Profiling and Functional Analysis of Long Noncoding RNAs and mRNAs during Porcine Skeletal Muscle Development
Abstract
1. Introduction
2. Materials and Methods
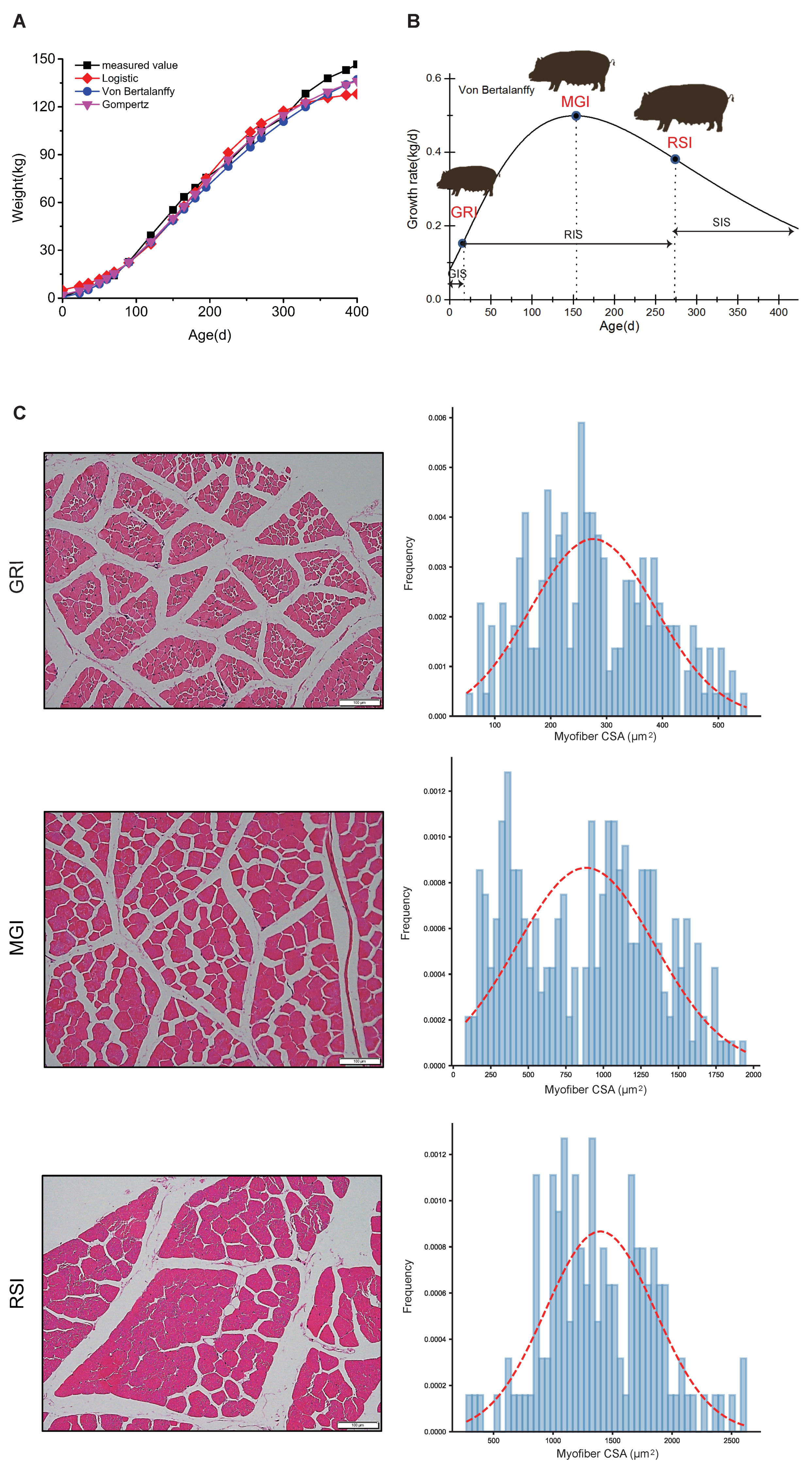
2.1. Growth Curve Model
2.2. Immunohistochemical Staining
2.3. Sample Collection and RNA Sequencing
2.4. Identification of lncRNAs
2.5. Differential Expression Analysis
2.6. Functional Enrichment Analysis
2.7. 5′ and 3′ Rapid Amplification of cDNA Ends (RACE)
2.8. Subcellular Localization
2.9. Cell Culture, Vector Construction, and Dual Luciferase Reporter Assay
2.10. Quantitative Real-Time PCR
2.11. Statistical Analysis
3. Results
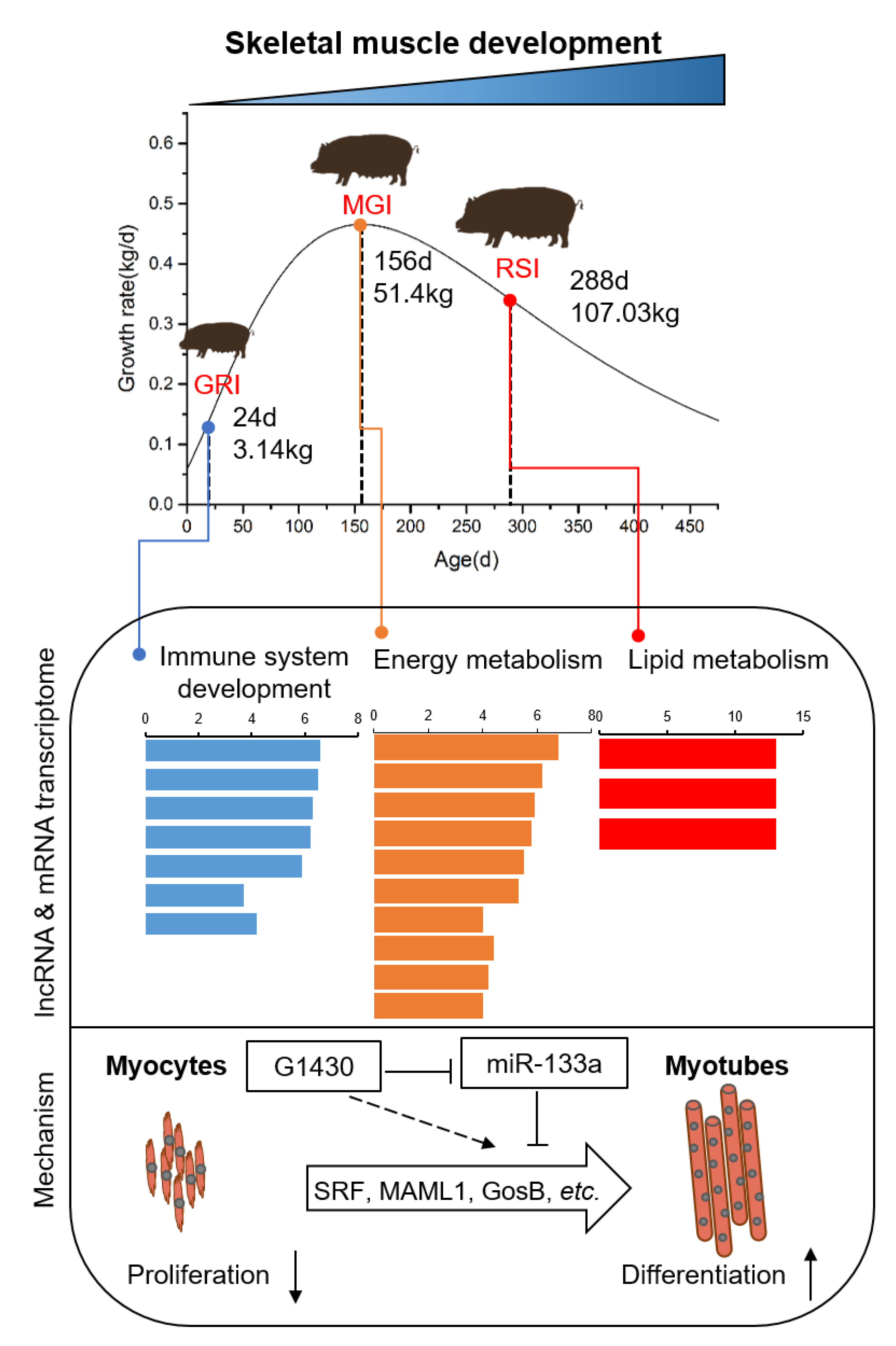
3.1. Growth Curves and Histological Analyses of Qingyu Pigs
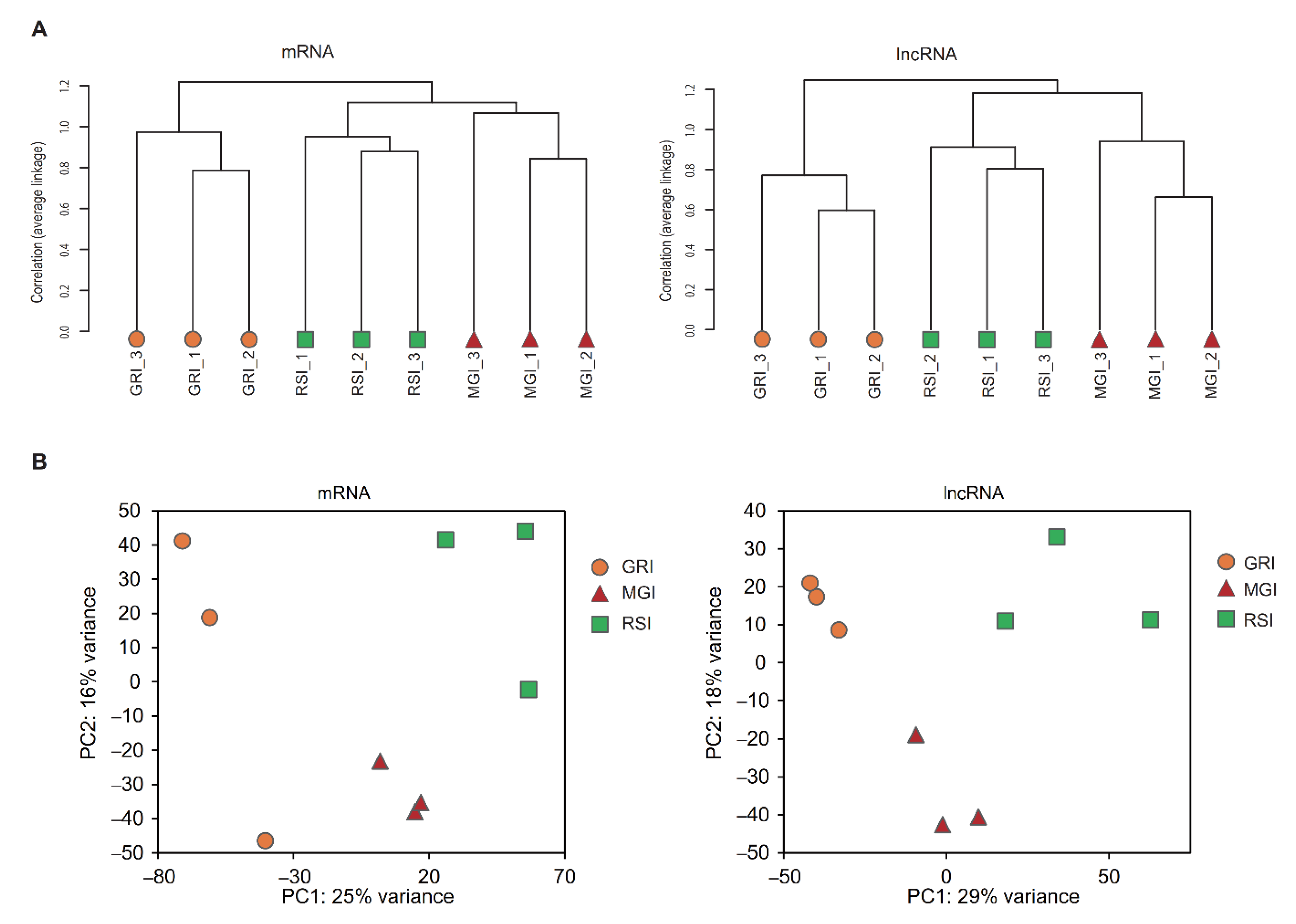
3.2. Expression Pattern of mRNAs and lncRNAs
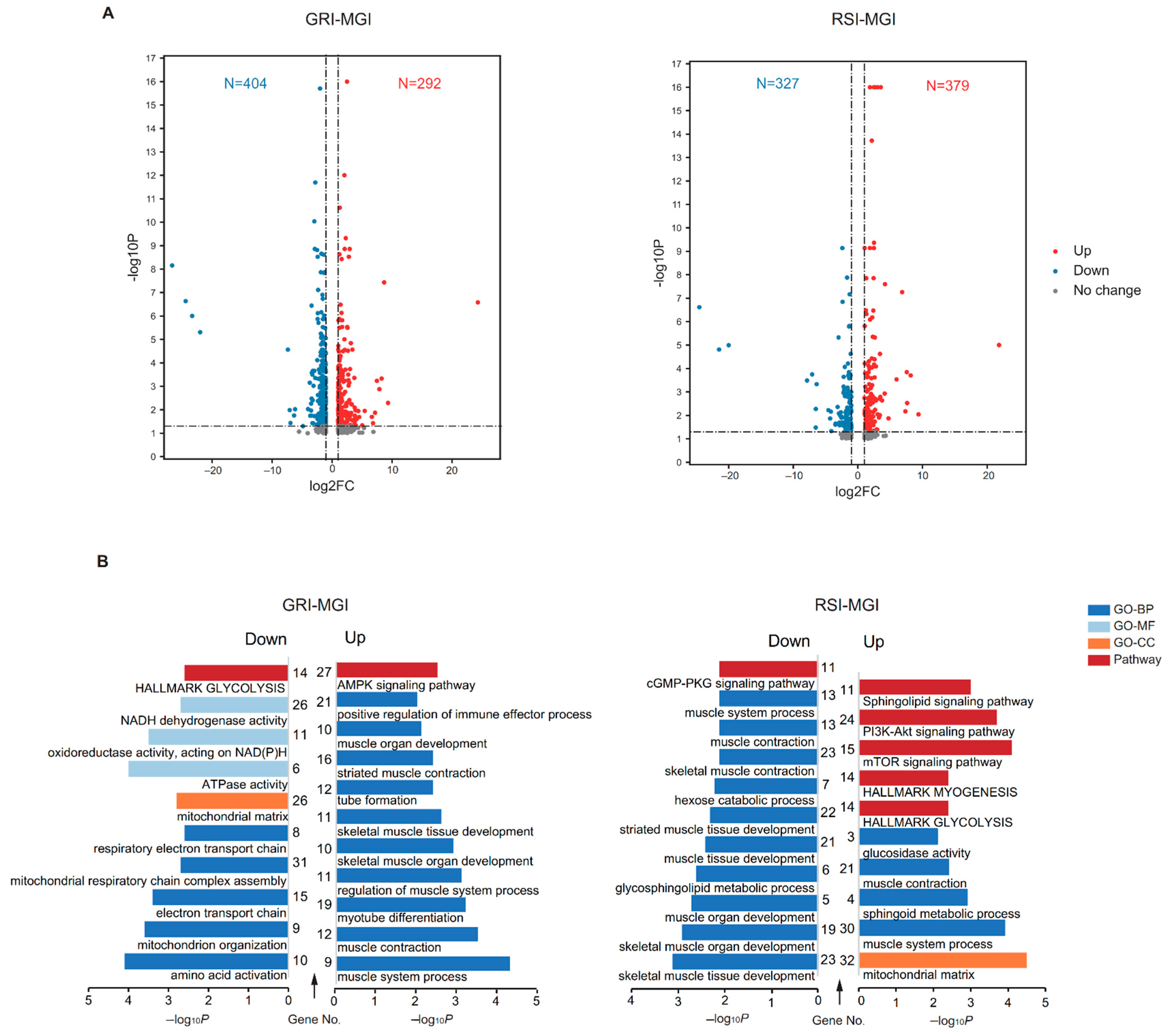
3.3. Functional Enrichment Analysis of Differentially Expressed mRNAs (DEGs)
3.4. Functional Enrichment Analysis of Differentially Expressed lncRNAs (DELs)
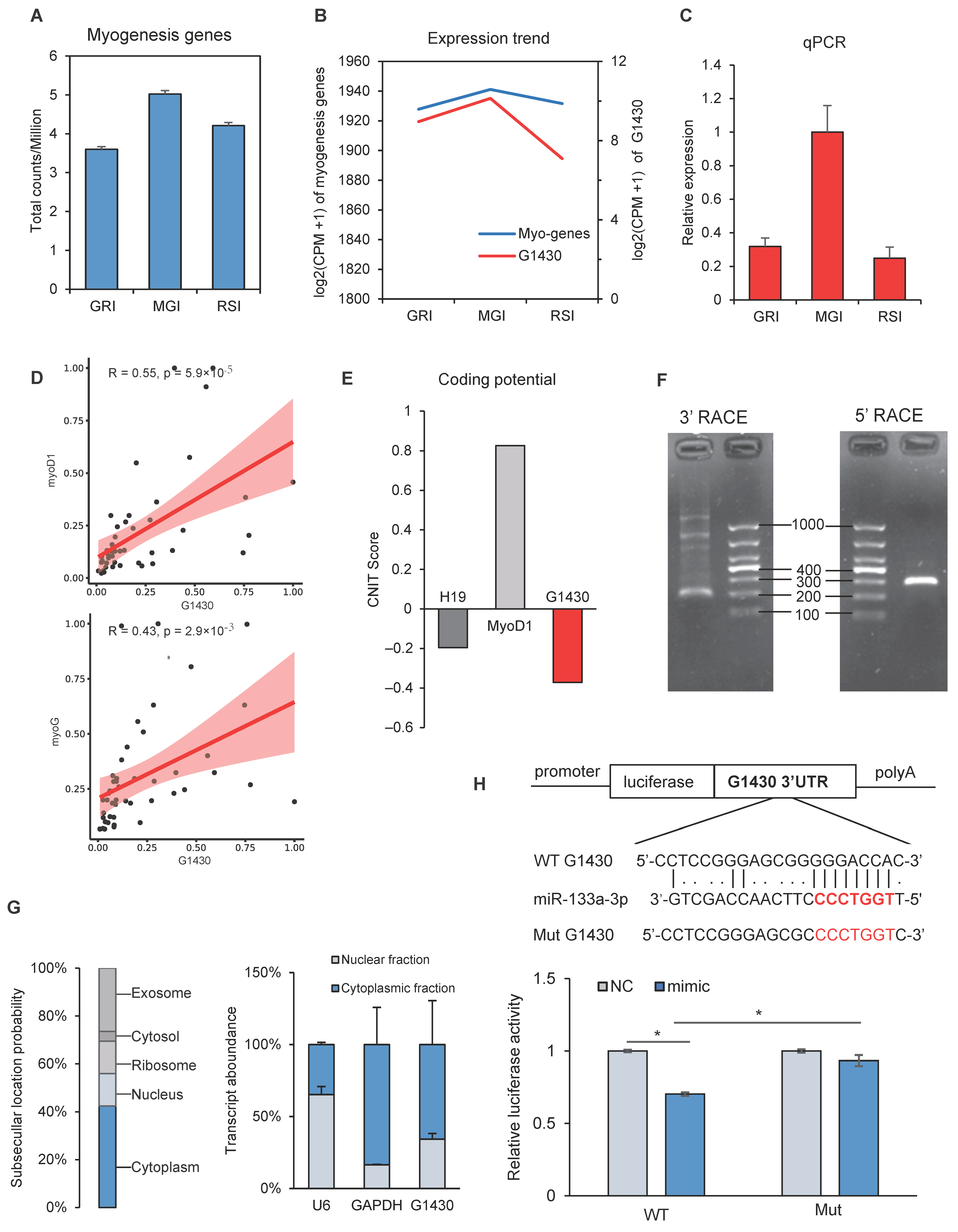
3.5. Dynamic Expression of Myogenesis Genes and lncRNAs
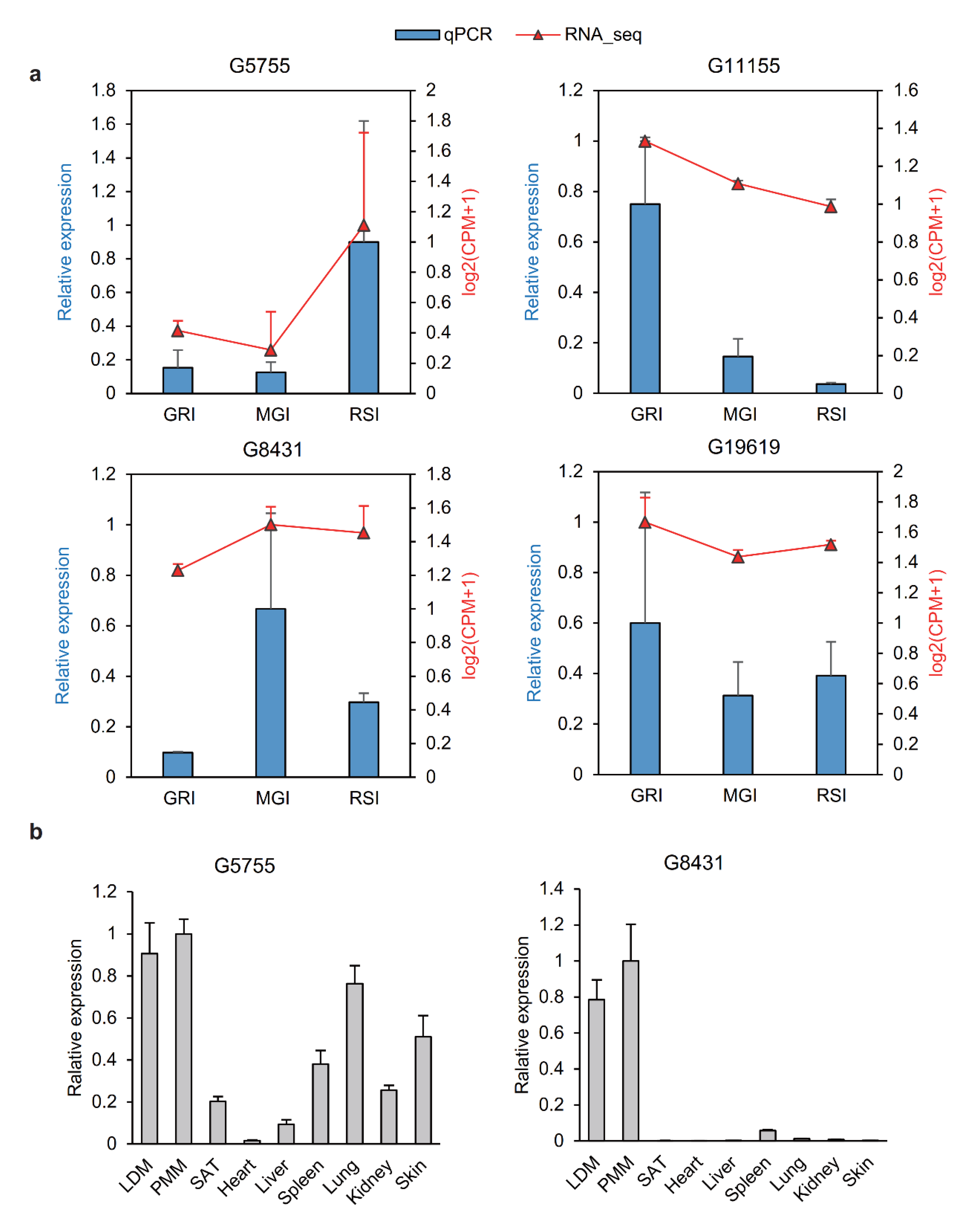
3.6. Validation of lncRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BW | Body Weight |
| MGI | The Inflection Point With The Maximum Growth Rate |
| GRI | The Inflection Point Of The Gradually Increasing Stage To The Rapidly Increasing Stage |
| RSI | The Inflection Point Of The Rapidly Increasing Stage To The Slowly Increasing Stage |
| DEGs | Differentially Expressed Genes |
| DELs | Differentially Expressed Lncrnas |
| RACE | Rapid Amplification Of Cdna Ends |
| HE | Hematoxylin-Eosin |
| GIS | Gradually Increasing Stage |
| RIS | Rapidly Increasing Stage |
| SIS | Slowly Increasing Stage |
| DMEM | Dulbecco’s Modified Eagle’S Medium |
| CPM | Counts Per Million |
| MSigDB | Molecular Signatures Database |
| CSA | Cross-Section Area |
| GO | Gene Ontology |
| BP | Biological Process |
| CC | Cellular Component |
| MF | Molecular Function |
| HSD | Honestly Significant Difference |
| qRT-PCR | Quantitative Real-Time Pcr |
| APOD | Apolipoprotein D |
| TNNT2 | Troponin T2 |
| MYBPH | Myosin Binding Protein H |
| MYL3 | Myosin Light Chain 3 |
| DAPK2 | Death Associated Protein Kinase 2 |
| RIT1 | Ras Like Without Caax 1 |
| TEAD4 | Tea Domain Transcription Factor 4 |
| OCEL1 | Occludin/Ell Domain Containing 1 |
| AKT2 | Akt Serine/Threonine Kinase 2 |
| APLNR | Apelin Receptor |
| myoD1 | Myogenic Differentiation 1 |
| myoG | Myogenin |
| DLEU1 | Lncrna Deleted In Lymphocytic Leukemia 1 |
| XIST | X-Inactive Specific Transcript |
| MDNCR | Muscle Differentiation-Associated Lncrna |
