CHD4 Predicts Aggressiveness in PTC Patients and Promotes Cancer Stemness and EMT in PTC Cells
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
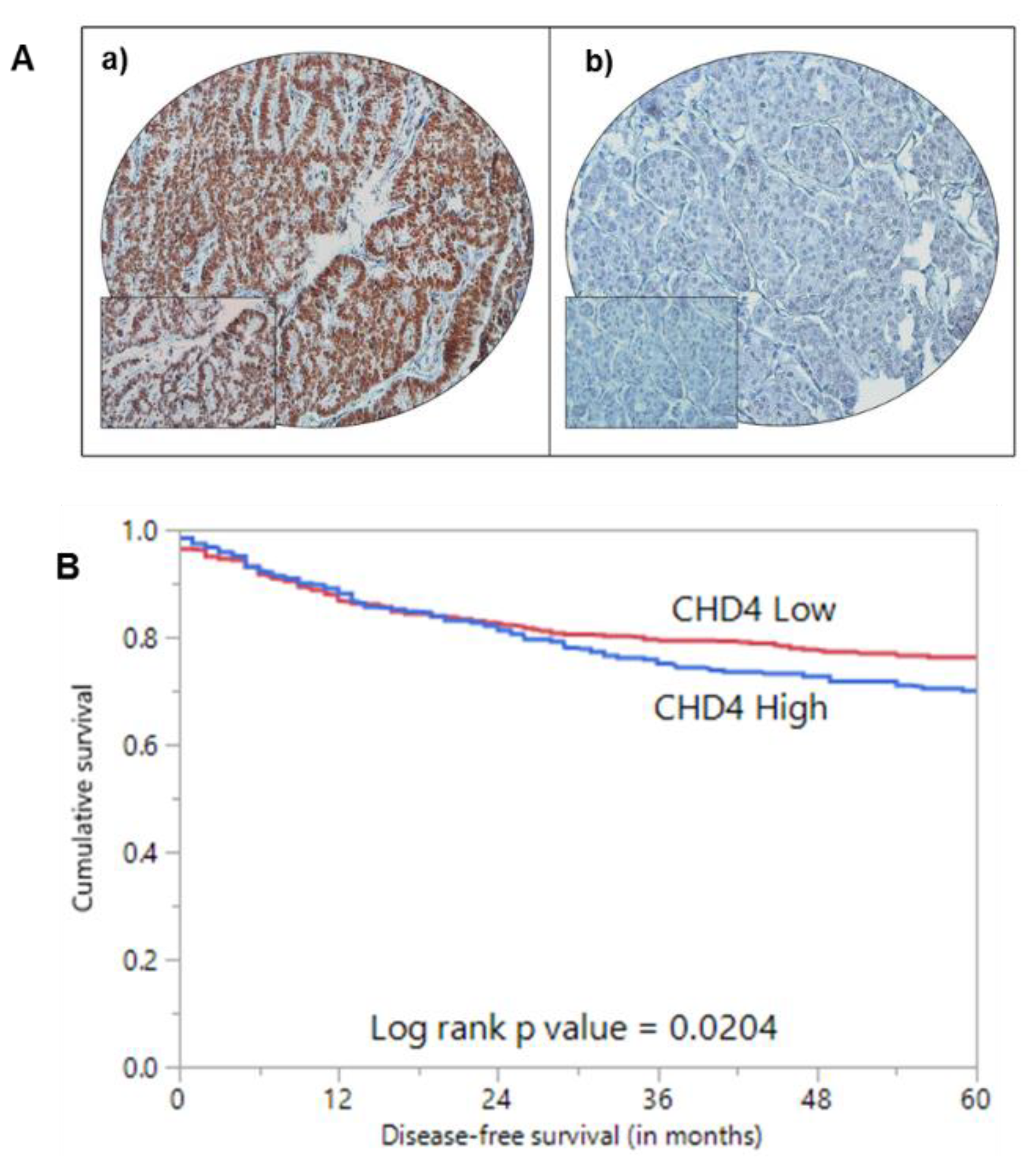
2.2. CHD4 Immunoexpression and Association with Clinico-Pathological Characteristics
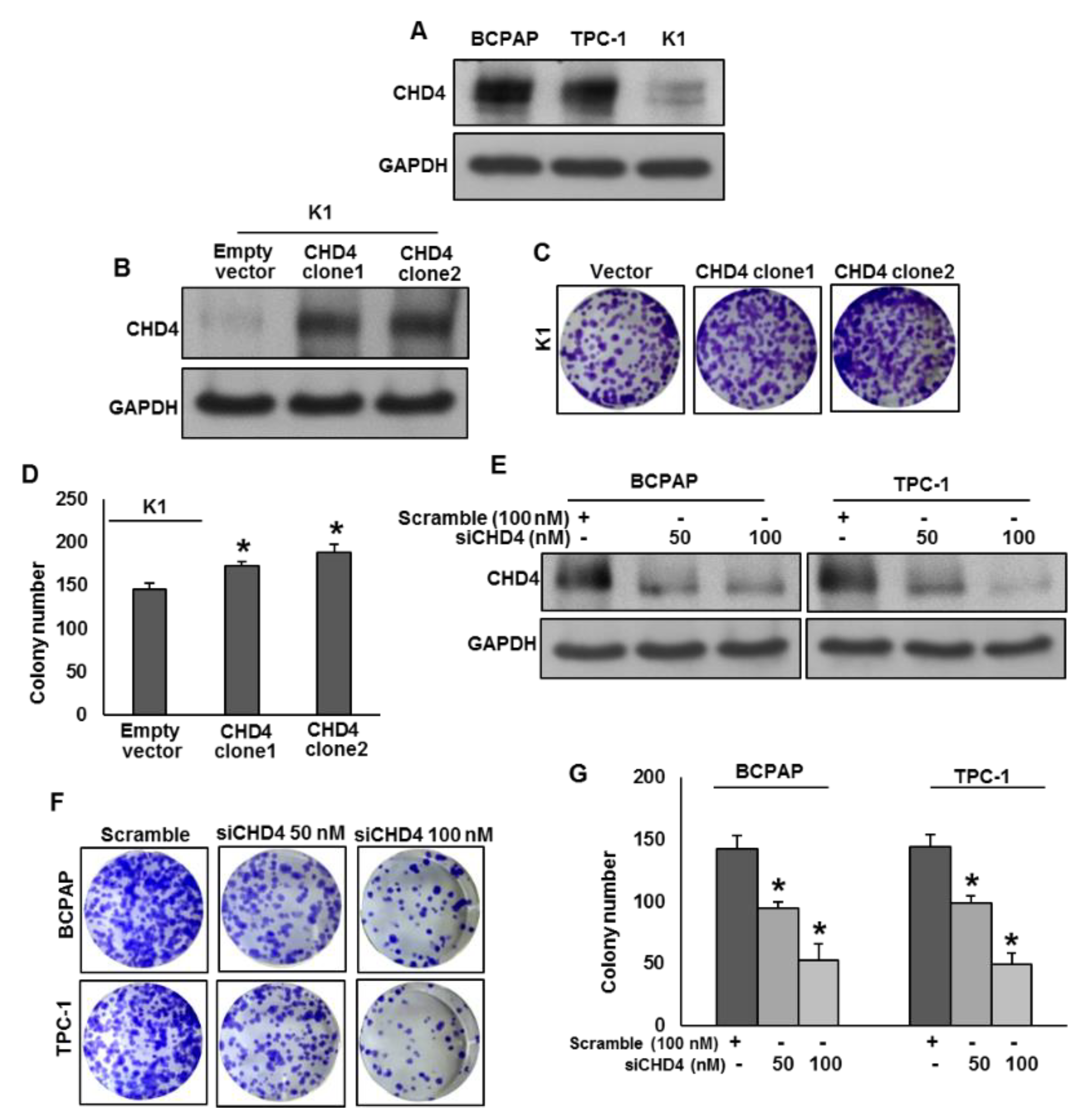
2.3. CHD4 Promotes PTC Cell Growth In Vitro
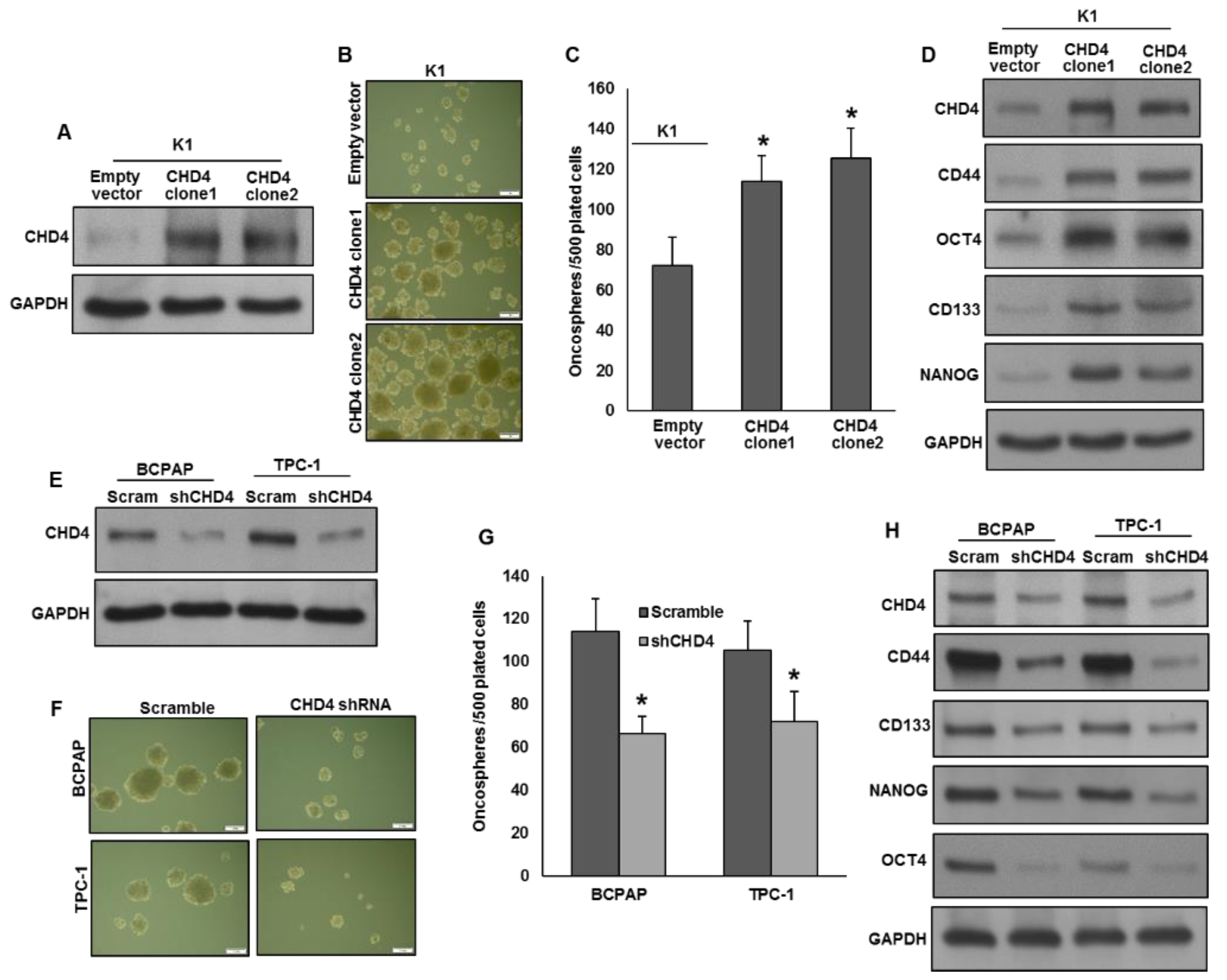
2.4. CHD4 Promotes the Self-Renewal Ability of Spheroids Generated from PTC Cells
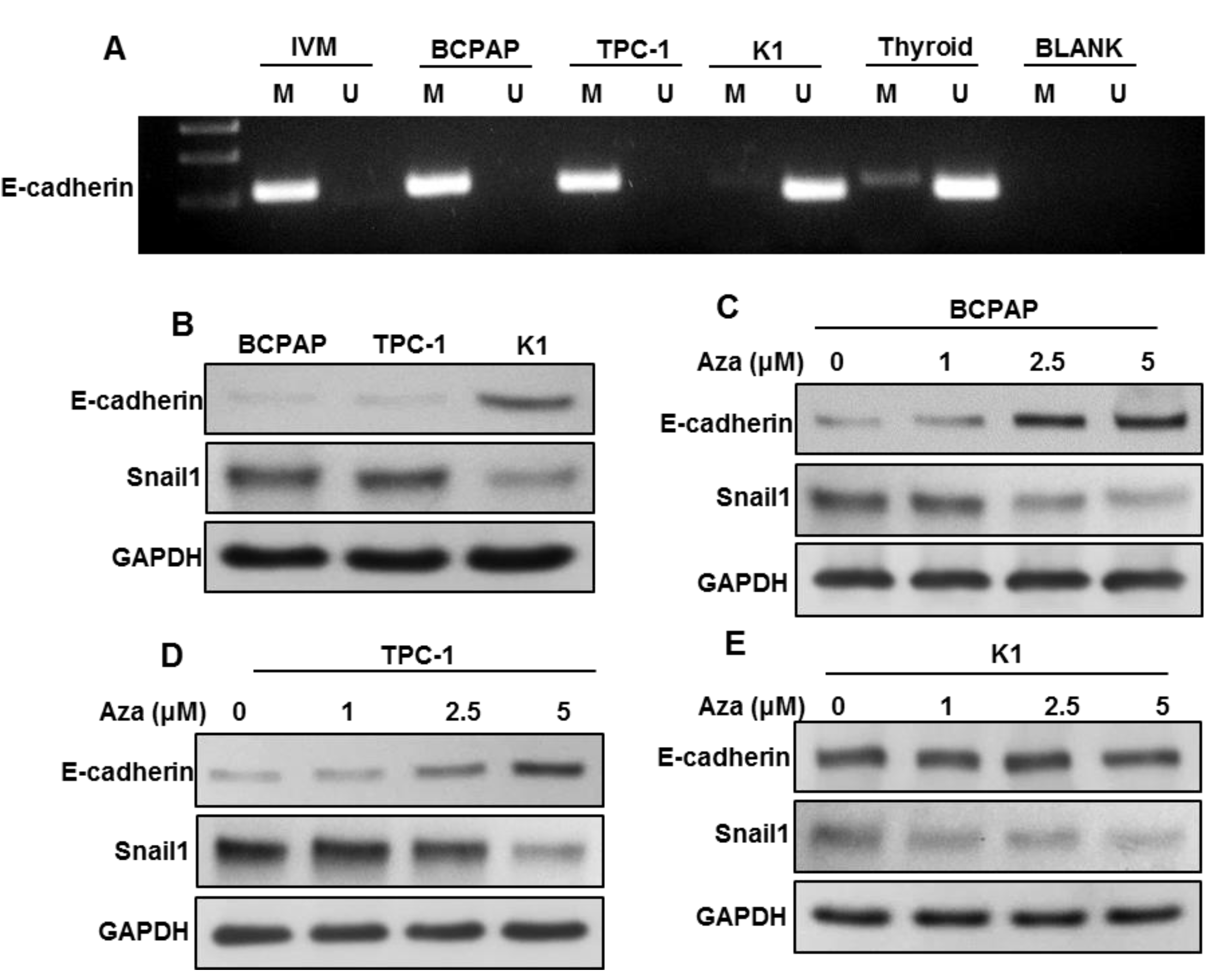
2.5. CHD4 Promotes Epithelial-to-Mesenchymal Transition in PTC Cells
2.6. DNA Methylation-Induced E-Cadherin Silencing Is Correlated with CHD4 Expression
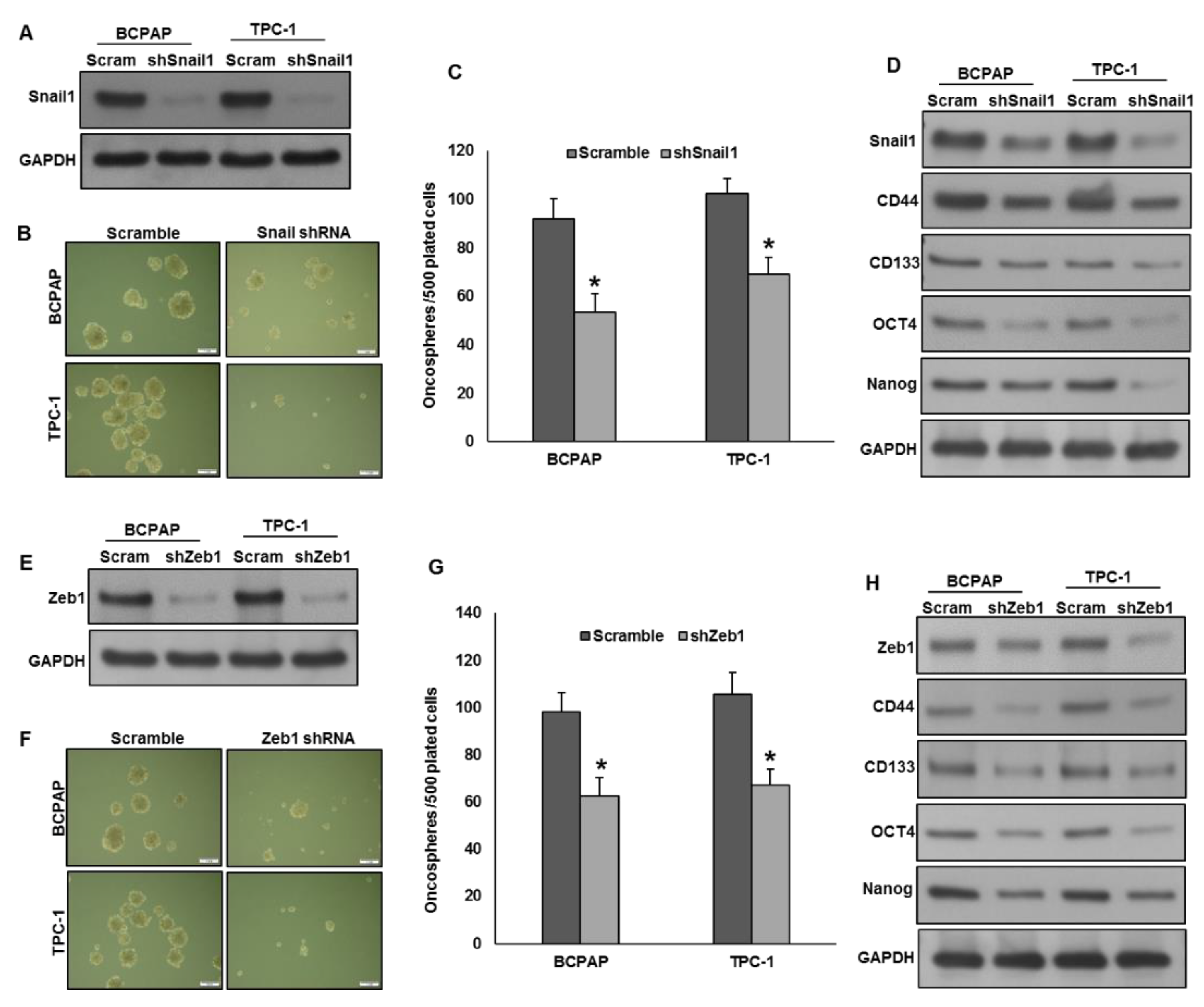
2.7. Silencing of Snail1 and Zeb1 Suppresses Self-Renewal Ability of Spheroids Generated from PTC Cells
3. Discussion
4. Materials and Methods
4.1. Patient Selection
4.2. Tissue Microarray (TMA) Construction and Immunohistochemistry
4.3. Bisulfite Modification and Methylation-Specific PCR
4.4. Cell Culture
4.5. Reagents and Antibodies
4.6. Clonogenic Assay
4.7. Gene Knockdown Using siRNA
4.8. Plasmid and Transfection
4.9. Trans-Well Cell Invasion and Migration Assays
4.10. Sphere Forming Assay
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Wiltshire, J.J.; Drake, T.M.; Uttley, L.; Balasubramanian, S.P. Systematic review of trends in the incidence rates of thyroid cancer. Thyroid 2016, 26, 1541–1552. [Google Scholar] [CrossRef]
- Powers, A.E.; Marcadis, A.R.; Lee, M.; Morris, L.G.; Marti, J.L. Changes in trends in thyroid cancer incidence in the United States, 1992 to 2016. JAMA 2019, 322, 2440–2441. [Google Scholar] [CrossRef]
- Henley, S.J.; Thomas, C.C.; Lewis, D.R.; Ward, E.M.; Islami, F.; Wu, M.; Weir, H.K.; Scott, S.; Sherman, R.L.; Ma, J. Annual report to the nation on the status of cancer, part II: Progress toward Healthy People 2020 objectives for 4 common cancers. Cancer 2020, 126, 2250–2266. [Google Scholar] [CrossRef]
- Bazarbashi, S.; Al Eid, H.; Minguet, J. Cancer incidence in Saudi Arabia: 2012 data from the Saudi cancer registry. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 2437. [Google Scholar]
