Effect of Structural Changes Induced by Deletion of 54FLRAPSWF61 Sequence in αB-crystallin on Chaperone Function and Anti-Apoptotic Activity
Abstract
:1. Introduction
2. Results
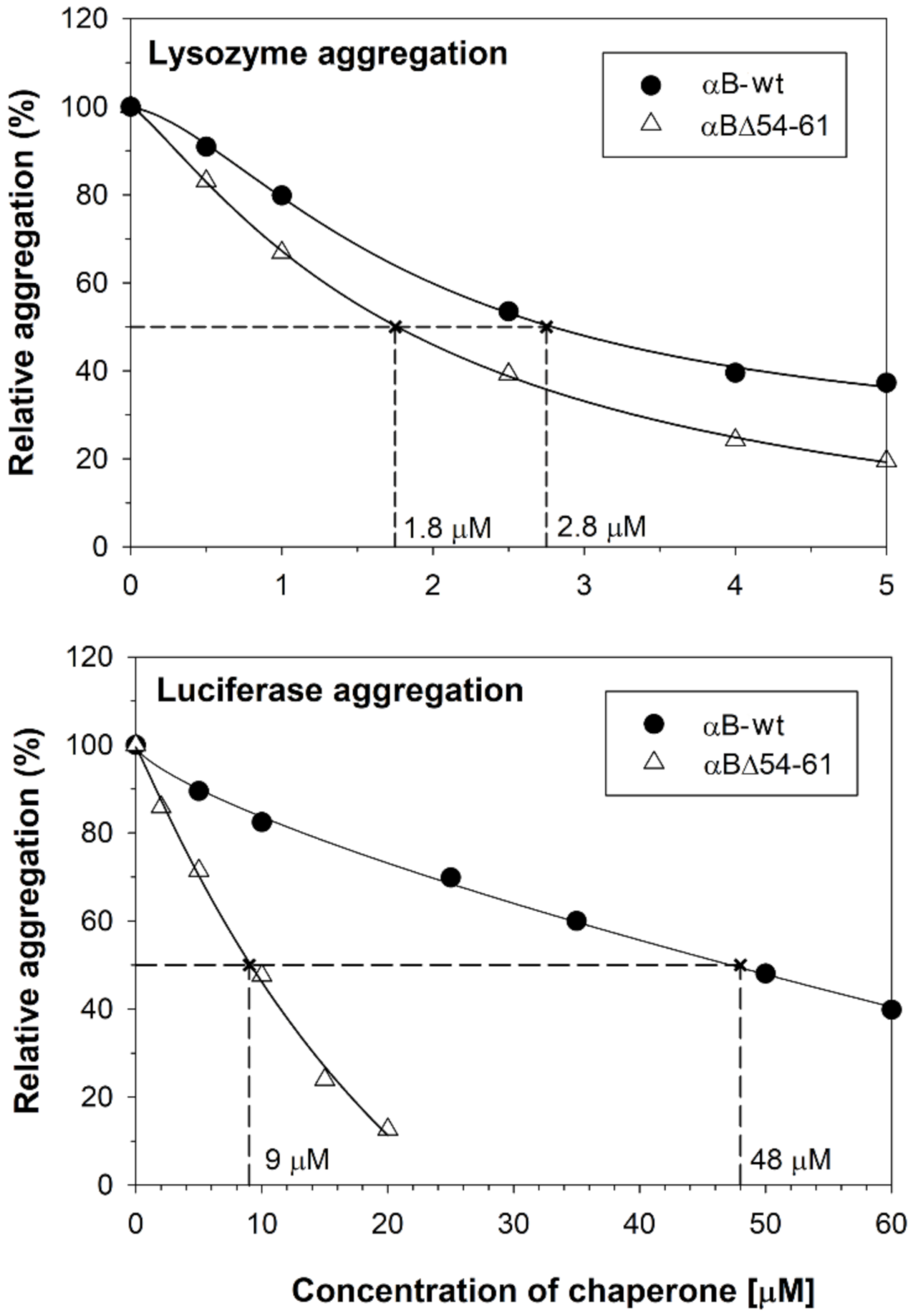
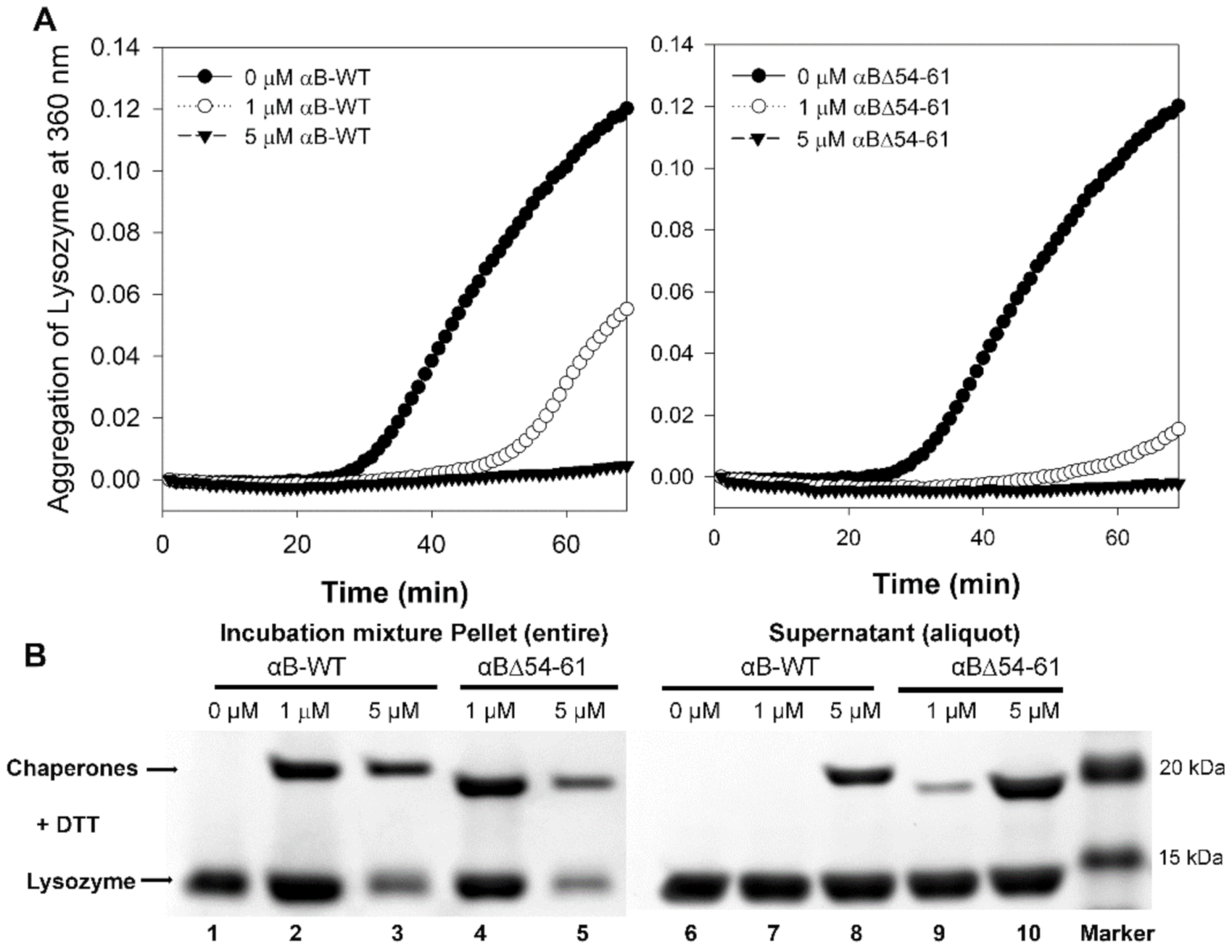
2.1. The Increase in Chaperone Activity of αBΔ54–61 Mutant Varies with the Substrate Protein
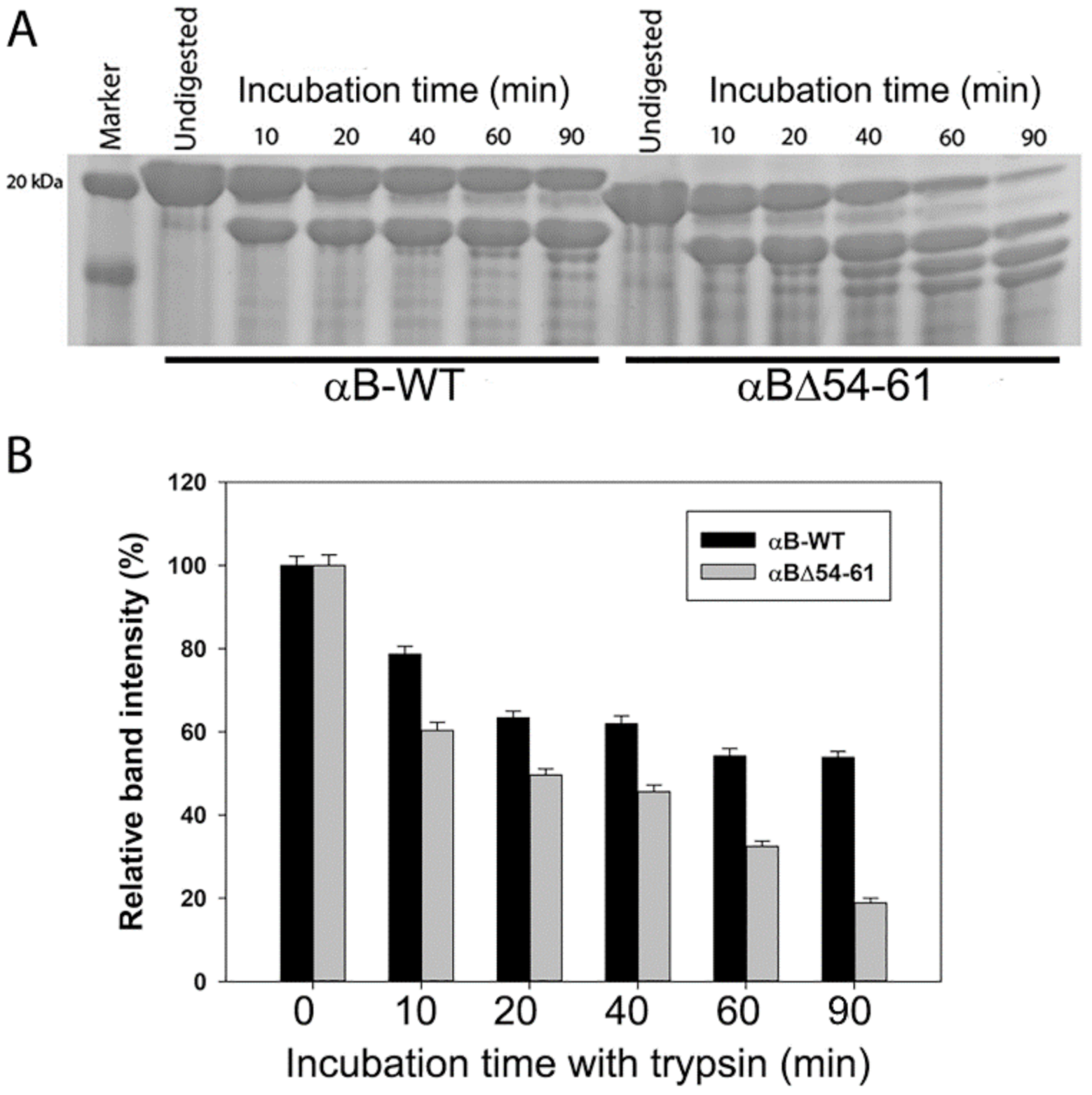
2.2. αBΔ54–61 Is More Susceptible to Cleavage by Trypsin
2.3. Deletion Mutant Forms Chaperone–Substrate Complex with Higher Mass
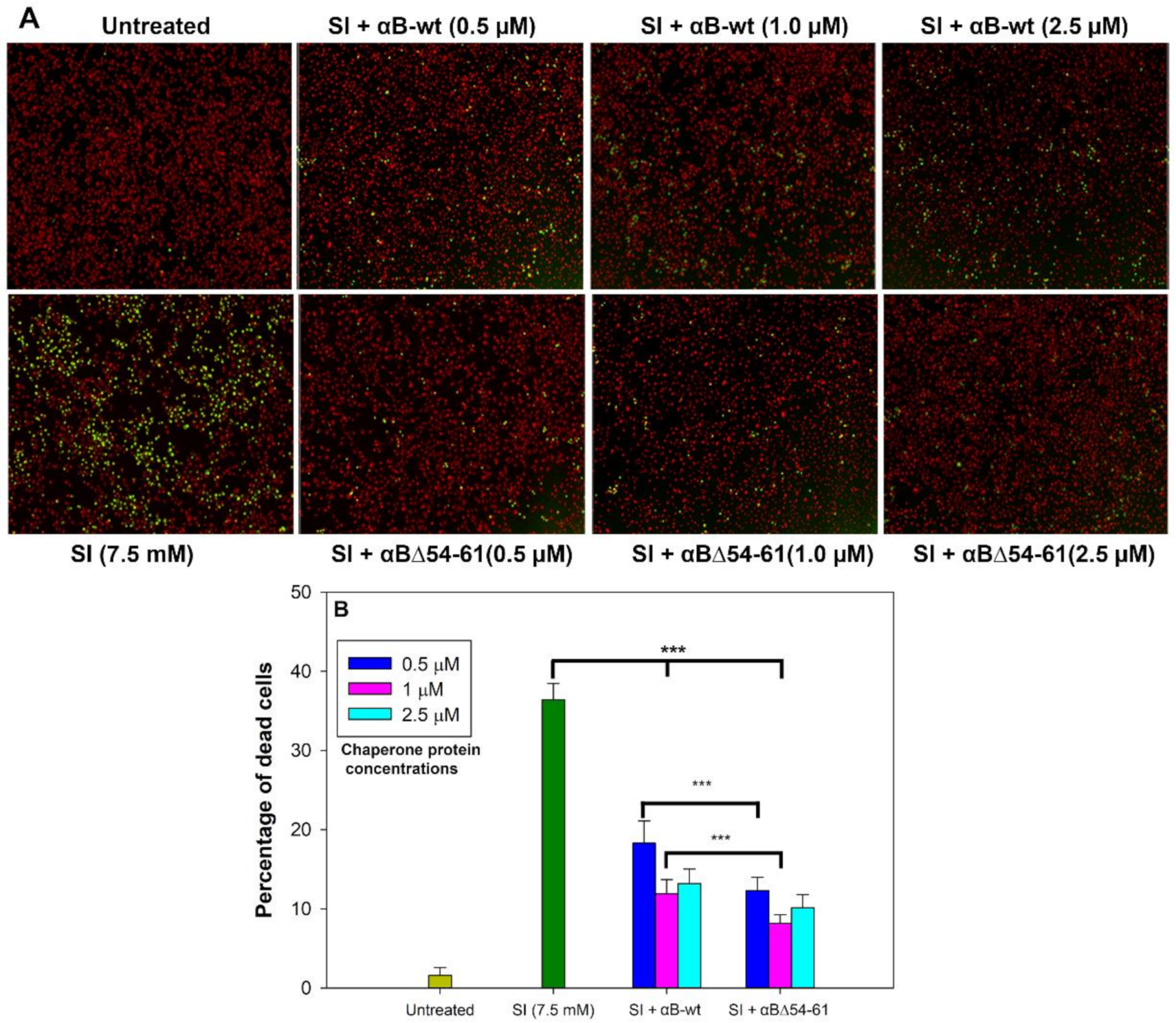
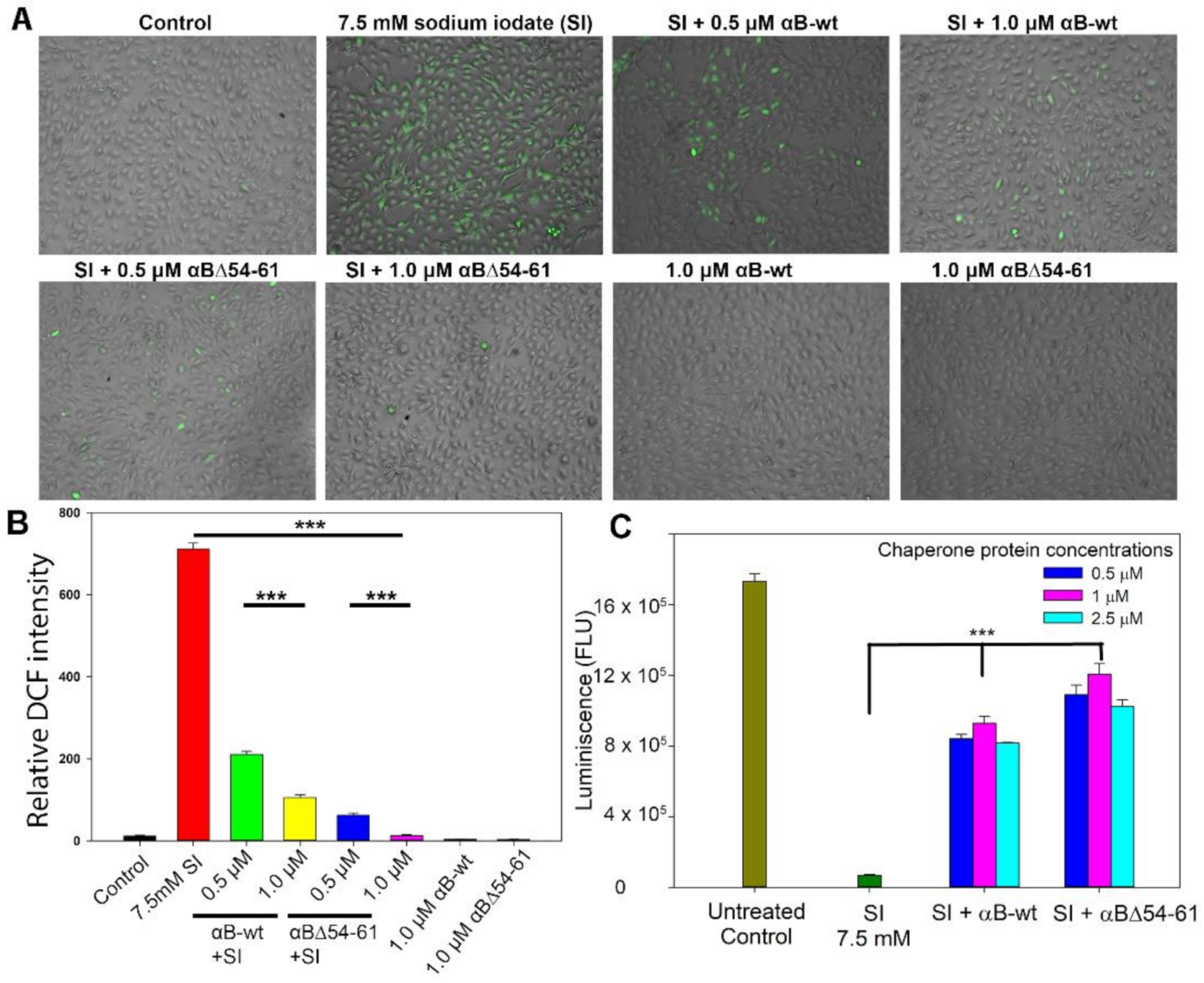
2.4. Deletion Mutant Showed Greater Anti-Oxidative and Anti-Apoptotic Activity Than αB-wt
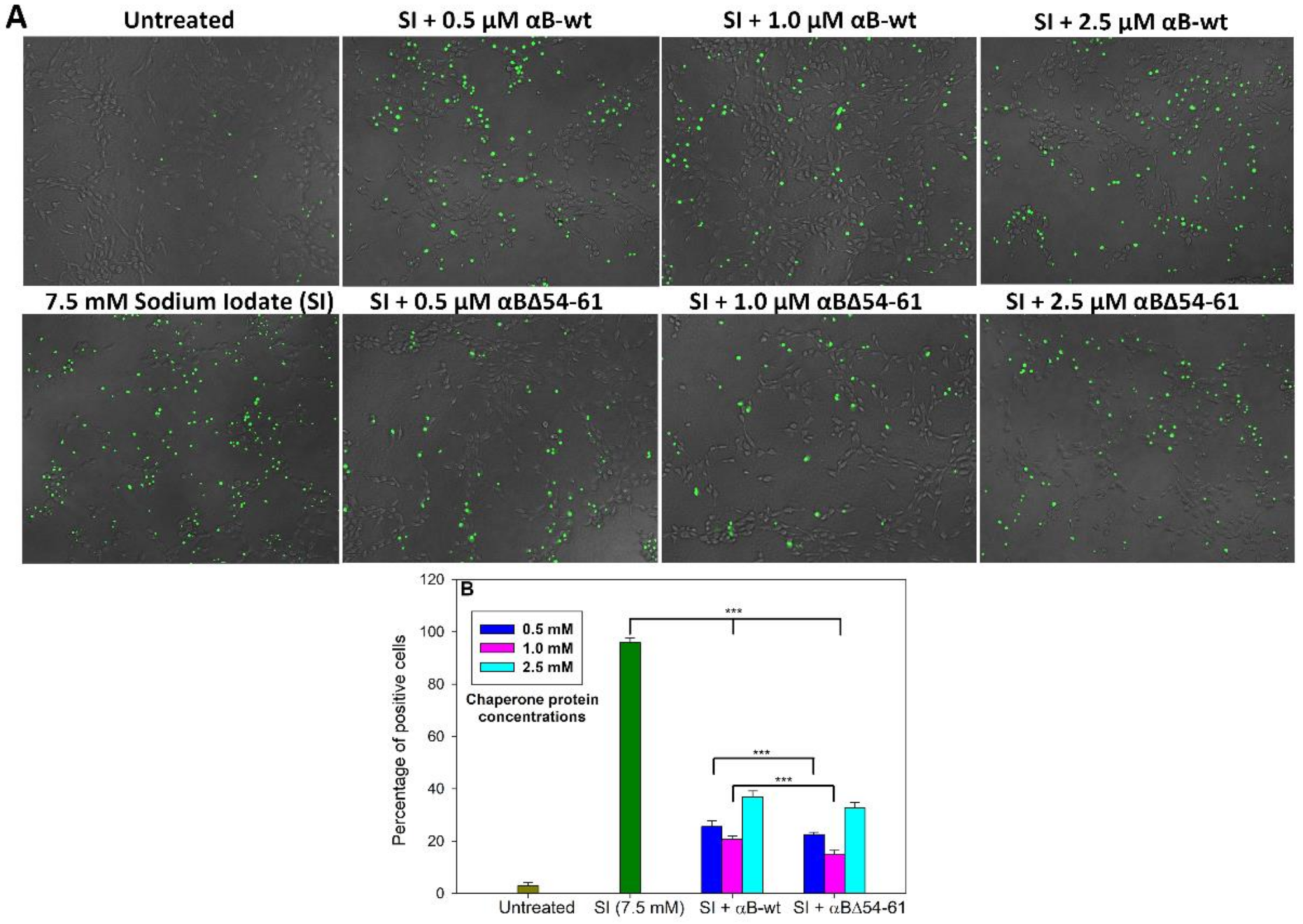
2.5. The Increased Anti-Apoptotic Activity of αBΔ54–61 Was Associated with Reduced Caspase Activation
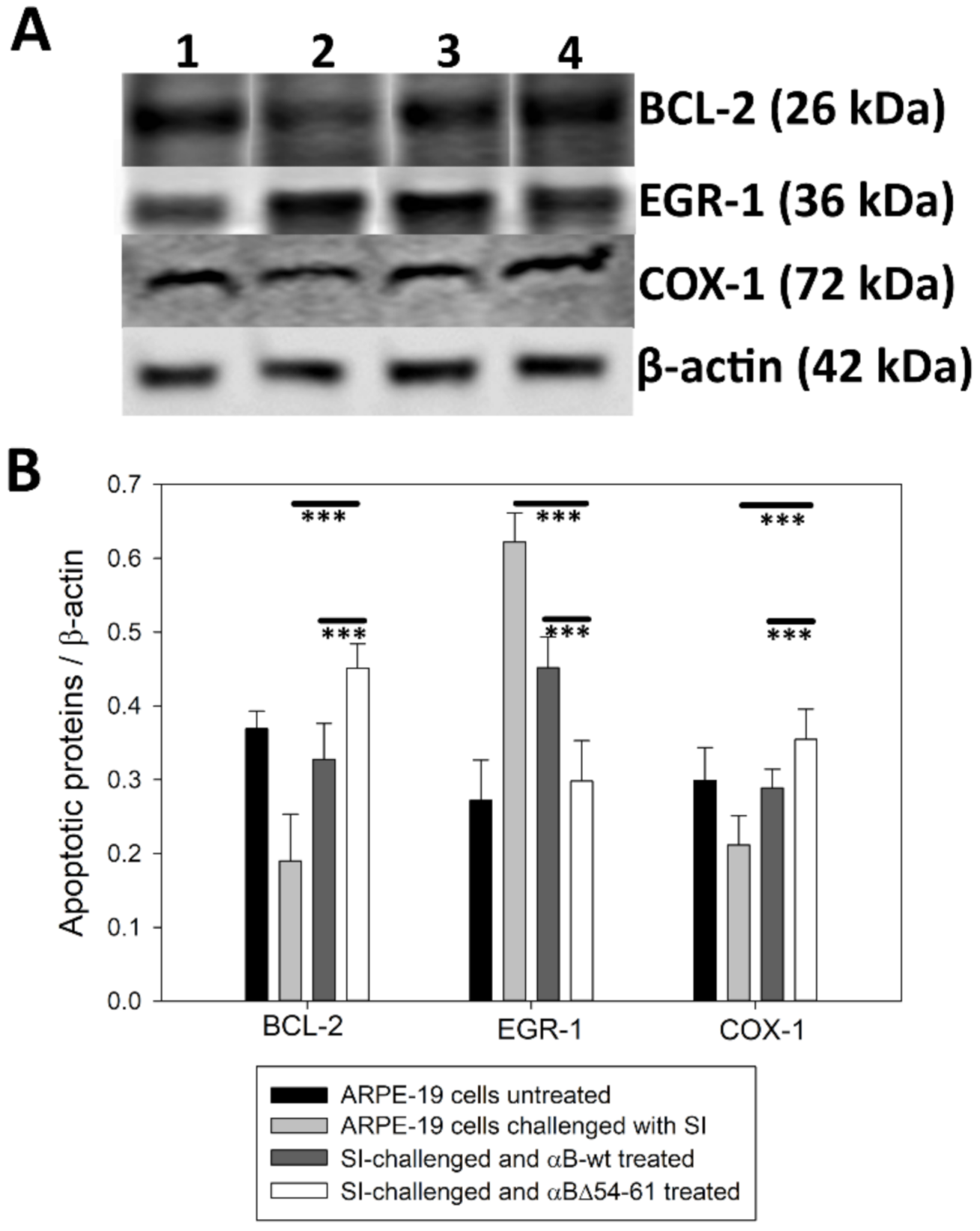
2.6. Treatment with αB-wt and αBΔ54–61 Alters the Levels of Apoptotic Cascade Components in SI-Treated ARPE-19 Cells
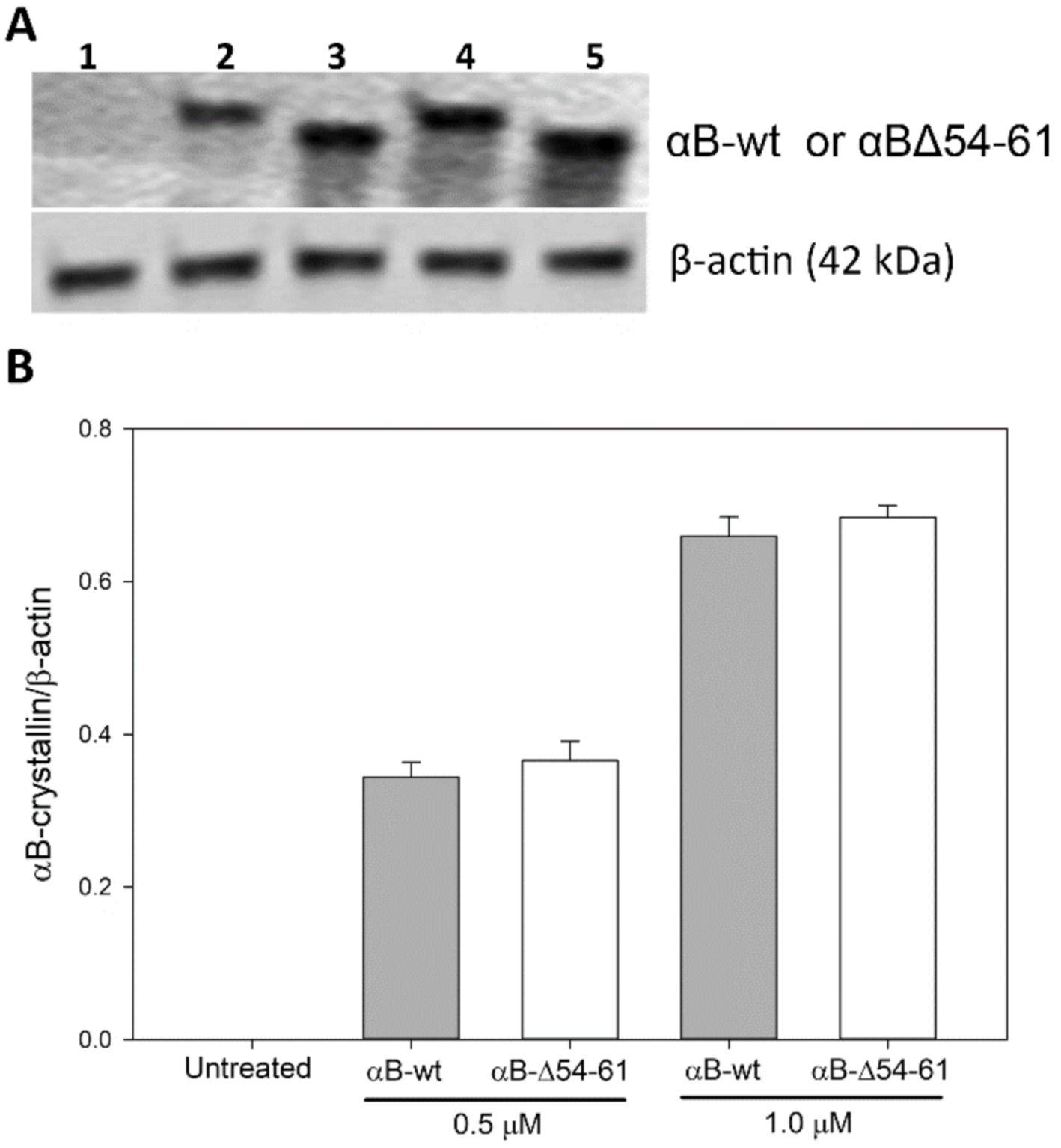
2.7. Transduction of αB-wt and αBΔ54–61 into ARPE-19 Cells
3. Discussion
4. Materials and Methods
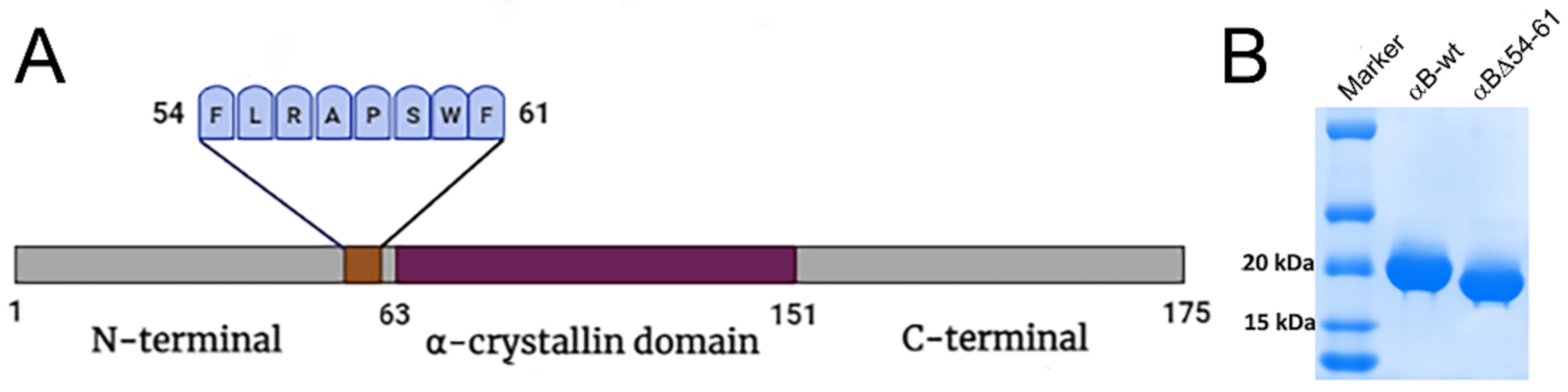
4.1. Overexpression and Purification of Wild-Type and Mutant αB-Crystallins
4.2. Chaperone Assays
4.3. Limited Proteolysis of Human αB-wt and αBΔ54–61 Using Trypsin
4.4. Chaperone–Substrate Complex Studies
4.5. Cytoprotective Effect of αB-wt and αBΔ54–61 in ARPE-19 Cells
4.6. Cell Viability Assay
4.7. Anti-Oxidative Potential of αB-wt and αBΔ54–61
4.8. Effect of Transduced αB-wt and αBΔ54–61 on Caspase Activation in ARPE-19 Cells
4.9. Regulatory Effect of αB-wt and αBΔ54–61 on Apoptotic Cascade Components in ARPE-19 Cells
4.10. Relative Amounts of Protein Transduced into the ARPE-19 Cells
4.11. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Narberhaus, F. α-Crystallin-type heat shock proteins: Socializing minichaperones in the context of a multichaperone network. Microbiol. Mol. Biol. Rev. 2002, 66, 64–93. [Google Scholar] [CrossRef] [Green Version]