| MD1 | Muscle Differentiation 1 |
| MEF2C | Myocyte Enhancer Factor 2c |
| MAML1 | Mastermind Like Transcriptional Coactivator 1 |
References
- La Berge, A.F. How the Ideology of Low Fat Conquered America. J. Hist. Med. Allied Sci. 2008, 63, 139–177. [Google Scholar] [CrossRef] [PubMed]
- Wernersson, R.; Schierup, M.H.; Jørgensen, F.G.; Gorodkin, J.; Panitz, F.; Staerfeldt, H.-H.; Christensen, O.F.; Mailund, T.; Hornshøj, H.; Klein, A.; et al. Pigs in sequence space: A 0.66X coverage pig genome survey based on shotgun sequencing. BMC Genom. 2005, 6, 70. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Rettenmeier, A.W.; Schmitz-Spanke, S. Recent advances in the use of Sus scrofa (pig) as a model system for proteomic studies. Proteomics 2011, 11, 776–793. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Gao, H.; Zhang, X.; Zhang, Y.; Zhao, Y. Effect of Sowing Date on Grain Filling Characters in Different Wheat Varieties. Acta Tritical Crops 2004, 24, 105–107. [Google Scholar]
- Luo, J.; Lei, H.; Shen, L.; Yang, R.; Pu, Q.; Zhu, K.; Li, M.; Tang, G.; Li, X.; Zhang, S.; et al. Estimation of Growth Curves and Suitable Slaughter Weight of the Liangshan Pig. Asian Australas. J. Anim. Sci. 2015, 28, 1252–1258. [Google Scholar] [CrossRef]
- Yang, X.R.; Yu, B.; Mao, X.B.; Zheng, P.; He, J.; Yu, J.; He, Y.; Reecy, J.M.; Chen, D.W. Lean and obese pig breeds exhibit differences in prenatal gene expression profiles of muscle development. Animal 2015, 9, 28–34. [Google Scholar] [CrossRef]
- Zou, C.; Li, J.; Luo, W.; Li, L.; Hu, A.; Fu, Y.; Hou, Y.; Li, C. Transcriptome analysis reveals long intergenic non-coding RNAs involved in skeletal muscle growth and development in pig. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Wu, G.; Ma, L.; Wang, L.; Zhou, J.; Ma, Y.; Yang, C. Analysis of Transcriptome and miRNAome in the Muscle of Bamei Pigs at Different Developmental Stages. Animal 2020, 10, 1198. [Google Scholar] [CrossRef]
- Qin, L.; Chen, Y.; Liu, X.; Ye, S.; Yu, K.; Huang, Z.; Yu, J.; Zhou, X.; Chen, H.; Mo, D. Integrative Analysis of Porcine microRNAome during Skeletal Muscle Development. PLoS ONE 2013, 8, e72418. [Google Scholar] [CrossRef]
- Yu, X.; Wang, Z.; Sun, H.; Yang, Y.; Li, K.; Tang, Z. Long non-coding MEG3 is a marker for skeletal muscle development and meat production traits in pigs. Anim. Genet. 2018, 49, 571–578. [Google Scholar] [CrossRef]
- Li, L.; Cheng, X.; Chen, L.; Li, J.; Luo, W.; Li, C. Long Noncoding Ribonucleic Acid MSTRG.59589 Promotes Porcine Skeletal Muscle Satellite Cells Differentiation by Enhancing the Function of PALLD. Front. Genet. 2019, 10, 1220. [Google Scholar] [CrossRef] [PubMed]
