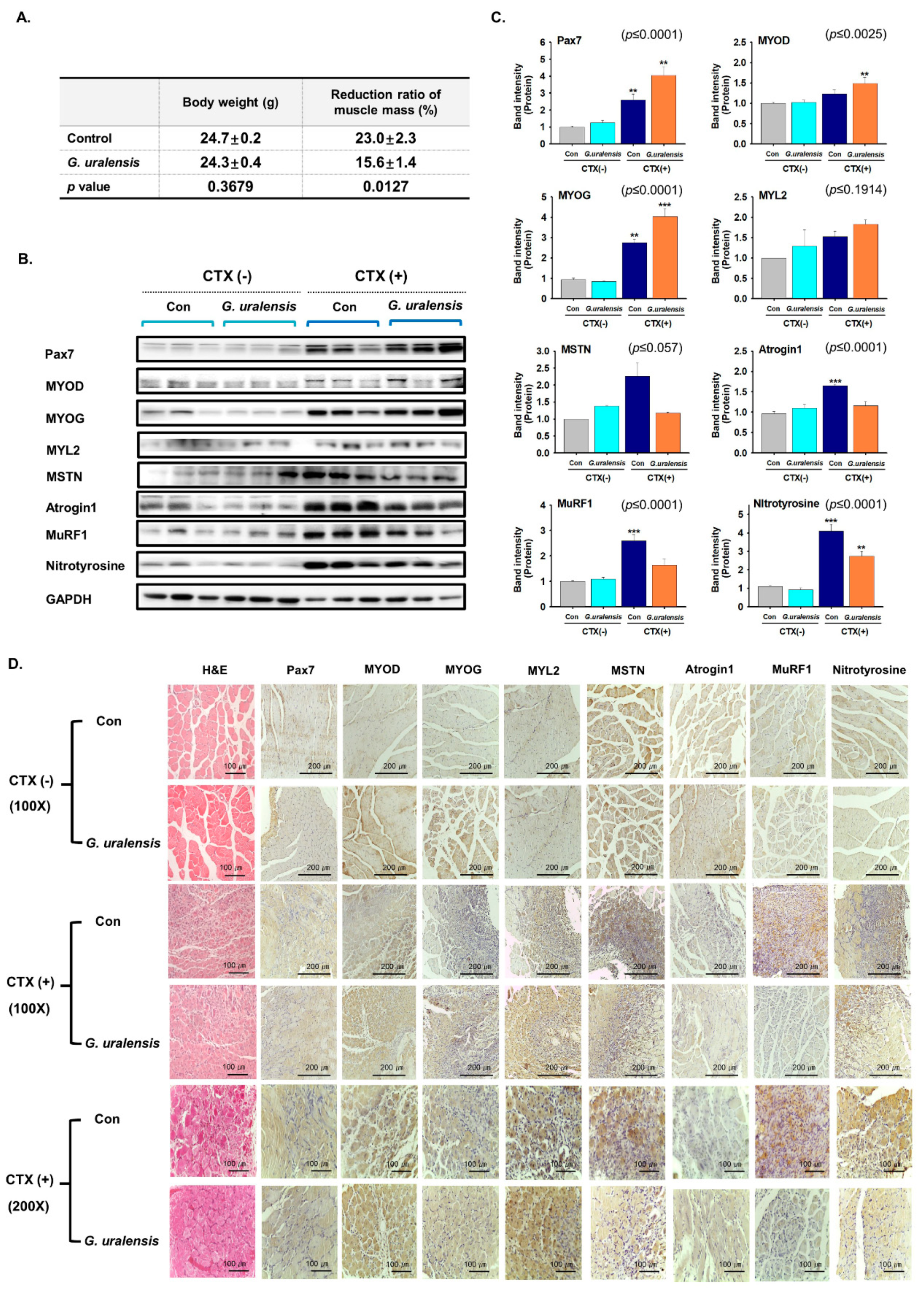
2.2. The CWE of G. uralensis Enhanced Muscle Regeneration
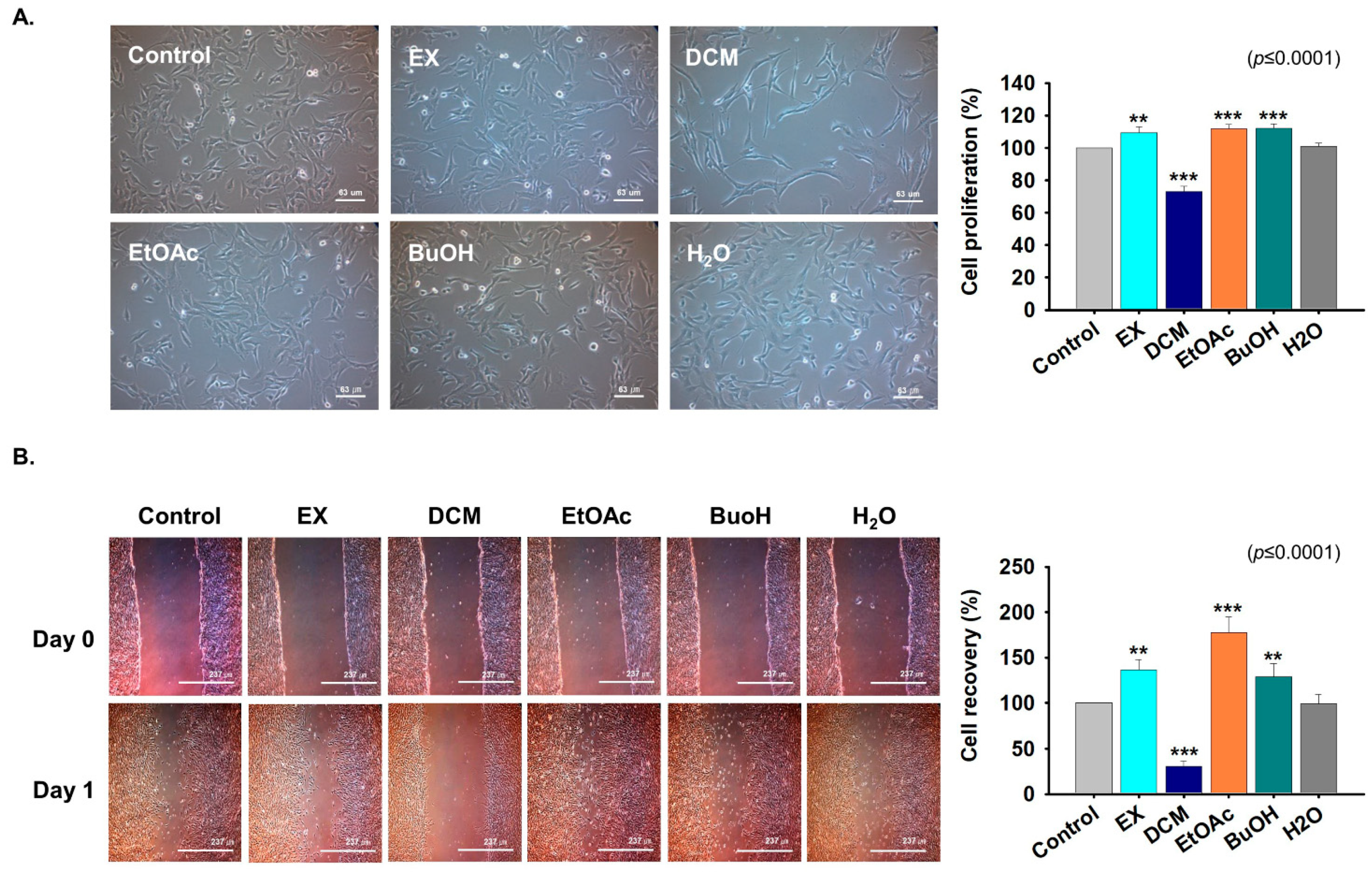
2.3. Fractions Derived from G. uralensis CWE Enhanced Myoblast Proliferation
2.4. Effects of G. uralensis CWE Derived Fractions on Myoblast Differentiation
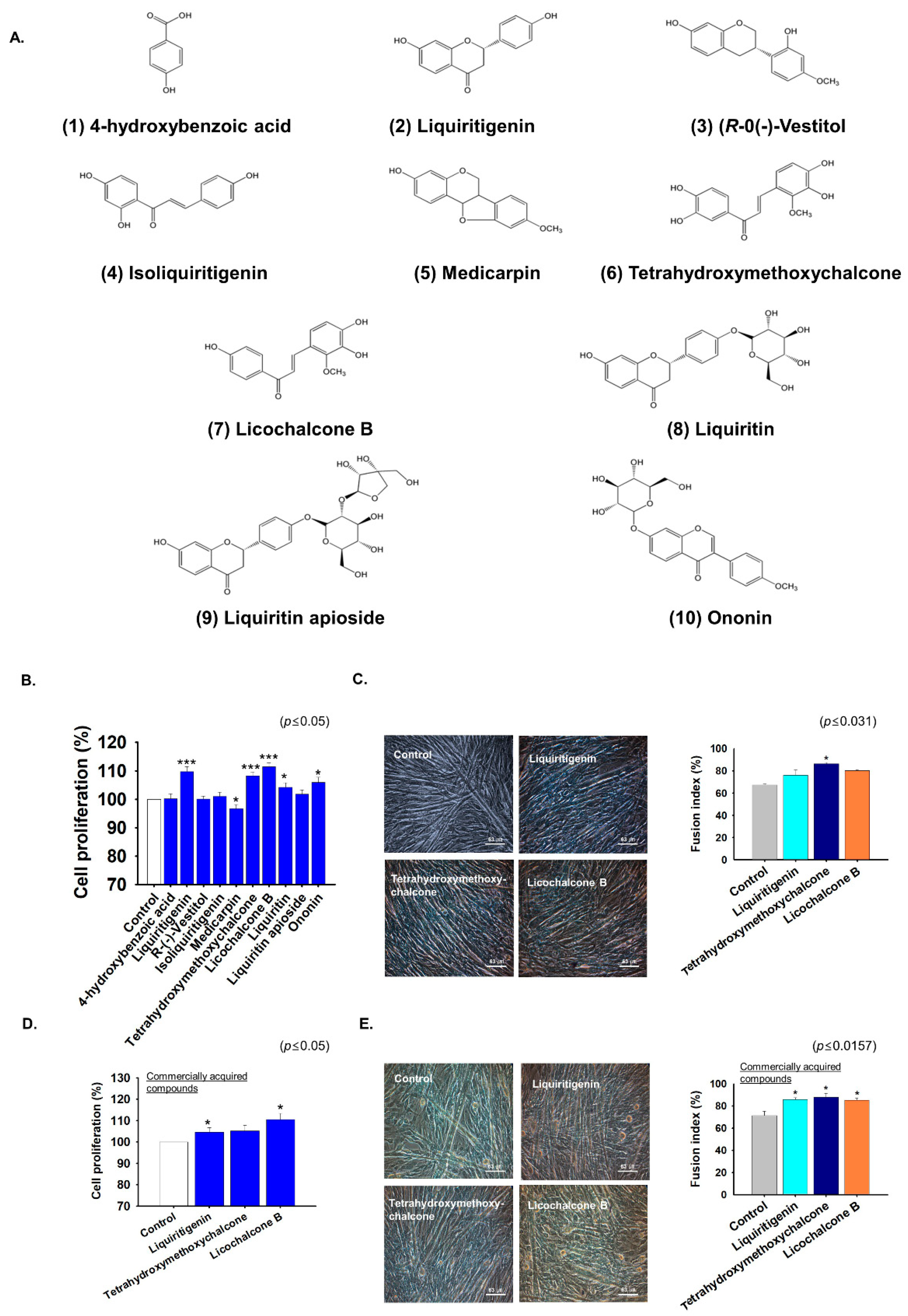
2.5. G. uralensis-EtOAc Derived Compounds Increased Myoblast Proliferation and Differentiation
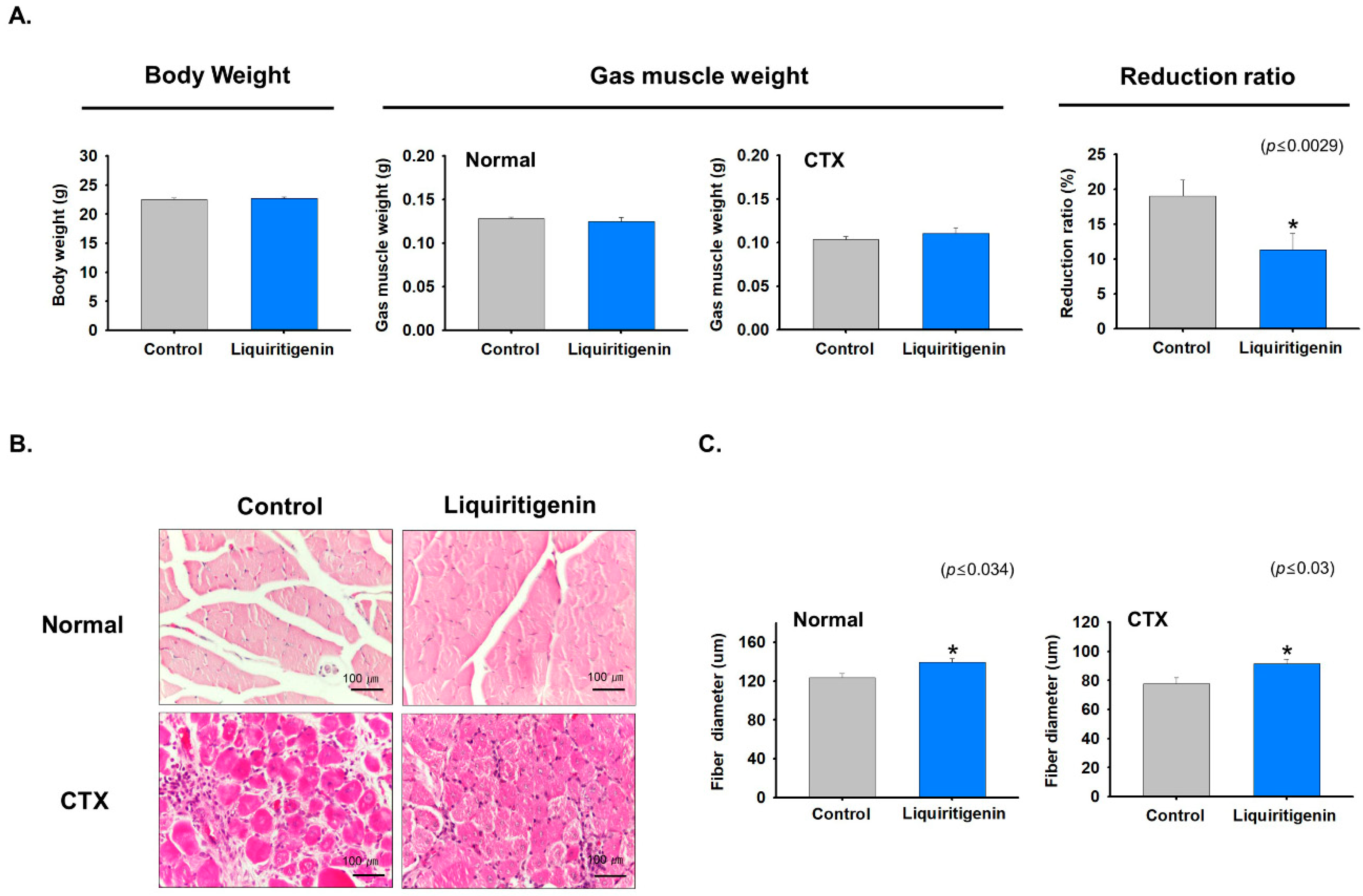
2.6. Liquiritigenin Enhanced Muscle Regeneration
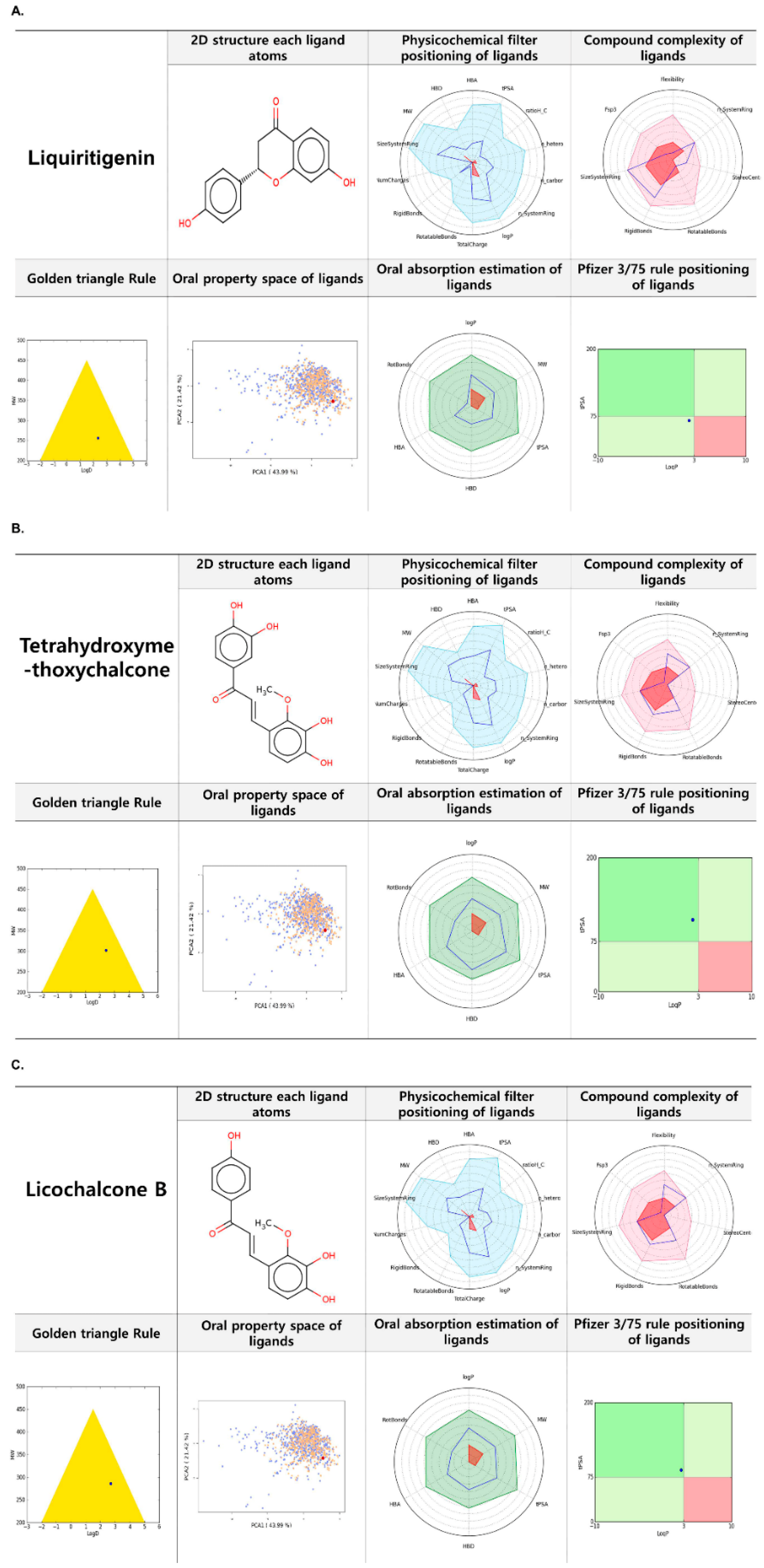
2.7. FAF-Drugs 4 Based Analysis of Liquiritigenin, Tetrahydroxymethoxychalcone, and Licochalcone B
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Plant Material and Preparation of G. uralensis Water Extract (CWE)
4.3. Extraction and Isolation
4.4. Animal Experiment
4.5. Cell Culture
4.6. Cell Proliferation Assay
4.7. Cell Differentiation
4.8. Scratch Experiment
4.9. Fusion Index
4.10. Metabolite Analysis
4.11. Real-Time RT-PCR
4.12. Western Blot
4.13. Immunohistochemistry
4.14. Analysis of Muscle Mass Reduction and Muscle Fiber Diameters
4.15. FAF-Drugs4 Analysis
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frontera, W.R.; Ochala, J. Skeletal muscle: A brief review of structure and function. Calcif. Tissue Int. 2015, 96, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Tidball, J.G.; Villalta, S.A. Regulatory interactions between muscle and the immune system during muscle regeneration. Am. J. Physiol Regul. Integr. Comp. Physiol. 2010, 298, R1173–R1187. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Rigby, P.W. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell 2014, 28, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Krauss, R.S. Muscle stem cells in developmental and regenerative myogenesis. Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Kuang, S.; Rudnicki, M.A. The emerging biology of satellite cells and their therapeutic potential. Trends Mol. Med. 2008, 14, 82–91. [Google Scholar] [CrossRef]
- Apponi, L.H.; Corbett, A.H.; Pavlath, G.K. RNA-binding proteins and gene regulation in myogenesis. Trends Pharmacol. Sci. 2011, 32, 652–658. [Google Scholar] [CrossRef]