- Horwitz, J. Alpha-crystallin. Exp. Eye Res. 2003, 76, 145–153. [Google Scholar] [CrossRef]
- Bhat, S.P.; Nagineni, C.N. αB subunit of lens-specific protein α crystallin is present in other ocular and non-ocular tissues. Biochem. Biophys. Res. Commun. 1989, 158, 319–325. [Google Scholar] [CrossRef]
- Nagaraj, R.H.; Nahomi, R.B.; Mueller, N.H.; Raghavan, C.T.; Ammar, D.A.; Petrash, J.M. Therapeutic potential of α-crystallin. Biochim. Biophys. Acta. 2016, 1860, 252–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwaki, T.; Wisniewski, T.; Iwaki, A.; Corbin, E.; Tomokane, N.; Tateishi, J.; Goldman, J. Accumulation of alpha B-crystallin in central nervous system glia and neurons in pathologic conditions. Am. J. Pathol. 1992, 140, 345–356. [Google Scholar] [PubMed]
- Clark, J.I.; Muchowski, P.J. Small heat-shock proteins and their potential role in human disease. Curr. Opin. Struct. Biol. 2000, 10, 52–59. [Google Scholar] [CrossRef]
- Sharma, K.K.; Santhoshkumar, P. Lens aging: Effects of crystallins. Biochim. Biophys. Acta. 2009, 1790, 1095–1108. [Google Scholar] [CrossRef] [Green Version]
- Jehle, S.; Vollmar, B.S.; Bardiaux, B.; Dove, K.K.; Rajagopal, P.; Gonen, T.; Oschkinat, H.; Klevit, R.E. N-terminal domain of alphaB-crystallin provides a conformational switch for multimerization and structural heterogeneity. Proc. Natl. Acad. Sci. USA 2011, 108, 6409–6414. [Google Scholar] [CrossRef] [Green Version]
- Carver, J.A.; Grosas, A.B.; Ecroyd, H.; Quinlan, R.A. The functional roles of the unstructured N- and C-terminal regions in αB-crystallin and other mammalian small heat-shock proteins. Cell Stress Chaperones 2017, 22, 627–638. [Google Scholar] [CrossRef]
- Laganowsky, A.; Benesch, J.L.P.; Landau, M.; Ding, L.; Sawaya, M.R.; Cascio, D.; Huang, Q.; Robinson, C.V.; Horwitz, J.; Eisenberg, D. Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Sci. 2010, 19, 1031–1043. [Google Scholar] [CrossRef] [Green Version]