- Hitachi, K.; Nakatani, M.; Funasaki, S.; Hijikata, I.; Maekawa, M.; Honda, M.; Tsuchida, K. Expression Levels of Long Non-Coding RNAs Change in Models of Altered Muscle Activity and Muscle Mass. Int. J. Mol. Sci. 2020, 21, 1628. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Mu, Y.; Ma, L.; Wang, C.; Tang, Z.; Yang, S.; Zhou, R.; Meng-Hua, L.; Li, M.-H.; Li, K. Systematic identification and characterization of long intergenic non-coding RNAs in fetal porcine skeletal muscle development. Sci. Rep. 2015, 5, 8957. [Google Scholar] [CrossRef] [PubMed]
- Nesetrilova, H. Multiphasic growth models for cattle. Czech J. Anim. Sci. 2005, 50, 347–354. [Google Scholar] [CrossRef]
- Kingsley, M.C.S. Fitting the von Bertalanffy growth equation to polar bear age–weight data. Can. J. Zool. 1979, 57, 1020–1025. [Google Scholar] [CrossRef]
- Mandal, A.; Huang, W.; Bhandari, S.; Basu, A. Goodness-of-fit testing in growth curve models: A general approach based on finite differences. Comput. Stat. Data Anal. 2011, 55, 1086–1098. [Google Scholar] [CrossRef]
- Shen, L.; Luo, J.; Du, J.; Liu, C.; Wu, X.; Pu, Q.; Fu, Y.; Tang, Q.; Liu, Y.; Li, Q.; et al. Transcriptome Analysis of Liangshan Pig Muscle Development at the Growth Curve Inflection Point and Asymptotic Stages Using Digital Gene Expression Profiling. PLoS ONE 2015, 10, e0135978. [Google Scholar] [CrossRef]
- Mehl, K.A.; Davis, J.M.; Berger, F.G.; Carson, J.A. Myofiber degeneration/regeneration is induced in the cachectic ApcMin/+ mouse. J. Appl. Physiol. 2005, 99, 2379–2387. [Google Scholar] [CrossRef]
- Méndez, J.; Stillman, B. Chromatin Association of Human Origin Recognition Complex, Cdc6, and Minichromosome Maintenance Proteins during the Cell Cycle: Assembly of Prereplication Complexes in Late Mitosis. Mol. Cell. Biol. 2000, 20, 8602–8612. [Google Scholar] [CrossRef]
- Iida, M.; Brand, T.M.; A Campbell, D.; Li, C.; Wheeler, D.L. Yes and Lyn play a role in nuclear translocation of the epidermal growth factor receptor. Oncogene 2012, 32, 759–767. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, X.; Qin, J.; Liu, S.; Zhao, R.; Yu, T.; Chu, G.; Yang, G.S.; Pang, W.; Wei-Jun, P. Comparative Analysis of Long Noncoding RNAs Expressed during Intramuscular Adipocytes Adipogenesis in Fat-Type and Lean-Type Pigs. J. Agric. Food Chem. 2018, 66, 12122–12130. [Google Scholar] [CrossRef]