- Ward, E.M.; Sherman, R.L.; Henley, S.J.; Jemal, A.; Siegel, D.A.; Feuer, E.J.; Firth, A.U.; Kohler, B.A.; Scott, S.; Ma, J. Annual report to the nation on the status of cancer, featuring cancer in men and women age 20–49 years. JNCI J. Natl. Cancer Inst. 2019, 111, 1279–1297. [Google Scholar] [CrossRef]
- Segal, R.; Miller, K.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Shi, X.; Liu, R.; Basolo, F.; Giannini, R.; Shen, X.; Teng, D.; Guan, H.; Shan, Z.; Teng, W.; Musholt, T.J. Differential clinicopathological risk and prognosis of major papillary thyroid cancer variants. J. Clin. Endocrinol. 2016, 101, 264–274. [Google Scholar] [CrossRef]
- Vigneri, R.; Malandrino, P.; Vigneri, P. The changing epidemiology of thyroid cancer: Why is incidence increasing? Curr. Opin. Oncol. 2015, 27, 1–7. [Google Scholar] [CrossRef]
- Lim, H.; Devesa, S.S.; Sosa, J.A.; Check, D.; Kitahara, C.M. Trends in thyroid cancer incidence and mortality in the United States, 1974–2013. JAMA 2017, 317, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Aliyev, E.; Ladra-González, M.J.; Sánchez-Ares, M.; Abdulkader-Nallib, I.; Piso-Neira, M.; Rodríguez-Carnero, G.; Vieiro-Balo, P.; Pérez-Becerra, R.; Gude-Sampedro, F.; Barreiro-Morandeira, F. Thyroid Papillary Microtumor: Validation of the (Updated) Porto Proposal Assessing Sex Hormone Receptor Expression and Mutational BRAF Gene Status. Am. J. Surg. Pathol. 2020, 44, 1161–1172. [Google Scholar] [CrossRef]
- Smith, S.M. PMC Versus PMT: Which Weighs More… A Pound of Rocks, or a Pound of Feathers? Am. J. Surg. Pathol. 2020. [Google Scholar] [CrossRef]
- Aliyev, E.; Ladra-González, M.J.; Sánchez-Ares, M.; Abdulkader-Nallib, I.; Piso-Neira, M.; Rodríguez-Carnero, G.; Vieiro-Balo, P.; Pérez-Becerra, R.; Gude-Sampedro, F.; Barreiro-Morandeira, F. The Authors Reply PMC and PMT: Real Medicine and Not Just Biology. Am. J. Surg. Pathol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.-R.; Yan, C.-X.; Mo, G.-Q.; Luo, Z.-Y.; Zhang, Y.; Wang, Y.; Huang, P.-T. Conventional US, elastography and contrast enhanced US features of papillary thyroid microcarcinoma predict central compartment lymph node metastases. Sci. Rep. 2015, 5, 7748. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-S.; Liang, J.; Li, J.-H.; Liu, X.; Jiang, L.; Long, J.-X.; Jiang, Y.-M.; Wei, Z.-X. The incidence and risk factors for central lymph node metastasis in cN0 papillary thyroid microcarcinoma: A meta-analysis. Eur. Arch. Otorhinolaryngol. 2017, 274, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.; Alzahrani, A.S.; Carson, K.A.; Shong, Y.K.; Kim, T.Y.; Viola, D.; Elisei, R.; Bendlová, B.; Yip, L.; Mian, C. Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J. Clin. Oncol. 2015, 33, 42. [Google Scholar] [CrossRef]
- Xing, M.; Liu, R.; Liu, X.; Murugan, A.K.; Zhu, G.; Zeiger, M.A.; Pai, S.; Bishop, J. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J. Clin. Oncol. 2014, 32, 2718. [Google Scholar] [CrossRef]
- Nixon, I.J.; Ganly, I.; Patel, S.G.; Palmer, F.L.; Di Lorenzo, M.M.; Grewal, R.K.; Larson, S.M.; Tuttle, R.M.; Shaha, A.; Shah, J.P. The results of selective use of radioactive iodine on survival and on recurrence in the management of papillary thyroid cancer, based on Memorial Sloan-Kettering Cancer Center risk group stratification. Thyroid 2013, 23, 683–694. [Google Scholar] [CrossRef]
- Ritter, A.; Mizrachi, A.; Bachar, G.; Vainer, I.; Shimon, I.; Hirsch, D.; Diker-Cohen, T.; Duskin-Bitan, H.; Robenshtok, E. Detecting recurrence following lobectomy for thyroid cancer: Role of thyroglobulin and thyroglobulin antibodies. J. Clin. Endocrinol. Metab. 2020, 105, dgaa152. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.A.; Mahajan, P.; Mertens, H.D.; Deery, M.J.; Zhang, W.; Pham, P.; Du, X.; Bartke, T.; Zhang, W.; Edlich, C. The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4. J. Mol. Biol. 2012, 422, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.Y.; Wade, P.A. Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer 2011, 11, 588. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, L.D.; LeClair, J.W.; Miller, K.N.; Strong, A.D.; Chan, H.L.; Oates, E.L.; Ligon, K.L.; Brennan, C.W.; Chheda, M.G. CHD4 regulates the DNA damage response and RAD51 expression in glioblastoma. Sci. Rep. 2019, 9, 4444. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.-W.; Wu, C.-C.; Chang, S.-J.; Chang, T.-M.; Chen, T.-Y.; Chai, C.-Y.; Chang, C.-L.; Hou, M.-F.; Pan, M.-R. CHD4-mediated loss of E-cadherin determines metastatic ability in triple-negative breast cancer cells. Exp. Cell Res. 2018, 363, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Huang, W.; Bellani, M.; Seidman, M.M.; Wu, K.; Fan, D.; Nie, Y.; Cai, Y.; Zhang, Y.W.; Yu, L.-R. CHD4 has oncogenic functions in initiating and maintaining epigenetic suppression of multiple tumor suppressor genes. Cancer Cell 2017, 31, 653–668.e657. [Google Scholar] [CrossRef]
- Xu, N.; Liu, F.; Wu, S.; Ye, M.; Ge, H.; Zhang, M.; Song, Y.; Tong, L.; Zhou, J.; Bai, C. CHD4 mediates proliferation and migration of non-small cell lung cancer via the RhoA/ROCK pathway by regulating PHF5A. BMC Cancer 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Xu, N.; Liu, F.; Zhou, J.; Bai, C. CHD4 is associated with poor prognosis of non-small cell lung cancer patients through promoting tumor cell proliferation. Eur. Respiratory Soc. 2016, 48, PA2862. [Google Scholar] [CrossRef]
- Wang, H.-C.; Chou, C.-L.; Yang, C.-C.; Huang, W.-L.; Hsu, Y.-C.; Luo, C.-W.; Chen, T.-J.; Li, C.-F.; Pan, M.-R. Over-expression of chd4 is an independent biomarker of poor prognosis in patients with rectal cancers receiving concurrent chemoradiotherapy. Int. J. Mol. Sci. 2019, 20, 4087. [Google Scholar] [CrossRef]
- D’Alesio, C.; Punzi, S.; Cicalese, A.; Fornasari, L.; Furia, L.; Riva, L.; Carugo, A.; Curigliano, G.; Criscitiello, C.; Pruneri, G. RNAi screens identify CHD4 as an essential gene in breast cancer growth. Oncotarget 2016, 7, 80901. [Google Scholar] [CrossRef]
- Nio, K.; Yamashita, T.; Okada, H.; Kondo, M.; Hayashi, T.; Hara, Y.; Nomura, Y.; Zeng, S.S.; Yoshida, M.; Hayashi, T. Defeating EpCAM+ liver cancer stem cells by targeting chromatin remodeling enzyme CHD4 in human hepatocellular carcinoma. J. Hepatol. 2015, 63, 1164–1172. [Google Scholar] [CrossRef]