- Jehle, S.; Rajagopal, P.; Bardiaux, B.; Markovic, S.; Kuhne, R.; Stout, J.R.; Higman, V.A.; Klevit, R.E.; van Rossum, B.-J.; Oschkinat, H. Solid-state NMR and SAXS studies provide a structural basis for the activation of [alpha]B-crystallin oligomers. Nat. Struct. Mol. Biol. 2010, 17, 1037–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagnéris, C.; Bateman, O.A.; Naylor, C.E.; Cronin, N.; Boelens, W.C.; Keep, N.H.; Slingsby, C. Crystal structures of [alpha]-crystallin domain dimers of [alpha]B-crystallin and Hsp20. J. Mol. Biol. 2009, 392, 1242–1252. [Google Scholar] [CrossRef] [PubMed]
- Aquilina, J.A.; Benesch, J.L.P.; Bateman, O.A.; Slingsby, C.; Robinson, C.V. Polydispersity of a mammalian chaperone: Mass spectrometry reveals the population of oligomers in αB-crystallin. Proc. Natl. Acad. Sci. USA 2003, 100, 10611–10616. [Google Scholar] [CrossRef] [Green Version]
- Jovcevski, B.; Andrew Aquilina, J.; Benesch, J.L.P.; Ecroyd, H. The influence of the N-terminal region proximal to the core domain on the assembly and chaperone activity of αB-crystallin. Cell Stress Chaperones. 2018, 5, 827–836. [Google Scholar] [CrossRef]
- Kundu, M.; Sen, P.C.; Das, K.P. Structure, stability, and chaperone function of αA-crystallin: Role of N-terminal region. Biopolymers 2007, 86, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Jaya, N.; Garcia, V.; Vierling, E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc. Natl. Acad. Sci. USA 2009, 106, 15604–15609. [Google Scholar] [CrossRef] [Green Version]
- Pasta, S.Y.; Raman, B.; Ramakrishna, T.; Rao, C.M. Role of the conserved SRLFDQFFG region of α-crystallin, a small heat shock protein: Effect on oligomeric size, subunit exchange, and chaperone-like activity. J. Biol. Chem. 2003, 278, 51159–51166. [Google Scholar] [CrossRef]
