Calmodulin as Ca2+-Dependent Interactor of FTO Dioxygenase
Abstract
:1. Introduction
2. Results
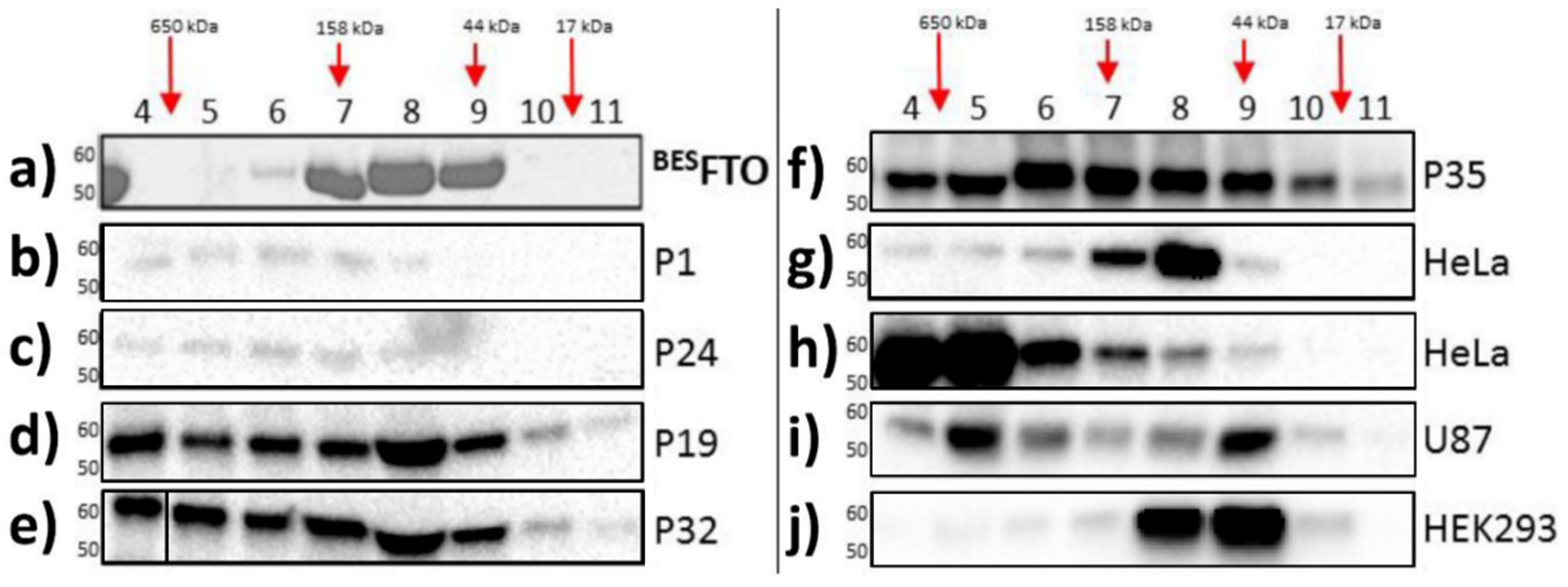
2.1. FTO Protein Forms Homo- and Heterocomplexes In Vivo
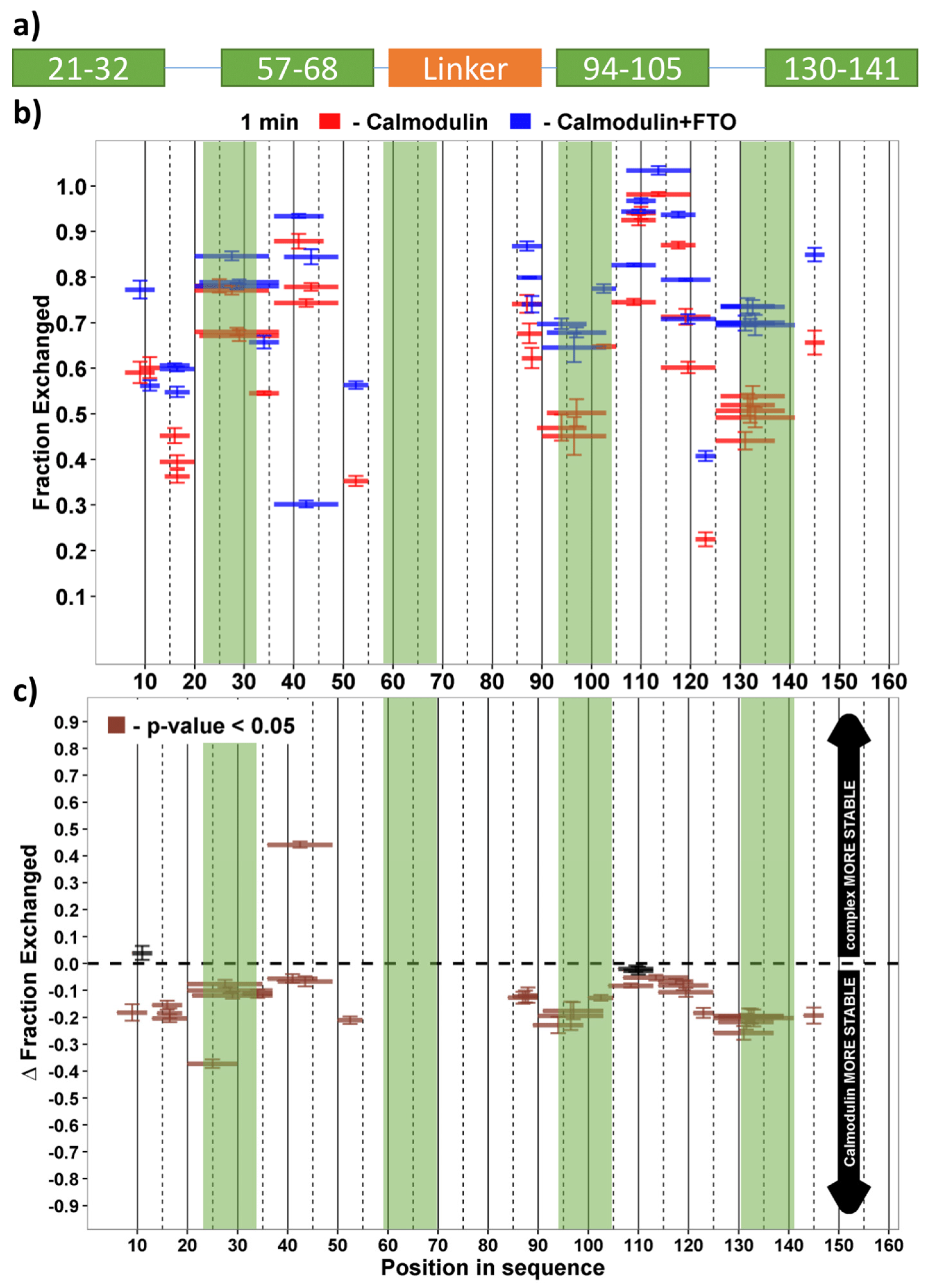
2.2. The Manner of FTO and CaM Interaction
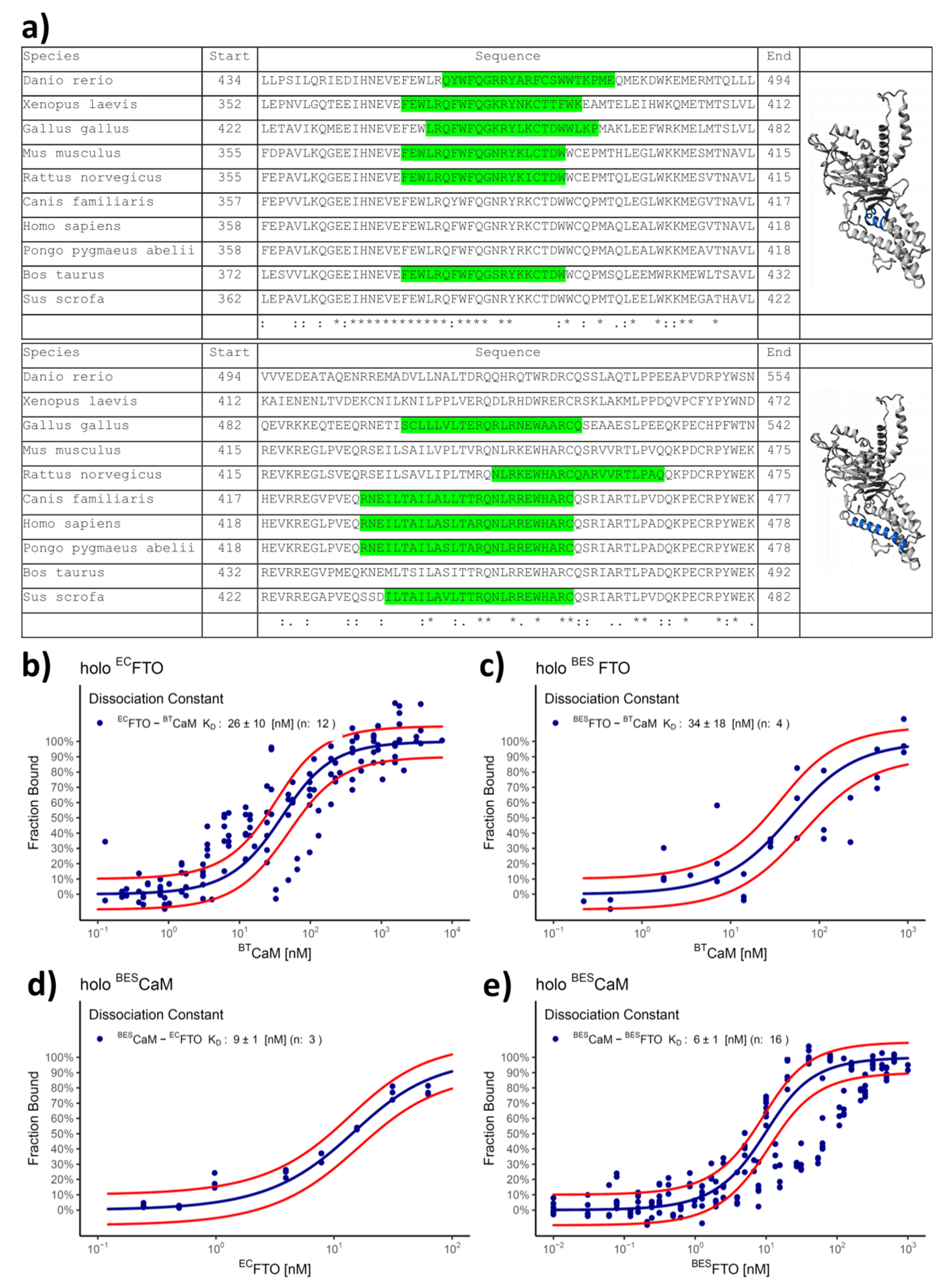
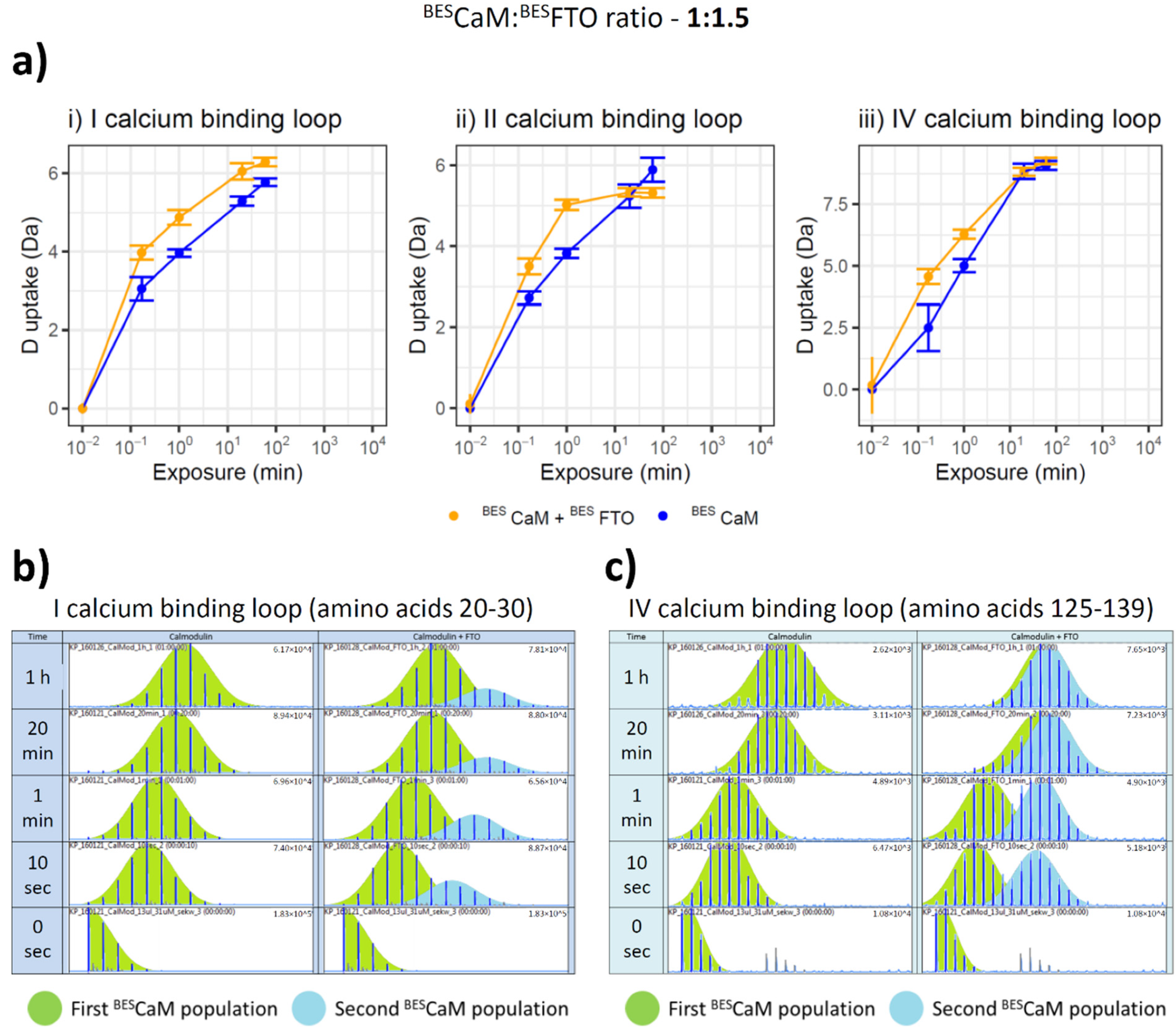
2.3. Both CaM Lobes Show the Ability to Interact with FTO