- Gil, N.; Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020, 21, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Kim, Y.; Adh, G.; Jo, B.; Gao, C.; Mcdowell, I.C.; Consortium, G.; Engelhardt, B.E.; Battle, A. Co-expression networks reveal the tissue-specific regulation of transcription and splicing. Genome Res. 2017, 27, 1843. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nat. Cell Biol. 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Huang, T.-H.; Zhu, M.-J.; Li, X.-Y.; Zhao, S. Discovery of Porcine microRNAs and Profiling from Skeletal Muscle Tissues during Development. PLoS ONE 2008, 3, e3225. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Yang, Y.; Wang, Z.; Zhao, S.; Mu, Y.; Li, K. Integrated analysis of miRNA and mRNA paired expression profiling of prenatal skeletal muscle development in three genotype pigs. Sci. Rep. 2015, 5, srep15544. [Google Scholar] [CrossRef] [PubMed]
- Strathe, A.B.; Danfaer, A.; Sørensen, H.; Kebreab, E. A multilevel nonlinear mixed-effects approach to model growth in pigs. J. Anim. Sci. 2010, 88, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Green, D.M.; Brotherstone, S.; Schofield, C.P.; Whittemore, C.T. Food intake and live growth performance of pigs measured automatically and continuously from 25 to 115 kg live weight. J. Sci. Food Agric. 2003, 83, 1150–1155. [Google Scholar] [CrossRef]
- Nam, J.; Bartel, D.P. Long noncoding RNAs in C. elegans. Genome Res. 2012, 22, 2529–2540. [Google Scholar] [CrossRef]
- Ling, Y.; Zheng, Q.; Sui, M.; Zhu, L.; Xu, L.; Zhang, Y.; Liu, Y.; Fang, F.; Chu, M.; Ma, Y. Comprehensive Analysis of LncRNA Reveals the Temporal-Specific Module of Goat Skeletal Muscle Development. Int. J. Mol. Sci. 2019, 20, 3950. [Google Scholar] [CrossRef]
- Ning, C.; Ma, T.; Hu, S.; Xu, Z.; Li, D. Long Non-coding RNA and mRNA Profile of Liver Tissue During Four Developmental Stages in the Chicken. Front. Genet. 2020, 11, 574. [Google Scholar] [CrossRef]
- Yang, L.; Li, P.; Yang, W.; Ruan, X.; Kiesewetter, K.; Zhu, J.; Cao, H. Integrative Transcriptome Analyses of Metabolic Responses in Mice Define Pivotal LncRNA Metabolic Regulators. Cell Metab. 2016, 24, 627–639. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, L.; Jia, J.; Chen, Q.; Yuan, Z.; Zhang, X.; Sun, W.; Ma, C.; Xu, F.; Zhan, S. Effects of Maternal Low-Protein Diet on Microbiota Structure and Function in the Jejunum of Huzhu Bamei Suckling Piglets. Animal 2019, 9, 713. [Google Scholar] [CrossRef] [PubMed]
- Schulfer, A.; Blaser, M.J. Risks of Antibiotic Exposures Early in Life on the Developing Microbiome. PLOS Pathog. 2015, 11, e1004903. [Google Scholar] [CrossRef]