- Chudnovsky, Y.; Kim, D.; Zheng, S.; Whyte, W.A.; Bansal, M.; Bray, M.-A.; Gopal, S.; Theisen, M.A.; Bilodeau, S.; Thiru, P. ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor-initiating cell state. Cell Rep. 2014, 6, 313–324. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Q.; McGrail, D.J.; Dai, H.; Li, K.; Lin, S.-Y. CHD4 mutations promote endometrial cancer stemness by activating TGF-beta signaling. Am. J. Cancer Res. 2018, 8, 903. [Google Scholar]
- Chanmee, T.; Ontong, P.; Mochizuki, N.; Kongtawelert, P.; Konno, K.; Itano, N. Excessive Hyaluronan Production Promotes Acquisition of Cancer Stem Cell Signatures Through the Coordinated Regulation of Twist and the TGF-β-Snail Signaling Axis. J. Biol. Chem. 2014, 289, 26038–26056. [Google Scholar] [CrossRef]
- Naber, H.P.; Drabsch, Y.; Snaar-Jagalska, B.E.; ten Dijke, P.; van Laar, T. Snail and Slug, key regulators of TGF-β-induced EMT, are sufficient for the induction of single-cell invasion. Biochem. Biophys. Res. Commun. 2013, 435, 58–63. [Google Scholar] [CrossRef]
- Kim, M.S.; Chung, N.G.; Kang, M.R.; Yoo, N.J.; Lee, S.H. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology 2011, 58, 660–668. [Google Scholar] [CrossRef]
- O’Shaughnessy, A.; Hendrich, B. CHD4 in the DNA-damage response and cell cycle progression: Not so NuRDy now. Biochem. Soc. Trans. 2013, 41, 777–782. [Google Scholar] [CrossRef]
- Sperlazza, J.; Rahmani, M.; Beckta, J.; Aust, M.; Hawkins, E.; Wang, S.Z.; Zu Zhu, S.; Podder, S.; Dumur, C.; Archer, K. Depletion of the chromatin remodeler CHD4 sensitizes AML blasts to genotoxic agents and reduces tumor formation. Blood J. Am. Soc. Hematol. 2015, 126, 1462–1472. [Google Scholar] [CrossRef]
- Loh, C.-Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: Signaling, therapeutic implications, and challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef]
- Lu, W.; Kang, Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell 2019, 49, 361–374. [Google Scholar] [CrossRef]
- Dong, C.; Yuan, T.; Wu, Y.; Wang, Y.; Fan, T.W.; Miriyala, S.; Lin, Y.; Yao, J.; Shi, J.; Kang, T. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 2013, 23, 316–331. [Google Scholar] [CrossRef]