- Ahmad, K.; Lee, E.J.; Moon, J.S.; Park, S.Y.; Choi, I. Multifaceted Interweaving Between Extracellular Matrix, Insulin Resistance, and Skeletal Muscle. Cells 2018, 7, 148. [Google Scholar] [CrossRef]
- Lee, E.J.; Jan, A.T.; Baig, M.H.; Ashraf, J.M.; Nahm, S.S.; Kim, Y.W.; Park, S.Y.; Choi, I. Fibromodulin: A master regulator of myostatin controlling progression of satellite cells through a myogenic program. FASEB J. 2016, 30, 2708–2719. [Google Scholar] [CrossRef]
- Thomas, K.; Engler, A.J.; Meyer, G.A. Extracellular matrix regulation in the muscle satellite cell niche. Connect. Tissue Res. 2015, 56, 1–8. [Google Scholar] [CrossRef]
- Murphy, M.M.; Lawson, J.A.; Mathew, S.J.; Hutcheson, D.A.; Kardon, G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 2011, 138, 3625–3637. [Google Scholar] [CrossRef]
- Bencze, M.; Negroni, E.; Vallese, D.; Yacoub-Youssef, H.; Chaouch, S.; Wolff, A.; Aamiri, A.; Di Santo, J.P.; Chazaud, B.; Butler-Browne, G.; et al. Proinflammatory macrophages enhance the regenerative capacity of human myoblasts by modifying their kinetics of proliferation and differentiation. Mol. Ther. 2012, 20, 2168–2179. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Ahmad, K.; Shaikh, S.; Jan, A.T.; Seo, M.G.; Lee, E.J.; Choi, I. Dermatopontin in Skeletal Muscle Extracellular Matrix Regulates Myogenesis. Cells 2019, 8, 332. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, K.; Shaikh, S.; Ahmad, S.S.; Lee, E.J.; Choi, I. Cross-Talk Between Extracellular Matrix and Skeletal Muscle: Implications for Myopathies. Front. Pharmacol. 2020, 11, 142. [Google Scholar] [CrossRef] [PubMed]
- Liguori, I.; Russo, G.; Aran, L.; Bulli, G.; Curcio, F.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D.; et al. Sarcopenia: Assessment of disease burden and strategies to improve outcomes. Clin. Interv. Aging 2018, 13, 913–927. [Google Scholar] [CrossRef] [PubMed]
- Vinciguerra, M.; Musaro, A.; Rosenthal, N. Regulation of muscle atrophy in aging and disease. Adv. Exp. Med. Biol. 2010, 694, 211–233. [Google Scholar] [CrossRef]
- Shefer, G.; Rauner, G.; Yablonka-Reuveni, Z.; Benayahu, D. Reduced satellite cell numbers and myogenic capacity in aging can be alleviated by endurance exercise. PLoS ONE 2010, 5, e13307. [Google Scholar] [CrossRef]