- Wellington, S.; Nag, P.P.; Michalska, K.; E Johnston, S.; Jedrzejczak, R.P.; Kaushik, V.K.; E Clatworthy, A.; Siddiqi, N.; McCarren, P.; Bajrami, B.; et al. A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase. Nat. Chem. Biol. 2017, 13, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Spaulding, C.N.; Klein, R.D.; Iv, H.L.S.; Janetka, J.W.; Hultgren, S.J. Precision antimicrobial therapeutics: The path of least resistance? NPJ Biofilms Microbiomes 2018, 4, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Han, F.; Chen, L.; Peng, X.; Chen, D.; Wu, D.; Feng, B.; Zhang, K. High nutrient intake during the early postnatal period accelerates skeletal muscle fiber growth and maturity in intrauterine growth-restricted pigs. Genes Nutr. 2018, 13, 23. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Peng, X.; Han, F.; Wu, F.; Chen, D.; Wu, D.; Feyera, T.; Zhang, K.; Che, L. Effects of Birth Weight and Postnatal Nutritional Restriction on Skeletal Muscle Development, Myofiber Maturation, and Metabolic Status of Early-Weaned Piglets. Animal 2020, 10, 156. [Google Scholar] [CrossRef] [PubMed]
- Swaim, L.E.; Connolly, L.E.; Volkman, H.E.; Humbert, O.; Born, D.E.; Ramakrishnan, L. Mycobacterium marinum Infection of Adult Zebrafish Causes Caseating Granulomatous Tuberculosis and Is Moderated by Adaptive Immunity. Infect. Immun. 2006, 74, 6108–6117. [Google Scholar] [CrossRef] [PubMed]
- Hess, I.; Boehm, T. Intravital Imaging of Thymopoiesis Reveals Dynamic Lympho-Epithelial Interactions. Immunity 2012, 36, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Neugebauer, N.; Luther, H.; Reinsch, N. Parent-of-origin effects cause genetic variation in pig performance traits. Animal 2010, 4, 672–681. [Google Scholar] [CrossRef]
- Wang, H.; Wang, J.; Yang, D.-D.; Liu, Z.-L.; Zeng, Y.; Chen, W. Expression of lipid metabolism genes provides new insights into intramuscular fat deposition in Laiwu pigs. Asian Australas. J. Anim. Sci. 2019, 33, 390–397. [Google Scholar] [CrossRef]
- Pannier, L.; Mullen, A.M.; Hamill, R.; Stapleton, P.; Sweeney, T. Association analysis of single nucleotide polymorphisms in DGAT1, TG and FABP4 genes and intramuscular fat in crossbred Bos taurus cattle. Meat Sci. 2010, 85, 515–518. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Mach, N.; Esteve-Codina, A.; Corominas, J.; Castelló, A.; Ballester, M.; Estellé, J.; Ibañez-Escriche, N.; Fernández, A.I.; Perezenciso, M.; et al. Liver transcriptome profile in pigs with extreme phenotypes of intramuscular fatty acid composition. BMC Genom. 2012, 13, 547. [Google Scholar] [CrossRef]