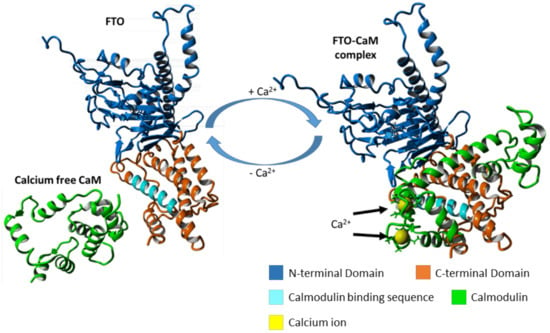
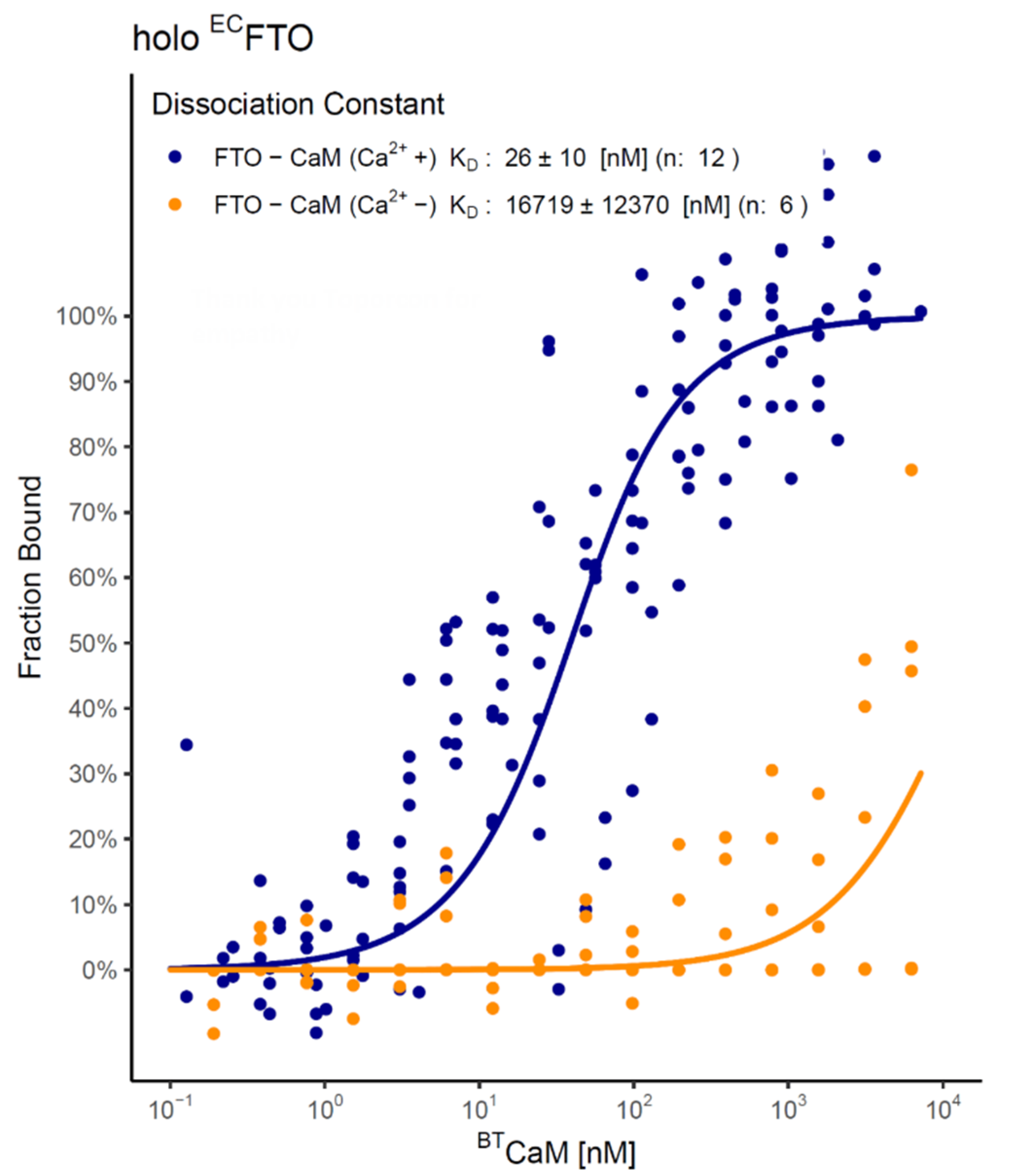
2.4. FTO–CaM Interaction Is Ca2+-Dependent
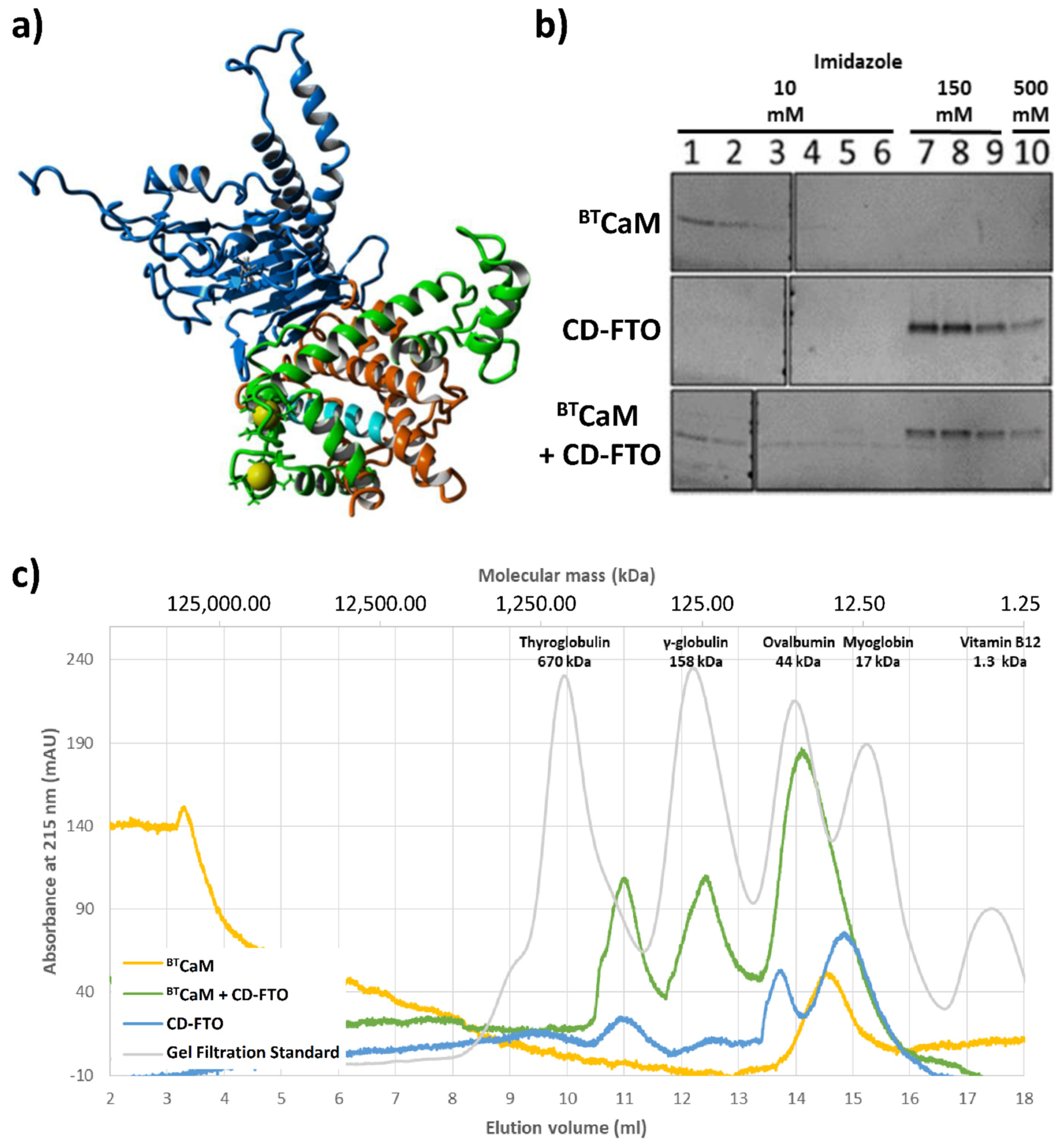
2.5. FTO Interacts with CaM via C-Terminal Domain
3. Discussion
4. Materials and Methods
4.1. Gel Filtration Chromatography
- For investigation of the molecular state of endogenous human FTO, fast protein liquid chromatography (FPLC) was performed. Recombinant FTO from baculovirus expression system (BESFTO) was expressed and purified as described previously [28]. Protein extracts from selected cancer tissues and HeLa, HEK293, and U87 cell lines were prepared as described previously [25]. A total of 900 µg of BESFTO and 200 µg of each protein lysate sample of a final volume of 500 µL were loaded onto a SEC650 column (#780-1650, BIO-RAD, Hercules, CA, USA) equilibrated and eluted with the PBS buffer. The eluate was monitored with NGC system ChromLab™ (BIO-RAD, Hercules, CA, USA) at 215, 260, and 280 nm. Protein standards (#1511901, BIO-RAD, Hercules, CA, USA) for calibration were analyzed using the same method. Next, fractions of 1 mL were precipitated by the addition of 20% pyrogallol red-molybdate (PRM, #611A, Sigma-Aldrich, Darmstadt, Germany) solution, subsequently re-dissolved in Laemmli buffer and analyzed by Western blot as before [25].