- Deutz, N.E.; Bauer, J.M.; Barazzoni, R.; Biolo, G.; Boirie, Y.; Bosy-Westphal, A.; Cederholm, T.; Cruz-Jentoft, A.; Krznaric, Z.; Nair, K.S.; et al. Protein intake and exercise for optimal muscle function with aging: Recommendations from the ESPEN Expert Group. Clin. Nutr. 2014, 33, 929–936. [Google Scholar] [CrossRef]
- Sakuma, K.; Yamaguchi, A. Inhibitors of myostatin-and proteasome-dependent signaling for attenuating muscle wasting. Recent Pat. Regen. Med. 2011, 1, 284–298. [Google Scholar] [CrossRef]
- McFarlane, C.; Plummer, E.; Thomas, M.; Hennebry, A.; Ashby, M.; Ling, N.; Smith, H.; Sharma, M.; Kambadur, R. Myostatin induces cachexia by activating the ubiquitin proteolytic system through an NF-kappaB-independent, FoxO1-dependent mechanism. J. Cell Physiol. 2006, 209, 501–514. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, Y.; Li, Y.; Wu, Z.; Zhu, D. Myostatin induces cyclin D1 degradation to cause cell cycle arrest through a phosphatidylinositol 3-kinase/AKT/GSK-3 beta pathway and is antagonized by insulin-like growth factor 1. J. Biol. Chem. 2007, 282, 3799–3808. [Google Scholar] [CrossRef]
- Dang, K.; Li, Y.Z.; Gong, L.C.; Xue, W.; Wang, H.P.; Goswami, N.; Gao, Y.F. Stable atrogin-1 (Fbxo32) and MuRF1 (Trim63) gene expression is involved in the protective mechanism in soleus muscle of hibernating Daurian ground squirrels (Spermophilus dauricus). Biol. Open 2016, 5, 62–71. [Google Scholar] [CrossRef]
- Cohen, S.; Brault, J.J.; Gygi, S.P.; Glass, D.J.; Valenzuela, D.M.; Gartner, C.; Latres, E.; Goldberg, A.L. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J. Cell Biol. 2009, 185, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Ischiropoulos, H. Biological tyrosine nitration: A pathophysiological function of nitric oxide and reactive oxygen species. Arch. Biochem. Biophys. 1998, 356, 1–11. [Google Scholar] [CrossRef]
- Zhang, Q.; Ye, M. Chemical analysis of the Chinese herbal medicine Gan-Cao (licorice). J. Chromatogr. A 2009, 1216, 1954–1969. [Google Scholar] [CrossRef]
- Asl, M.N.; Hosseinzadeh, H. Review of pharmacological effects of Glycyrrhiza sp. and its bioactive compounds. Phytother. Res. 2008, 22, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Hwang, H.J.; Ha, J.S.; Jeong, H.S.; Kim, J.H. Screening of medicinal plant extracts for antioxidant activity. Life Sci. 2003, 73, 167–179. [Google Scholar] [CrossRef]