- Gao, J.; Yang, P.; Cui, Y.; Meng, Q.; Feng, Y.; Hao, Y.; Liu, J.; Piao, X.; Gu, X. Identification of Metabonomics Changes in Longissimus Dorsi Muscle of Finishing Pigs Following Heat Stress through LC-MS/MS-Based Metabonomics Method. Animal 2020, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. The mitochondrial transporter family SLC25: Identification, properties and physiopathology. Mol. Asp. Med. 2013, 34, 465–484. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F.; Monné, M. Discoveries, metabolic roles and diseases of mitochondrial carriers: A review. Biochim. Biophys. Acta Bioenerg. 2016, 1863, 2362–2378. [Google Scholar] [CrossRef] [PubMed]
- Goubert, E.; Mircheva, Y.; Lasorsa, F.M.; Melon, C.; Profilo, E.; Sutera, J.; Becq, H.; Palmieri, F.; Palmieri, L.; Aniksztejn, L.; et al. Inhibition of the Mitochondrial Glutamate Carrier SLC25A22 in Astrocytes Leads to Intracellular Glutamate Accumulation. Front. Cell. Neurosci. 2017, 11, 149. [Google Scholar] [CrossRef] [PubMed]
- Granchi, D.; Baldini, N.; Ulivieri, F.M.; Caudarella, R. Role of Citrate in Pathophysiology and Medical Management of Bone Diseases. Nutrients 2019, 11, 2576. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, J.; Ma, H. Molecular characterization, tissue expression profile, and SNP analysis of porcine SLC13A5. Genet. Mol. Res. 2015, 14, 16090–16101. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Cohen, D.E. Functional characterization of thioesterase superfamily member 1/Acyl-CoA thioesterase 11: Implications for metabolic regulation. J. Lipid Res. 2012, 53, 2620–2631. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Niepel, M.W.; Kawano, Y.; Han, S.; Liu, S.; Marsili, A.; Larsen, P.R.; Lee, C.-H.; Cohen, D.E. Targeted deletion of thioesterase superfamily member 1 promotes energy expenditure and protects against obesity and insulin resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 5417–5422. [Google Scholar] [CrossRef]
- Okada, K.; LeClair, K.B.; Zhang, Y.; Li, Y.; Ozdemir, C.; Krisko, T.I.; Hagen, S.J.; Betensky, R.A.; Banks, A.S.; Cohen, D.E. Thioesterase superfamily member 1 suppresses cold thermogenesis by limiting the oxidation of lipid droplet-derived fatty acids in brown adipose tissue. Mol. Metab. 2016, 5, 340–351. [Google Scholar] [CrossRef]
- Jeong, J.; Kwon, E.G.; Im, S.K.; Seo, K.S.; Baik, M. Expression of fat deposition and fat removal genes is associated with intramuscular fat content in longissimus dorsi muscle of Korean cattle steers1. J. Anim. Sci. 2012, 90, 2044–2053. [Google Scholar] [CrossRef]