- In the case of the CaM and FTO C-terminal domain interaction study, protein samples: 1.5 nmol of bovine testicle CaM (BTCaM, supply by Medicago Company; Uppsala, Sweden, 7.6 µM) and/or 3.0 nmol of recombinant His-tagged C-terminal ECFTO domain (CD-FTO), (15.2. µM) were incubated for 10 min at 4 °C in SEC1 buffer of a final volume of 200 µL and loaded onto a SEC650 equilibrated and eluted with the SEC1 buffer. The eluate was monitored with NGC system ChromLab™ (BIO-RAD) at 215, 260 and 280 nm. Protein standards for calibration were analyzed by the same method.
4.2. In Silico Analysis of FTO Sequence
4.3. cDNA Cloning and Plasmid Constructs
4.4. Expression and Purification of ECFTO and BESFTO
4.5. Expression and Purification of BESCaM in Baculovirus Expression System (BES)
4.6. Expression and Purification of FTO C-Terminal Domain in E. coli
4.7. Preparation of Protein Samples
4.8. Pull-Down Assay
4.9. Microscale Thermophoresis (MST)
- HisTag labeling: For each labeling, 125 pmol of protein (50 µL of 2.5 µM solution) and 125 pmol of the dye were used. After labeling, the protein was transferred to experimental buffer MST2 alone or supplemented with 0.5 mM CaCl2 (MST3).
- Lysine labeling: The procedure was performed as before [28]. Briefly, for each labeling, 1 nanomole of protein (50 µL of 20 µM solution) and 2.5 nanomoles of the dye were used. The labeling efficiency was determined according to the manufacturer’s protocol. Only samples with the labeling efficiency in the range of 65–90% were used in further experiments to make sure that in most cases only one molecule of the dye would be attached to the protein. After labeling, the protein was transferred to experimental buffer supplemented with 0.5 mM (NH4)2Fe(SO4)2, 0.5 mM CaCl2 and 1 mM 2-OG (MST1). For the case of BESFTO labeling, the procedure perturbed the interaction: a ligand-induced fluorescence change occurred and only one weak and probably unspecific dissociation constant was visible, with a value of 24.7 ± 3.6 µM for the BESFTO–BESCaM complex (Supplementary Figure S1). This suggests that protein labeling may occur on lysines proximal to the FTO and CaM interface and such labeling strategy for this type of experiment should be avoided.
4.10. Hydrogen–Deuterium Exchange (HDX)
4.11. Molecular Modeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BDNF | Brain-derived neurotrophic factor |
| BES | Baculovirus Expression System |
| BESFTO | FTO protein expressed in baculovirus expression system |
| BTCaM | Bovine testicle calmodulin |
| CaM | Calmodulin |
| CaMKII | Calcium/calmodulin-dependent protein kinase II |
| CD-FTO | Recombinant His-tagged C-terminal ECFTO domain |
| CRE | cAMP response element |
| CREB | cAMP response element-binding protein |
| DBHS | Drosophila behavior/human splicing |
| ECFTO | FTO protein expressed in E. coli |
| FTO | Fat mass and obesity-associated proteins |
| GSK-3 | Glycogen syntase kinase 3 |
| HEK293 | Human embryonic kidney cell culture |
| HeLa | Henrietta Lacks cells |
| HNSCC | Head and neck cancer |
| KD | Dissociation constant |
| MRS | Methionyl tRNA synthetaze |
| MST | MicroScale Thermophoresis |
| HDX | Hydrogen–Deuterium Exchange |
| N6-meA | N6-methyladenosine |
| NLS | Nuclear Localization Signal |
| NPY1R | Neuropeptide Y receptor type 1 |
| p54nrb | Non-POU domain-containing octamer-binding protein |
| PIRT | Phosphoinositide-interacting regulator of Transient receptor potential |
| PRM | Pyrogallol red-molybdate |
| PSPC1 | Paraspeckle component 1 |
| SFPQ | Splicing factor, proline- and glutamine-rich |
| SK2-A | Small conductance potassium channel |
| SK-N-SH | Human neuroblastoma cells |
| U87 | Human primary glioblastoma cell line |
| XPO2 | Exportin-2 |
References
- Jia, G.; Yang, C.G.; Yang, S.; Jian, X.; Yi, C.; Zhou, Z.; He, C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008, 582, 3313–3319. [Google Scholar] [CrossRef] [Green Version]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Toh, J.D.W.; Sun, L.; Lau, L.Z.M.; Tan, J.; Low, J.J.A.; Tang, C.W.Q.; Cheong, E.J.Y.; Tan, M.J.H.; Chen, Y.; Hong, W.; et al. A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor of N6-methyladenosine demethylase FTO. Chem. Sci. 2015, 6, 112–122. [Google Scholar] [CrossRef]
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m 6 A, m 6 A m, and m 1 A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985. [Google Scholar] [CrossRef] [Green Version]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.J.; Chen, Q.; et al. Reversible methylation of m6Amin the 5′ cap controls mRNA stability. Nature 2017, 7637, 371–375. [Google Scholar] [CrossRef] [Green Version]
- Hirayama, M.; Wei, F.Y.; Chujo, T.; Oki, S.; Yakita, M.; Kobayashi, D.; Araki, N.; Takahashi, N.; Yoshida, R.; Nakayama, H.; et al. FTO Demethylates Cyclin D1 mRNA and Controls Cell-Cycle Progression. Cell Rep. 2020, 31, 107464. [Google Scholar] [CrossRef]
- Gulati, P.; Avezov, E.; Ma, M.; Antrobus, R.; Lehner, P.; O’Rahilly, S.; Yeo, G.S.H. Fat mass and obesity-related (FTO) shuttles between the nucleus and cytoplasm. Biosci. Rep. 2014, 34, 621–628. [Google Scholar] [CrossRef]