- Cho, J.Y.; Moon, J.H.; Seong, K.Y.; Park, K.H. Antimicrobial activity of 4-hydroxybenzoic acid and trans 4-hydroxycinnamic acid isolated and identified from rice hull. Biosci. Biotechnol. Biochem. 1998, 62, 2273–2276. [Google Scholar] [CrossRef]
- Lee, Y.; Lee, D.-E.; Lee, H.-S.; Kim, S.-K.; Lee, W.S.; Kim, S.-H.; Kim, M.-W. Influence of auxins, cytokinins, and nitrogen on production of rutin from callus and adventitious roots of the white mulberry tree (Morusalba L.). Plant. Cell Tissue Organ. Cult. (PCTOC) 2011, 105, 9–19. [Google Scholar] [CrossRef]
- Ma, C.J.; Li, G.S.; Zhang, D.L.; Liu, K.; Fan, X. One step isolation and purification of liquiritigenin and isoliquiritigenin from Glycyrrhiza uralensis Risch. using high-speed counter-current chromatography. J. Chromatogr. A 2005, 1078, 188–192. [Google Scholar] [CrossRef]
- Na, I.-S.; Park, M.-J.; Noh, C.-H.; Min, J.-W.; Bang, M.-H.; Yang, D.-C. Production of flavonoid aglycone from Korean Glycyrrhizae radix by biofermentation process. J. Physiol. Pathol. Korean Med. 2008, 22, 569–574. [Google Scholar]
- Hamburger, M.O.; Cordell, G.A.; Tantivatana, P.; Ruangrungsi, N. Traditional medicinal plants of Thailand, VIII. Isoflavonoids of Dalbergia candenatensis. J. Nat. Prod. 1987, 50, 696–699. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sun, X.; Yu, Z.; Pan, X.; Gu, F.; Chen, J.; Dong, W.; Zhao, L.; Zhong, C. Exposure to pyrithiamine increases beta-amyloid accumulation, Tau hyperphosphorylation, and glycogen synthase kinase-3 activity in the brain. Neurotox Res. 2011, 19, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.K.; Chun, H.S. Isoliquiritigenin isolated from licorice Glycyrrhiza uralensis prevents 6-hydroxydopamine-induced apoptosis in dopaminergic neurons. Biosci. Biotechnol. Biochem. 2012, 76, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Le Bail, J.C.; Champavier, Y.; Chulia, A.J.; Habrioux, G. Effects of phytoestrogens on aromatase, 3beta and 17beta-hydroxysteroid dehydrogenase activities and human breast cancer cells. Life Sci. 2000, 66, 1281–1291. [Google Scholar] [CrossRef]
- Hatano, T.; Takagi, M.; Ito, H.; Yoshida, T. Acylated flavonoid glycosides and accompanying phenolics from licorice. Phytochemistry 1998, 47, 287–293. [Google Scholar] [CrossRef]