- Chu, M.; Wu, X.; Guo, X.; Pei, J.; Jiao, F.; Fang, H.; Liang, C.; Ding, X.; Bao, P.; Yan, P. Association between single-nucleotide polymorphisms of fatty acid synthase gene and meat quality traits in Datong Yak (Bos grunniens). Genet. Mol. Res. 2015, 14, 2617–2625. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-S.; Jin, S.-K.; Jeong, Y.-H.; Jung, Y.-C.; Jung, J.-H.; Shim, K.-S.; Choi, Y.I. Relationships between Single Nucleotide Polymorphism Markers and Meat Quality Traits of Duroc Breeding Stocks in Korea. Asian Australas. J. Anim. Sci. 2016, 29, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Piórkowska, K.; Małopolska, M.M.; Ropka-Molik, K.; Szyndler-Nędza, M.; Wiechniak, A.; Żukowski, K.; Lambert, B.D.; Tyra, M. Evaluation of SCD, ACACA and FASN Mutations: Effects on Pork Quality and other Production Traits in Pigs Selected Based on RNA-Seq Results. Animal 2020, 10, 123. [Google Scholar] [CrossRef]
- Lu, Z.; He, X.; Ma, B.; Zhang, L.; Li, J.; Jiang, Y.; Zhou, G.; Gao, F. Chronic Heat Stress Impairs the Quality of Breast-Muscle Meat in Broilers by Affecting Redox Status and Energy-Substance Metabolism. J. Agric. Food Chem. 2017, 65, 11251–11258. [Google Scholar] [CrossRef]
- Moisá, S.J.; Shike, D.W.; Graugnard, D.E.; Rodriguez-Zas, S.L.; Everts, R.E.; Lewin, H.A.; Faulkner, D.B.; Berger, L.L.; Loor, J.J. Bioinformatics Analysis of Transcriptome Dynamics During Growth in Angus Cattle Longissimus Muscle. Bioinform. Biol. Insights 2013, 7, 253–270. [Google Scholar] [CrossRef]
- Guo, W.; Wang, S.H.; Cao, H.; Xu, K.; Zhang, J.; Du, Z.; Lu, W.; Feng, J.D.; Li, N.; Wu, C.H.; et al. Gene Microarray Analysis for Porcine Adipose Tissue: Comparison of Gene Expression between Chinese Xiang Pig and Large White. Asian Australas. J. Anim. Sci. 2008, 21, 11–18. [Google Scholar] [CrossRef]
- Samant, S.A.; Kanwal, A.; Pillai, V.B.; Bao, R.; Gupta, M.P. The histone deacetylase SIRT6 blocks myostatin expression and development of muscle atrophy. Sci. Rep. 2017, 7, 11877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, S.; Liu, K.; Liu, Y. Long non-coding RNA deleted in lymphocytic leukaemia 1 promotes hepatocellular carcinoma progression by sponging miR-133a to regulate IGF-1R expression. J. Cell. Mol. Med. 2019, 23, 5154–5164. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Liu, Y.; Lu, Y.; Yang, B.; Tang, L. LncRNA XIST Promotes Pancreatic Cancer Proliferation Through miR-133a/EGFR. J. Cell. Biochem. 2017, 118, 3349–3358. [Google Scholar] [CrossRef]
- Reddy, A.M.; Zheng, Y.; Jagadeeswaran, G.; Macmil, S.L.; Wiley, G.B.; A Roe, B.; DeSilva, U.; Zhang, W.; Sunkar, R. Cloning, characterization and expression analysis of porcine microRNAs. BMC Genom. 2009, 10, 15–65. [Google Scholar] [CrossRef]
- Deng, Z.-L.; Chen, J.; Wang, D.-Z. Transgenic overexpression of miR-133a in skeletal muscle. BMC Musculoskelet. Disord. 2011, 12, 115. [Google Scholar] [CrossRef] [PubMed]