- Sreelakshmi, Y.; Santhoshkumar, P.; Bhattacharyya, J.; Sharma, K.K. αA-crystallin interacting regions in the small heat shock protein, αB-crystallin. Biochemistry 2004, 43, 15785–15795. [Google Scholar] [CrossRef]
- Sreelakshmi, Y.; Sharma, K.K. Recognition sequence 2 (Residues 60−71) plays a role in oligomerization and exchange dynamics of αB-crystallin. Biochemistry 2005, 44, 12245–12252. [Google Scholar] [CrossRef]
- Santhoshkumar, P.; Murugesan, R.; Sharma, K.K. Deletion of 54FLRAPSWF61 residues decreases the oligomeric size and enhances the chaperone function of αB-crystallin. Biochemistry 2009, 48, 5066–5073. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, J.G.; Clark, J.I. Insights into the domains required for dimerization and assembly of human αB crystallin. Protein Sci. 2005, 14, 684–695. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, J.G.; Estrada, M.R.; Clark, J.I. Interactive domains for chaperone activity in the small heat shock protein, human αB crystallin. Biochemistry 2005, 44, 14854–14869. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Goshe, J.; Miller, A.; Santhoshkumar, P.; Luckey, C.; Bhat, M.B.; Nagaraj, R.H. Paradoxical effects of substitution and deletion mutation of arg56 on the structure and chaperone function of human αB-crystallin. Biochemistry 2007, 46, 1117–1127. [Google Scholar] [CrossRef]
- Ahmad, M.F.; Raman, B.; Ramakrishna, T.; Rao, C.M. Effect of phosphorylation on αB-crystallin: Differences in stability, subunit exchange and chaperone activity of homo and mixed oligomers of αB-crystallin and its phosphorylation-mimicking mutant. J. Mol. Biol. 2008, 375, 1040–1051. [Google Scholar] [CrossRef] [PubMed]
- Bakthisaran, R.; Akula, K.K.; Tangirala, R.; Rao, C.M. Phosphorylation of αB-crystallin: Role in stress, aging and patho-physiological conditions. Biochim. Biophys. Acta. 2016, 1860, 167–182. [Google Scholar] [CrossRef] [Green Version]
- Muraleva, N.A.; Kolosova, N.G.; Stefanova, N.A. p38 MAPK–dependent alphaB-crystallin phosphorylation in Alzheimer’s disease–like pathology in OXYS rats. Exp. Gerontol. 2019, 119, 45–52. [Google Scholar] [CrossRef]