- Bravo, S.B.; Pampín, S.; Cameselle-Teijeiro, J.; Carneiro, C.; Domínguez, F.; Barreiro, F.; Alvarez, C.V. TGF-β-induced apoptosis in human thyrocytes is mediated by p27 kip1 reduction and is overridden in neoplastic thyrocytes by NF-κB activation. Oncogene 2003, 22, 7819–7830. [Google Scholar] [CrossRef][Green Version]
- Garcia-Rendueles, A.; Rodrigues, J.; Garcia-Rendueles, M.; Suarez-Fariña, M.; Perez-Romero, S.; Barreiro, F.; Bernabeu, I.; Rodriguez-Garcia, J.; Fugazzola, L.; Sakai, T. Rewiring of the apoptotic TGF-β-SMAD/NFκB pathway through an oncogenic function of p27 in human papillary thyroid cancer. Oncogene 2017, 36, 652–666. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Geutjes, E.-J.; De Lint, K.; Roepman, P.; Bruurs, L.; Yu, L.; Wang, W.; Van Blijswijk, J.; Mohammad, H.; De Rink, I. The NuRD complex cooperates with DNMTs to maintain silencing of key colorectal tumor suppressor genes. Oncogene 2014, 33, 2157. [Google Scholar] [CrossRef] [PubMed]
- Bavi, P.; Jehan, Z.; Atizado, V.; Al-Dossari, H.; Al-Dayel, F.; Tulbah, A.; Amr, S.S.; Sheikh, S.S.; Ezzat, A.; El-Solh, H. Prevalence of fragile histidine triad expression in tumors from Saudi Arabia: A tissue microarray analysis. Cancer Epidemiol. Prev. Biomark. 2006, 15, 1708–1718. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Uddin, S.; Siraj, A.K.; Al-Rasheed, M.; Ahmed, M.; Bu, R.; Myers, J.N.; Al-Nuaim, A.; Al-Sobhi, S.; Al-Dayel, F.; Bavi, P. Fatty acid synthase and AKT pathway signaling in a subset of papillary thyroid cancers. J. Clin. Endocrinol. Metab. 2008, 93, 4088–4097. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, T.; Suzuki, J.; Wang, Y.V.; Menendez, S.; Morera, L.B.; Raya, A.; Wahl, G.M.; Izpisua Belmonte, J.C. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2009, 460, 1140–1144. [Google Scholar] [CrossRef]
- Zhao, M.; Sano, D.; Pickering, C.R.; Jasser, S.A.; Henderson, Y.C.; Clayman, G.L.; Sturgis, E.M.; Ow, T.J.; Lotan, R.; Carey, T.E. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin. Cancer Res. 2011, 17, 7248–7264. [Google Scholar] [CrossRef]
- Bu, R.; Siraj, A.K.; Divya, S.P.; Kong, Y.; Parvathareddy, S.K.; Al-Rasheed, M.; Al-Obaisi, K.A.; Victoria, I.G.; Al-Sobhi, S.S.; Al-Dawish, M. Telomerase reverse transcriptase mutations are independent predictor of disease-free survival in Middle Eastern papillary thyroid cancer. Int. J. Cancer 2018, 142, 2028–2039. [Google Scholar] [CrossRef]