- Koutsoulidou, A.; Mastroyiannopoulos, N.P.; Furling, D.; Uney, J.B.; Phylactou, L.A. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev. Biol. 2011, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Niu, L.L.; Wei, W.; Zhang, W.; Li, X.; Cao, J.; Zhao, S.H. A feedback circuit between miR-133 and the ERK1/2 pathway involving an exquisite mechanism for regulating myoblast proliferation and differentiation. Cell Death Dis. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, J.; Jiang, R.; Wei, X.; Song, C.; Huang, Y.; Lan, X.; Lei, C.; Ma, Y.; Hu, L.; et al. Long Non-coding RNA Profiling Reveals an Abundant MDNCR that Promotes Differentiation of Myoblasts by Sponging miR-133a. Mol. Ther. Nucleic Acids 2018, 12, 610–625. [Google Scholar] [CrossRef]


| Samples | Raw Yield (G) | Raw Reads (M) | Clean Yield (G) | Clean Reads (M) | Clean Q20 (%) | Clean GC (%) |
|---|---|---|---|---|---|---|
| GRI-1 | 13.708 | 91.38 | 13.347 | 89.96 | 98.04 | 50.19 |
| GRI-2 | 14.745 | 98.3 | 14.271 | 95.79 | 97.79 | 49.62 |
| GRI-3 | 14.26 | 95.06 | 13.814 | 92.58 | 97.85 | 50.46 |
| MGI-1 | 17.549 | 117 | 17.103 | 114.87 | 98.01 | 50.67 |
| MGI-2 | 16.739 | 111.59 | 16.341 | 109.99 | 98.06 | 50.46 |
| MGI-3 | 14.605 | 97.37 | 14.237 | 95.48 | 97.84 | 50.16 |
| RSI-1 | 14.72 | 98.13 | 14.28 | 96.15 | 97.97 | 50.13 |
| RSI-2 | 12.683 | 84.56 | 12.332 | 82.72 | 97.9 | 50.63 |
| RSI-3 | 14.789 | 98.6 | 14.4 | 96.74 | 97.9 | 51.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Y.; Gan, M.; Shen, L.; Li, L.; Fan, Y.; Chen, Y.; Chen, L.; Niu, L.; Zhao, Y.; Jiang, A.; et al. Profiling and Functional Analysis of Long Noncoding RNAs and mRNAs during Porcine Skeletal Muscle Development. Int. J. Mol. Sci. 2021, 22, 503. https://doi.org/10.3390/ijms22020503
Tan Y, Gan M, Shen L, Li L, Fan Y, Chen Y, Chen L, Niu L, Zhao Y, Jiang A, et al. Profiling and Functional Analysis of Long Noncoding RNAs and mRNAs during Porcine Skeletal Muscle Development. International Journal of Molecular Sciences. 2021; 22(2):503. https://doi.org/10.3390/ijms22020503
Chicago/Turabian StyleTan, Ya, Mailin Gan, Linyuan Shen, Liang Li, Yuan Fan, Ying Chen, Lei Chen, Lili Niu, Ye Zhao, Anan Jiang, and et al. 2021. "Profiling and Functional Analysis of Long Noncoding RNAs and mRNAs during Porcine Skeletal Muscle Development" International Journal of Molecular Sciences 22, no. 2: 503. https://doi.org/10.3390/ijms22020503
APA StyleTan, Y., Gan, M., Shen, L., Li, L., Fan, Y., Chen, Y., Chen, L., Niu, L., Zhao, Y., Jiang, A., Jiang, D., Zhang, S., & Zhu, L. (2021). Profiling and Functional Analysis of Long Noncoding RNAs and mRNAs during Porcine Skeletal Muscle Development. International Journal of Molecular Sciences, 22(2), 503. https://doi.org/10.3390/ijms22020503