- Lin, L.; Hales, C.M.; Garber, K.; Jin, P. Fat mass and obesity-associated (FTO) protein interacts with CaMKII and modulates the activity of CREB signaling pathway. Hum. Mol. Genet. 2014, 23, 3299–3306. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.G.; Kennedy, M.B. Distinct forebrain and cerebellar isozymes of type II Ca2+/calmodulin-dependent protein kinase associate differently with the postsynaptic density fraction. J. Biol. Chem. 1985, 260, 9039–9046. [Google Scholar] [CrossRef]
- Hudmon, A.; Schulman, H. Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem. J. 2002, 364, 593–611. [Google Scholar] [CrossRef]
- Hudmon, A.; Schulman, H. Neuronal Ca2+/calmodulin-dependent protein kinase II: The role of structure and autoregulation in cellular function. Annu. Rev. Biochem. 2002, 71, 473–510. [Google Scholar] [CrossRef]
- Giese, K.P.; Fedorov, N.B.; Filipkowski, R.K.; Silva, A.J. Autophosphorylation at Thr286 of the α calcium-calmodulin kinase II in LTP and learning. Science 1998, 279, 870–873. [Google Scholar] [CrossRef]
- Soderling, T.R.; Derkach, V.A. Postsynaptic protein phosphorylation and LTP. Trends Neurosci. 2000, 23, 75–80. [Google Scholar] [CrossRef]
- Skelding, K.A.; Rostas, J.A.P. Regulation of CaMKII in vivo: The importance of targeting and the intracellular microenvironment. Neurochem. Res. 2009, 34, 1792–1804. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, L.; Wong, C.C.L.; Li, G.; Xu, T.; Pajvani, U.; Park, S.K.R.; Wronska, A.; Chen, B.X.; Marks, A.R.; Fukamizu, A.; et al. Calcium signaling through CaMKII regulates hepatic glucose production in fasting and obesity. Cell Metab. 2012, 15, 739–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brindle, P.K.; Montminy, M.R. The CREB family of transcription activators. Curr. Opin. Genet. Dev. 1992, 2, 199–204. [Google Scholar] [CrossRef]
- Knott, G.J.; Bond, C.S.; Fox, A.H. The DBHS proteins SFPQ, NONO and PSPC1: A multipurpose molecular scaffold. Nucleic Acids Res. 2016, 44, 3989–4004. [Google Scholar] [CrossRef]
- Song, H.; Wang, Y.; Wang, R.; Zhang, X.; Liu, Y.; Jia, G.; Chen, P.R. SFPQ Is an FTO-Binding Protein that Facilitates the Demethylation Substrate Preference. Cell Chem. Biol. 2020, 27, 283–291.e6. [Google Scholar] [CrossRef]
- Gulati, P.; Cheung, M.K.; Antrobus, R.; Church, C.D.; Harding, H.P.; Tung, Y.C.L.; Rimmington, D.; Ma, M.; Ron, D.; Lehner, P.J.; et al. Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proc. Natl. Acad. Sci. USA 2013, 110, 2557–2562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faulds, K.J.; Egelston, J.N.; Sedivy, L.J.; Mitchell, M.K.; Garimella, S.; Kozlowski, H.; D’Alessandro, A.; Hansen, K.C.; Balsbaugh, J.L.; Phiel, C.J. Glycogen synthase kinase-3 (gsk-3) activity regulates mRNA methylation in mouse embryonic stem cells. J. Biol. Chem. 2018, 293, 10731–10743. [Google Scholar] [CrossRef] [Green Version]
- Tai, H.; Wang, X.; Zhou, J.; Han, X.; Fang, T.; Gong, H.; Huang, N.; Chen, H.; Qin, J.; Yang, M.; et al. Protein kinase Cβ activates fat mass and obesity-associated protein by influencing its ubiquitin/proteasome degradation. FASEB J. 2017, 31, 4396–4406. [Google Scholar] [CrossRef] [Green Version]
- Yap, K.L.; Kim, J.; Truong, K.; Sherman, M.; Yuan, T.; Ikura, M. Calmodulin Target Database. J. Struct. Funct. Genomics 2000, 1, 8–14. [Google Scholar] [CrossRef]
- Lai, M.; Brun, D.; Edelstein, S.J.; Le Novère, N. Modulation of Calmodulin Lobes by Different Targets: An Allosteric Model with Hemiconcerted Conformational Transitions. PLoS Comput. Biol. 2015, 11, e1004063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berchtold, M.W.; Villalobo, A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim. Biophys. Acta—Mol. Cell Res. 2014, 1843, 398–435. [Google Scholar] [CrossRef] [PubMed]
- Pilžys, T.; Marcinkowski, M.; Kukwa, W.; Garbicz, D.; Dylewska, M.; Ferenc, K.; Mieczkowski, A.; Kukwa, A.; Migacz, E.; Wołosz, D.; et al. ALKBH overexpression in head and neck cancer: Potential target for novel anticancer therapy. Sci. Rep. 2019, 9, 13249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sachse, G.; Church, C.; Stewart, M.; Cater, H.; Teboul, L.; Cox, R.D.; Ashcroft, F.M. FTO demethylase activity is essential for normal bone growth and bone mineralization in mice. Biochim. Biophys. Acta—Mol. Basis Dis. 2018, 1864, 843–850. [Google Scholar] [CrossRef]
- Cao, Y.; Zhuang, Y.; Chen, J.; Xu, W.; Shou, Y.; Huang, X.; Shu, Q.; Li, X. Dynamic effects of Fto in regulating the proliferation and differentiation of adult neural stem cells of mice. Hum. Mol. Genet. 2020, 29, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Marcinkowski, M.; Pilžys, T.; Garbicz, D.; Piwowarski, J.; Mielecki, D.; Nowaczyk, G.; Taube, M.; Gielnik, M.; Kozak, M.; Winiewska-szajewska, M.; et al. Effect of posttranslational modifications on the structure and activity of FTO demethylase. Int. J. Mol. Sci. 2021, 22, 4512. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Stabels, M.; Eggertson, M.; Fadgen, K.; Yu, Y.Q. Conformational Characterization of Calmodulin by Hydrogen Deuterium Exchange Mass Spectrometry. Available online: https://www.waters.com/webassets/cms/library/docs/720003864en.pdf (accessed on 20 July 2021).