- Bartelt-Kirbach, B.; Wiegreffe, C.; Birk, S.; Baur, T.; Moron, M.; Britsch, S.; Golenhofen, N. HspB5/αB-crystallin phosphorylation at S45 and S59 is essential for protection of the dendritic tree of rat hippocampal neurons. J. Neurochem. 2021, 157, 2055–2069. [Google Scholar] [CrossRef]
- Bai, F.; Xi, J.; Higashikubo, R.; Andley, U.P. A comparative analysis of αA- and αB-crystallin expression during the cell cycle in primary mouse lens epithelial cultures. Exp. Eye Res. 2004, 79, 795–805. [Google Scholar] [CrossRef]
- Adhikari, A.S.; Singh, B.N.; Rao, K.S.; Rao, C.M. αB-crystallin, a small heat shock protein, modulates NF-κB activity in a phosphorylation-dependent manner and protects muscle myoblasts from TNF-α induced cytotoxicity. Biochim. Biophys. Acta 2011, 1813, 1532–1542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamradt, M.C.; Lu, M.; Werner, M.E.; Kwan, T.; Chen, F.; Strohecker, A.; Oshita, S.; Wilkinson, J.C.; Yu, C.; Oliver, P.G.; et al. The small heat shock protein αB-crystallin is a novel inhibitor of trail-induced apoptosis that suppresses the activation of caspase-3. J. Biol. Chem. 2005, 280, 11059–11066. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.W.; Liu, J.P.; Xiang, H.; Li, D.W.C. Human [alpha]A- and [alpha]B-crystallins bind to Bax and Bcl-XS to sequester their translocation during staurosporine-induced apoptosis. Cell Death Differ. 2004, 11, 512–526. [Google Scholar] [CrossRef]
- Pasupuleti, N.; Matsuyama, S.; Voss, O.; Doseff, A.I.; Song, K.; Danielpour, D.; Nagaraj, R.H. The anti-apoptotic function of human αA-crystallin is directly related to its chaperone activity. Cell Death Dis. 2010, 1, e31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, H.; Okamoto, K.; Nakayama, H.; Isobe, T.; Kato, K. Phosphorylation of αB-crystallin in response to various types of stress. J. Biol. Chem. 1997, 272, 