| Clinico-Pathological Variables | n (%) |
|---|---|
| Age | |
| Median | 38.0 |
| Range (IQR) ^ | 29.0–50.1 |
| Gender | |
| Female | 1090 (75.9) |
| Male | 346 (24.1) |
| Histopathology | |
| Classical Variant | 936 (65.2) |
| Follicular Variant | 256 (17.8) |
| Tall Cell Variant | 131 (9.1) |
| Hobnail Variant | 1 (0.1) |
| Others | 112 (7.8) |
| Extra Thyroidal Extension | |
| Present | 619 (43.1) |
| Absent | 817 (56.9) |
| Tumor size | |
| ≤1 cm | 137 (9.5) |
| >1 cm | 1246 (86.8) |
| Unknown | 53 (3.7) |
| pT | |
| T1 | 396 (27.6) |
| T2 | 289 (20.1) |
| T3 | 588 (40.9) |
| T4 | 110 (7.7) |
| Unknown | 53 (3.7) |
| pN | |
| N0 | 582 (40.6) |
| N1 | 710 (49.4) |
| Nx | 144 (10.0) |
| pM | |
| M0 | 1291 (89.9) |
| M1 | 55 (3.8) |
| Mx | 90 (6.3) |
| Stage | |
| I | 1169 (81.4) |
| II | 156 (10.9) |
| III | 20 (1.4) |
| IVA | 19 (1.3) |
| IVB | 28 (1.9) |
| Unknown | 44 (3.1) |
| Total | CHD4 Over-Expression | CHD4 Low Expression | p Value | |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | ||
| No. of patients | 1436 | 650 (45.3) | 786 (54.7) | |
| Age (Yrs) | ||||
| <55 | 1169 (81.4) | 524 (44.8) | 645 (55.2) | 0.4837 |
| ≥55 | 267 (18.6) | 126 (47.2) | 141 (52.8) | |
| Sex | ||||
| Female | 1090 (75.9) | 476 (43.7) | 614 (56.3) | 0.0314 |
| Male | 346 (24.1) | 174 (50.3) | 172 (49.7) | |
| Histology Type | ||||
| Classical Variant | 936 (65.2) | 437 (46.7) | 499 (53.3) | <0.0001 |
| Follicular Variant | 256 (17.8) | 88 (34.4) | 168 (65.6) | |
| Tall-Cell Variant | 131 (9.1) | 78 (59.5) | 53 (40.5) | |
| Other variants | 113 (7.9) | 47 (41.6) | 66 (58.4) | |
| Extrathyroidal extension | ||||
| Present | 619 (43.1) | 297 (48.0) | 322 (52.0) | 0.0720 |
| Absent | 817 (56.9) | 353 (43.2) | 464 (56.8) | |
| Tumor size | ||||
| ≤1 cm | 137 (9.9) | 57 (41.6) | 80 (58.4) | 0.3451 |
| >1 cm | 1246 (90.1) | 571 (45.8) | 675 (54.2) | |
| pT | ||||
| pT1 | 396 (28.6) | 175 (44.2) | 221 (55.8) | 0.2923 |
| pT2 | 289 (20.9) | 122 (42.2) | 167 (57.8) | |