- Ferenc, K.; Pilžys, T.; Garbicz, D.; Marcinkowski, M.; Skorobogatov, O.; Dylewska, M.; Gajewski, Z.; Grzesiuk, E.; Zabielski, R. Intracellular and tissue specific expression of FTO protein in pig: Changes with age, energy intake and metabolic status. Sci. Rep. 2020, 10, 13029. [Google Scholar] [CrossRef]
- Black, D.J.; Tran, Q.K.; Persechini, A. Monitoring the total available calmodulin concentration in intact cells over the physiological range in free Ca2+. Cell Calcium 2004, 35, 415–425. [Google Scholar] [CrossRef]
- Shen, Y.; Lee, Y.S.; Soelaiman, S.; Bergson, P.; Lu, D.; Chen, A.; Beckingham, K.; Grabarek, Z.; Mrksich, M.; Tang, W.J. Physiological calcium concentrations regulate calmodulin binding and catalysis of adenylyl cyclase exotoxins. EMBO J. 2002, 21, 6721–6732. [Google Scholar] [CrossRef] [Green Version]
- Tansey, M.G.; Luby-Phelps, K.; Kamm, K.E.; Stull, J.T. Ca2+-dependent phosphorylation of myosin light chain kinase decreases the Ca2+ sensitivity of light chain phosphorylation within smooth muscle cells. J. Biol. Chem. 1994, 269, 9912–9920. [Google Scholar] [CrossRef]
- Gerendasy, D. Homeostatic tuning of Ca2+ signal transduction by members of the calpacitin protein family. J. Neurosci. Res. 1999, 58, 107–119. [Google Scholar] [CrossRef]
- Jurado, L.A.; Chockalingam, P.S.; Jarrett, H.W. Apocalmodulin. Physiol. Rev. 1999, 79, 661–682. [Google Scholar] [CrossRef]
- Seeger, C.; Talibov, V.O.; Danielson, U.H. Biophysical analysis of the dynamics of calmodulin interactions with neurogranin and Ca2+/calmodulin-dependent kinase II. J. Mol. Recognit. 2017, 30, e2621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sisco, N.J.; Luu, D.D.; Kim, M.; Van Horn, W.D. PIRT the TRP channel regulating protein binds calmodulin and cholesterol-like ligands. Biomolecules 2020, 10, 478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tidow, H.; Nissen, P. Structural diversity of calmodulin binding to its target sites. FEBS J. 2013, 280, 5551–5565. [Google Scholar] [CrossRef]
- Crivici, A.; Ikura, M. Molecular and structural basis of target recognition by calmodulin. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 85–116. [Google Scholar] [CrossRef]
- Chagot, B.; Chazin, W.J. Solution NMR structure of apo-calmodulin in complex with the IQ motif of human cardiac sodium channel NaV1.5. J. Mol. Biol. 2011, 406, 106–119. [Google Scholar] [CrossRef] [Green Version]
- Houdusse, A.; Gaucher, J.F.; Krementsova, E.; Mui, S.; Trybus, K.M.; Cohen, C. Crystal structure of apo-calmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features. Proc. Natl. Acad. Sci. USA 2006, 103, 19326–19331. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, M.A.; Rivard, A.F.; Bächinger, H.P.; Adelman, J.P. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature 2001, 410, 1120–1124. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Crum, M.; Miller, M.C. Crystal structures of apocalmodulin and an apocalmodulin/SK potassium channel gating domain complex. Structure 2004, 12, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Behnen, P.; Davis, E.; Delaney, E.; Frohm, B.; Bauer, M.; Cedervall, T.; O’Connell, D.; Akerfeldt, K.S.; Linse, S. Calcium-Dependent Interaction of Calmodulin with Human 80S Ribosomes and Polyribosomes. Biochemistry 2012, 51, 6718–6727. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: http://www.R-project.org/ (accessed on 7 April 2021).