29934–29941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Wang, W.; Shah, A.; Yu, M.; Liu, Y.; He, L.; Dang, J.; Yang, L.; Yan, M.; Ying, Y.; et al. Sodium iodate induces ferroptosis in human retinal pigment epithelium ARPE-19 cells. Cell Death Dis. 2021, 12, 230. [Google Scholar] [CrossRef]
- Takahashi, A.; Masuda, A.; Sun, M.; Centonze, V.E.; Herman, B. Oxidative stress-induced apoptosis is associated with alterations in mitochondrial caspase activity and Bcl-2-dependent alterations in mitochondrial pH (pHm). Brain Res. Bull. 2004, 62, 497–504. [Google Scholar] [CrossRef]
- Sharpe, J.C.; Arnoult, D.; Youle, R.J. Control of mitochondrial permeability by Bcl-2 family members. Biochim. Biophys. Acta. 2004, 1644, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Nakajima, E.; Fukiage, C.; Azuma, M.; Shearer, T.R. Differential gene expression in the lens epithelial cells from selenite injected rats. Exp. Eye Res. 2002, 74, 231–236. [Google Scholar] [CrossRef]
- Horwitz, J. proctor lecture: The function of α-Crystallin. Invest. Opthalmol. Vis. Sci. 1993, 34, 10–22. [Google Scholar]
- Peschek, J.; Braun, N.; Rohrberg, J.; Back, K.C.; Kriehuber, T.; Kastenmüller, A.; Weinkauf, S.; Buchner, J. Regulated structural transitions unleash the chaperone activity of αB-crystallin. Proc. Natl. Acad. Sci. USA 2013, 110, E3780–E3789. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.K.; Kim, R.; Kim, S.-H. Crystal structure of a small heat-shock protein. Nature 1998, 394, 595–599. [Google Scholar] [CrossRef]
- Merck, K.B.; De Haard-Hoekman, W.A.; Oude Essink, B.B.; Bloemendal, H.; De Jong, W.W. Expression and aggregation of recombinant αA-crystallin and its two domains. Biochim. Biophys. Acta 1992, 1130, 267–276. [Google Scholar] [CrossRef]
- Aquilina, J.A.; Watt, S.J. The N-terminal domain of αB-crystallin is protected from proteolysis by bound substrate. Biochem. Biophys. Res. Commun. 2007, 353, 1115–1120. [Google Scholar] [CrossRef]