- Saitoh, T.; Shibata, S.; Sankawa, U.; Furuya, T.; Ayabe, S. Biosynthesis of echinatin. A new biosynthetical scheme of retrochalcone. Tetrahedron Lett. 1975, 4463–4466. [Google Scholar] [CrossRef]
- Kajiyama, K.; Demizu, S.; Hiraga, Y.; Kinoshita, K.; Koyama, K.; Takahashi, K.; Tamura, Y.; Okada, K.; Kinoshita, T. Two prenylated retrochalcones from Glycyrrhiza inflata. Phytochemistry 1992, 31, 3229–3232. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, J.; Li, Y.J.; Zheng, Y.F.; Li, P. Antioxidant and anti-inflammatory activities of six flavonoids separated from licorice. Food Chem. 2013, 141, 1063–1071. [Google Scholar] [CrossRef]
- Fu, B.; Li, H.; Wang, X.; Lee, F.S.; Cui, S. Isolation and identification of flavonoids in licorice and a study of their inhibitory effects on tyrosinase. J. Agric. Food Chem. 2005, 53, 7408–7414. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, J.H.; Park, J.H.; Kim, S.Y.; Choi, J.Y.; Lee, S.H.; Kim, Y.S.; Kang, S.S.; Jang, E.C.; Han, Y. Liquiritigenin, a licorice flavonoid, helps mice resist disseminated candidiasis due to Candida albicans by Th1 immune response, whereas liquiritin, its glycoside form, does not. Int. Immunopharmacol. 2009, 9, 632–638. [Google Scholar] [CrossRef]
- Montoro, P.; Maldini, M.; Russo, M.; Postorino, S.; Piacente, S.; Pizza, C. Metabolic profiling of roots of liquorice (Glycyrrhiza glabra) from different geographical areas by ESI/MS/MS and determination of major metabolites by LC-ESI/MS and LC-ESI/MS/MS. J. Pharm. Biomed. Anal. 2011, 54, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Fedoreyev, S.A.; Bulgakov, V.P.; Grishchenko, O.V.; Veselova, M.V.; Krivoschekova, O.E.; Kulesh, N.I.; Denisenko, V.A.; Tchernoded, G.K.; Zhuravlev, Y.N. Isoflavonoid composition of a callus culture of the relict tree Maackia amurensis Rupr. et Maxim. J. Agric. Food Chem. 2008, 56, 7023–7031. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Yang, Y.; Zhao, Y.; Gu, D.; He, D.; Yili, A.; Ma, Q.; Cheng, Z.; Gao, Y.; Aisa, H.A.; et al. Comparative Study on Separation and Purification of Isoflavones from the Seeds and Sprouts of Chickpea by HSCCC. J. Liq. Chromatogr. Relat. Technol. 2009, 32, 2879–2892. [Google Scholar] [CrossRef]