| pT3 | 588 (42.5) | 284 (48.3) | 304 (51.7) | |
| pT4 | 110 (8.0) | 47 (42.7) | 63 (57.3) | |
| pN | ||||
| pN0 | 582 (45.1) | 237 (40.7) | 345 (59.3) | 0.0085 |
| pN1 | 710 (54.9) | 341 (48.0) | 369 (52.0) | |
| pM | ||||
| pM0 | 1291 (95.9) | 575 (44.5) | 716 (55.5) | 0.3498 |
| pM1 | 55 (4.1) | 21 (38.2) | 34 (61.8) | |
| Stage | ||||
| I | 1169 (84.0) | 524 (44.8) | 645 (55.2) | 0.4698 |
| II | 156 (11.2) | 74 (47.4) | 82 (52.6) | |
| III | 20 (1.4) | 11 (55.0) | 9 (45.0) | |
| IVA | 19 (1.4) | 10 (52.6) | 9 (47.4) | |
| IVB | 28 (2.0) | 9 (32.1) | 19 (67.9) | |
| BRAFmutation | ||||
| Positive | 704 (56.5) | 407 (57.8) | 297 (42.2) | <0.0001 |
| Negative | 543 (43.5) | 167 (30.8) | 376 (69.2) | |
| Disease Free Survival | ||||
| 5 years | 70.0 | 76.2 | 0.0204 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pratheeshkumar, P.; Siraj, A.K.; Divya, S.P.; Parvathareddy, S.K.; Alobaisi, K.; Al-Sobhi, S.S.; Al-Dayel, F.; Al-Kuraya, K.S. CHD4 Predicts Aggressiveness in PTC Patients and Promotes Cancer Stemness and EMT in PTC Cells. Int. J. Mol. Sci. 2021, 22, 504. https://doi.org/10.3390/ijms22020504
Pratheeshkumar P, Siraj AK, Divya SP, Parvathareddy SK, Alobaisi K, Al-Sobhi SS, Al-Dayel F, Al-Kuraya KS. CHD4 Predicts Aggressiveness in PTC Patients and Promotes Cancer Stemness and EMT in PTC Cells. International Journal of Molecular Sciences. 2021; 22(2):504. https://doi.org/10.3390/ijms22020504
Chicago/Turabian StylePratheeshkumar, Poyil, Abdul K. Siraj, Sasidharan Padmaja Divya, Sandeep Kumar Parvathareddy, Khadija Alobaisi, Saif S. Al-Sobhi, Fouad Al-Dayel, and Khawla S. Al-Kuraya. 2021. "CHD4 Predicts Aggressiveness in PTC Patients and Promotes Cancer Stemness and EMT in PTC Cells" International Journal of Molecular Sciences 22, no. 2: 504. https://doi.org/10.3390/ijms22020504
APA StylePratheeshkumar, P., Siraj, A. K., Divya, S. P., Parvathareddy, S. K., Alobaisi, K., Al-Sobhi, S. S., Al-Dayel, F., & Al-Kuraya, K. S. (2021). CHD4 Predicts Aggressiveness in PTC Patients and Promotes Cancer Stemness and EMT in PTC Cells. International Journal of Molecular Sciences, 22(2), 504. https://doi.org/10.3390/ijms22020504