- Sharma, K.K.; Kaur, H.; Kester, K. Functional elements in molecular chaperone alpha-crystallin: Identification of binding sites in alpha B-crystallin. Biochem. Biophys. Res. Commun. 1997, 239, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Koteiche, H.A.; McHaourab, H.S.; Stewart, P.L. Cryoelectron microscopy and EPR analysis of engineered symmetric and polydisperse Hsp16.5 assemblies reveals determinants of polydispersity and substrate binding. J. Biol. Chem. 2006, 281, 40420–40428. [Google Scholar] [CrossRef] [Green Version]
- Mishra, S.; Chandler, S.A.; Williams, D.; Claxton, D.P.; Koteiche, H.A.; Stewart, P.L.; Benesch, J.L.P.; McHaourab, H.S. Engineering of a Polydisperse Small Heat-Shock Protein Reveals Conserved Motifs of Oligomer Plasticity. Structure 2018, 26, 1116–1126.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogalla, T.; Ehrnsperger, M.; Preville, X.; Kotlyarov, A.; Lutsch, G.; Ducasse, C.; Paul, C.; Wieske, M.; Arrigo, A.-P.; Buchner, J.; et al. Regulation of Hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor α by phosphorylation. J. Biol. Chem. 1999, 274, 18947–18956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garvey, M.; Griesser, S.S.; Griesser, H.J.; Thierry, B.; Nussio, M.R.; Shapter, J.G.; Ecroyd, H.; Giorgetti, S.; Bellotti, V.; Gerrard, J.A.; et al. Enhanced molecular chaperone activity of the small heat-shock protein alphaB-cystallin following covalent immobilization onto a solid-phase support. Biopolymers 2011, 95, 376–389. [Google Scholar] [CrossRef]
- Rao, C.M.; Raman, B.; Ramakrishna, T.; Rajaraman, K.; Ghosh, D.; Datta, S.; Trivedi, V.D.; Sukhaswami, M.B. Structural perturbation of alpha-crystallin and its chaperone-like activity. Int. J. Biol. Macromol. 1998, 22, 271–281. [Google Scholar] [CrossRef]