- Saddala, M.S.; Huang, H. Identification of novel inhibitors for TNFalpha, TNFR1 and TNFalpha-TNFR1 complex using pharmacophore-based approaches. J. Transl Med. 2019, 17, 215. [Google Scholar] [CrossRef] [PubMed]
- Sarti, S.; Ruggiero, E.; Coin, A.; Toffanello, E.D.; Perissinotto, E.; Miotto, F.; Pintore, G.; Inelmen, E.M.; Manzato, E.; Sergi, G. Dietary intake and physical performance in healthy elderly women: A 3-year follow-up. Exp. Gerontol. 2013, 48, 250–254. [Google Scholar] [CrossRef]
- Takeda, T.; Tsuiji, K.; Li, B.; Tadakawa, M.; Yaegashi, N. Proliferative effect of Hachimijiogan, a Japanese herbal medicine, in C2C12 skeletal muscle cells. Clin. Interv. Aging 2015, 10, 445–451. [Google Scholar] [CrossRef]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building muscle: Molecular regulation of myogenesis. Cold Spring Harb. Perspect Biol. 2012, 4. [Google Scholar] [CrossRef]
- Peixoto da Silva, S.; Santos, J.M.O.; Costa, E.S.M.P.; Gil da Costa, R.M.; Medeiros, R. Cancer cachexia and its pathophysiology: Links with sarcopenia, anorexia and asthenia. J. Cachexia Sarcopenia Muscle 2020. [Google Scholar] [CrossRef]
- Bowen, T.S.; Schuler, G.; Adams, V. Skeletal muscle wasting in cachexia and sarcopenia: Molecular pathophysiology and impact of exercise training. J. Cachexia Sarcopenia Muscle 2015, 6, 197–207. [Google Scholar] [CrossRef]
- Cohen, S.; Nathan, J.A.; Goldberg, A.L. Muscle wasting in disease: Molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 2015, 14, 58–74. [Google Scholar] [CrossRef]
- Baig, M.H.; Jan, A.T.; Rabbani, G.; Ahmad, K.; Ashraf, J.M.; Kim, T.; Min, H.S.; Lee, Y.H.; Cho, W.K.; Ma, J.Y.; et al. Methylglyoxal and Advanced Glycation End products: Insight of the regulatory machinery affecting the myogenic program and of its modulation by natural compounds. Sci Rep. 2017, 7, 5916. [Google Scholar] [CrossRef] [PubMed]
- Chiappalupi, S.; Sorci, G.; Vukasinovic, A.; Salvadori, L.; Sagheddu, R.; Coletti, D.; Renga, G.; Romani, L.; Donato, R.; Riuzzi, F. Targeting RAGE prevents muscle wasting and prolongs survival in cancer cachexia. J. Cachexia Sarcopenia Muscle 2020. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.R.; Jeong, C.K.; Park, K.K.; Choi, J.H.; Park, J.H.; Lim, S.S.; Chung, W.Y. Anti-inflammatory effects of licorice and roasted licorice extracts on TPA-induced acute inflammation and collagen-induced arthritis in mice. J. Biomed. Biotechnol. 2010, 2010, 709378. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Tao, W.; Huang, H.; Du, Y.; Chu, X.; Chen, G. Protective effect of liquiritigenin on depressive-like behavior in mice after lipopolysaccharide administration. Psychiatry Res. 2016, 240, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, S.J.; Yun, K.J.; Cho, Y.W.; Park, H.J.; Lee, K.T. Isoliquiritigenin isolated from the roots of Glycyrrhiza uralensis inhibits LPS-induced iNOS and COX-2 expression via the attenuation of NF-kappaB in RAW 264.7 macrophages. Eur. J. Pharmacol. 2008, 584, 175–184. [Google Scholar] [CrossRef]
- Kim, J.K.; Shin, E.K.; Park, J.H.; Kim, Y.H.; Park, J.H. Antitumor and antimetastatic effects of licochalcone A in mouse models. J. Mol. Med. (Berl) 2010, 88, 829–838. [Google Scholar] [CrossRef]
- Tanemoto, R.; Okuyama, T.; Matsuo, H.; Okumura, T.; Ikeya, Y.; Nishizawa, M. The constituents of licorice (Glycyrrhiza uralensis) differentially suppress nitric oxide production in interleukin-1beta-treated hepatocytes. Biochem. Biophys. Rep. 2015, 2, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Velica, P.; Bunce, C.M. A quick, simple and unbiased method to quantify C2C12 myogenic differentiation. Muscle Nerve 2011, 44, 366–370. [Google Scholar] [CrossRef]
- Picard, B.; Depreux, F.; Geay, Y. Muscle differentiation of normal and double-muscled bovine foetal myoblasts in primary culture. BAM-PADOVA 1998, 8, 197–204. [Google Scholar]
- Lee, A.R.; Nam, K.; Lee, B.J.; Lee, S.W.; Baek, S.M.; Bang, J.S.; Choi, S.K.; Park, S.J.; Kim, T.H.; Jeong, K.S.; et al. Hepatic Cellular Distribution of Silica Nanoparticles by Surface Energy Modification. Int. J. Mol. Sci. 2019, 20, 3812. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Lagorce, D.; Bouslama, L.; Becot, J.; Miteva, M.A.; Villoutreix, B.O. FAF-Drugs4: Free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics 2017, 33, 3658–3660. [Google Scholar] [CrossRef] [PubMed]
- Lagarde, N.; Goldwaser, E.; Pencheva, T.; Jereva, D.; Pajeva, I.; Rey, J.; Tuffery, P.; Villoutreix, B.O.; Miteva, M.A. A Free Web-Based Protocol to Assist Structure-Based Virtual Screening Experiments. Int. J. Mol. Sci. 2019, 20, 4648. [Google Scholar] [CrossRef] [PubMed]