- Bartoschik, T.; Galinec, S.; Kleusch, C.; Walkiewicz, K.; Breitsprecher, D.; Weigert, S.; Muller, Y.A.; You, C.; Piehler, J.; Vercruysse, T.; et al. Near-native, site-specific and purification-free protein labeling for quantitative protein interaction analysis by MicroScale Thermophoresis. Sci. Rep. 2018, 8, 4977. [Google Scholar] [CrossRef] [PubMed]
- Winiewska, M.; Bugajska, E.; Poznañski, J. ITC-derived binding affinity may be biased due to titrant (nano)-aggregation. Binding of halogenated benzotriazoles to the catalytic domain of human protein kinase CK2. PLoS ONE 2017, 12, e0173260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Technique | Experiment | Buffer Composition | |
|---|---|---|---|
| PBS | Gel filtration chromatography | Estimation of the molecular size of protein sample | 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl |
| SEC1 | Gel filtration chromatography | Estimation of the molecular size of protein sample | 50 mM MES, pH 6.5, 150 mM NaCl, 10% (v/v) glycerol, 1 mM 2-OG, 0.5 mM MnCl2, 0.5 mM CaCl2 |
| CON1 | Construct preparation | Preparation of plasmids carrying C-terminal domain of FTO | 1x buffer stock solution for KOD Hot Start DNA Polymerase, 1.5 mM MgCl2, 0.2 mM each dNTP, 0.5 µM each primer, 1 U of KOD Hot Start DNA Polymerase (Sigma-Aldrich, #71086), 1 ng of pET-28a(+) carrying hFTO cDNA |
| SOC | Bacteria preparation | Transformation | 10 mM NaCl, 2.5 mM KCl, 2% (w/v) tryptone, 0.5% (w/v) yeast extract, 20 mM glucose, 10 mM MgCl2, 10 mM MgSO4 |
| PRP1 | Protein purification | Lysis of the bacteria | 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl, 10% (v/v) glycerol, 0.1% (v/v) TWEEN® 20, 0.1% (w/v) lysozyme, 5 mM imidazole, 5 mM β-mercaptoethanol |
| PRP2 | Protein purification | Elution from Ni-Charged Resin | 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl, 10% (v/v) glycerol, 0.1% (v/v) TWEEN® 20, 150 mM imidazole, 5 mM β-mercaptoethanol |
| PRP3 | Protein purification | Dialysis | 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 50% (v/v) glycerol, 5mM β-mercaptoethanol |
| PRP4 | Protein purification | Lysis of the BES Insect cells | 50 mM HEPES, pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 5 mM β-mercaptoethanol |
| PDA1 | Pull-down assay | Examination of protein–protein interaction | 50 mM MES, pH 6.5, 150 mM NaCl, 10% glycerol (v/v), 2 mM L-ascorbic acid, 1 mM 2-OG, 0.5 mM (NH4)2Fe(SO4)2, 0.5 mM CaCl2 |
| MST1 | Microscale thermophoresis | Experimental buffer for ECFTO and BESFTO with the lysine labeling | 50 mM MES pH 6.1, 150 mM NaCl, 0.05% (v/v) TWEEN® 20, 2 mM L-ascorbic acid, 1 mM 2-OG, 0.5 mM (NH4)2Fe(SO4)2, 0.5 mM CaCl2 |
| MST2 | Microscale thermophoresis | Experimental buffer for ECFTO and BESFTO with the HisTag labeling | 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl, 10% (v/v) glycerol, 2 mM L-ascorbic acid, 0.5 mM (NH4)2Fe(SO4)2, 1 mM 2-OG |
| MST3 | Microscale thermophoresis | Experimental buffer for ECFTO and BESFTO with the HisTag labeling | 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl, 10% (v/v) glycerol, 2 mM L-ascorbic acid, 1 mM 2-OG, 0.5 mM (NH4)2Fe(SO4)2, 0.5 mM CaCl2 |
| HDX1 | Hydrogen–deuterium exchange | Sample incubation | 50 mM MES pH 6.1, 150 mM NaCl, 0.004% (v/v) TWEEN® 20, 2 mM L-ascorbic acid, 1 mM 2-OG, 0.5 mM (NH4)2Fe(SO4)2, 0.5 mM CaCl2 |
| HDX2 | Hydrogen–deuterium exchange | HDX reaction | 30 mM Tris-DCl, pD 7.5 (pHREAD+0.4), 150 mM NaCl in D2O (#DLM-4DR-99.8-PK, Cambridge Isotope Laboratories, Inc.) |
| HDX3 | Hydrogen–deuterium exchange | HDX stopping | 2 M glycine, pH 2.4, 107 mM NaCl |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcinkowski, M.; Pilžys, T.; Garbicz, D.; Piwowarski, J.; Przygońska, K.; Winiewska-Szajewska, M.; Ferenc, K.; Skorobogatov, O.; Poznański, J.; Grzesiuk, E. Calmodulin as Ca2+-Dependent Interactor of FTO Dioxygenase. Int. J. Mol. Sci. 2021, 22, 10869. https://doi.org/10.3390/ijms221910869
Marcinkowski M, Pilžys T, Garbicz D, Piwowarski J, Przygońska K, Winiewska-Szajewska M, Ferenc K, Skorobogatov O, Poznański J, Grzesiuk E. Calmodulin as Ca2+-Dependent Interactor of FTO Dioxygenase. International Journal of Molecular Sciences. 2021; 22(19):10869. https://doi.org/10.3390/ijms221910869
Chicago/Turabian StyleMarcinkowski, Michał, Tomaš Pilžys, Damian Garbicz, Jan Piwowarski, Kaja Przygońska, Maria Winiewska-Szajewska, Karolina Ferenc, Oleksandr Skorobogatov, Jarosław Poznański, and Elżbieta Grzesiuk. 2021. "Calmodulin as Ca2+-Dependent Interactor of FTO Dioxygenase" International Journal of Molecular Sciences 22, no. 19: 10869. https://doi.org/10.3390/ijms221910869
APA StyleMarcinkowski, M., Pilžys, T., Garbicz, D., Piwowarski, J., Przygońska, K., Winiewska-Szajewska, M., Ferenc, K., Skorobogatov, O., Poznański, J., & Grzesiuk, E. (2021). Calmodulin as Ca2+-Dependent Interactor of FTO Dioxygenase. International Journal of Molecular Sciences, 22(19), 10869. https://doi.org/10.3390/ijms221910869