| Isolated Compounds | Weight (mg) | Yield (%) a | NMR Spectra | |
|---|---|---|---|---|
| 1H | 13C | |||
| 4-hydroxybenzoic acid | 23.90 | 0.0006 | 600 MHz | 150 MHz |
| Liquiritigenin | 108.5 | 0.0026 | 600 MHz | 150 MHz |
| R-(-)-Vestitol | 51.70 | 0.0012 | 600 MHz | 150 MHz |
| Isoliquiritigenin | 145.5 | 0.0035 | 600 MHz | 150 MHz |
| Medicarpin | 11.20 | 0.0003 | 600 MHz | 150 MHz |
| Tetrahydroxymethoxychalcone | 35.20 | 0.0008 | 600 MHz | 150 MHz |
| Licochalcone B | 5.600 | 0.0001 | 600 MHz | 150 MHz |
| Liquiritin | 969.0 | 0.0231 | 600 MHz | 150 MHz |
| Liquiritin apioside | 113.5 | 0.0027 | 600 MHz | 150 MHz |
| Ononin | 22.20 | 0.0005 | 600 MHz | 150 MHz |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, E.J.; Shaikh, S.; Ahmad, K.; Ahmad, S.S.; Lim, J.H.; Park, S.; Yang, H.J.; Cho, W.-K.; Park, S.-J.; Lee, Y.-H.; et al. Isolation and Characterization of Compounds from Glycyrrhiza uralensis as Therapeutic Agents for the Muscle Disorders. Int. J. Mol. Sci. 2021, 22, 876. https://doi.org/10.3390/ijms22020876
Lee EJ, Shaikh S, Ahmad K, Ahmad SS, Lim JH, Park S, Yang HJ, Cho W-K, Park S-J, Lee Y-H, et al. Isolation and Characterization of Compounds from Glycyrrhiza uralensis as Therapeutic Agents for the Muscle Disorders. International Journal of Molecular Sciences. 2021; 22(2):876. https://doi.org/10.3390/ijms22020876
Chicago/Turabian StyleLee, Eun Ju, Sibhghatulla Shaikh, Khurshid Ahmad, Syed Sayeed Ahmad, Jeong Ho Lim, Soyoung Park, Hye Jin Yang, Won-Kyung Cho, Sang-Joon Park, Yong-Ho Lee, and et al. 2021. "Isolation and Characterization of Compounds from Glycyrrhiza uralensis as Therapeutic Agents for the Muscle Disorders" International Journal of Molecular Sciences 22, no. 2: 876. https://doi.org/10.3390/ijms22020876
APA StyleLee, E. J., Shaikh, S., Ahmad, K., Ahmad, S. S., Lim, J. H., Park, S., Yang, H. J., Cho, W.-K., Park, S.-J., Lee, Y.-H., Park, S.-Y., Ma, J.-Y., & Choi, I. (2021). Isolation and Characterization of Compounds from Glycyrrhiza uralensis as Therapeutic Agents for the Muscle Disorders. International Journal of Molecular Sciences, 22(2), 876. https://doi.org/10.3390/ijms22020876