- Reddy, G.B.; Das, K.P.; Petrash, J.M.; Surewicz, W.K. Temperature-dependent chaperone activity and structural properties of human alphaA- and alphaB-crystallins. J. Biol. Chem. 2000, 275, 4565–4570. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.H.; Kim, S.W.; Lim, C.M.; Jeong, J.Y.; Piao, C.S.; Lee, J.K. alphaB-crystallin suppresses oxidative stress-induced astrocyte apoptosis by inhibiting caspase-3 activation. Neurosci. Res. 2009, 64, 355–361. [Google Scholar] [CrossRef]
- Li, R.; Reiser, G. Phosphorylation of Ser45 and Ser59 of αB-crystallin and p38/extracellular regulated kinase activity determine αB-crystallin-mediated protection of rat brain astrocytes from C2-ceramide- and staurosporine-induced cell death. J. Neurochem. 2011, 118, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Launay, N.; Tarze, A.; Vicart, P.; Lilienbaum, A. Serine 59 phosphorylation of αB-crystallin down-regulates its anti-apoptotic function by binding and sequestering Bcl-2 in breast cancer cells. J. Biol. Chem. 2010, 285, 37324–37332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamradt, M.C.; Chen, F.; Sam, S.; Cryns, V.L. The small heat shock protein alpha B-crystallin negatively regulates apoptosis during myogenic differentiation by inhibiting caspase-3 activation. J. Biol. Chem. 2002, 277, 38731–38736. [Google Scholar] [CrossRef] [Green Version]
- Stegh, A.H.; Kesari, S.; Mahoney, J.E.; Jenq, H.T.; Forloney, K.L.; Protopopov, A.; Louis, D.N.; Chin, L.; DePinho, R.A. Bcl2L12-mediated inhibition of effector caspase-3 and caspase-7 via distinct mechanisms in glioblastoma. Proc. Natl. Acad. Sci. USA 2008, 105, 10703–10708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakajima, T.; Belusko, P.B.; Walkup, R.D.; Azuma, M.; Shearer, T.R. Involvement of Egr-1 in lens epithelial cell death induced by selenite. Exp. Eye Res. 2006, 82, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.B.; Sun, Y.; Smith, D.L.; Green, B. Identification of the posttranslational modifications of bovine lens alpha B-crystallins by mass spectrometry. Protein Sci. 1992, 1, 601–608. [Google Scholar] [CrossRef] [PubMed]
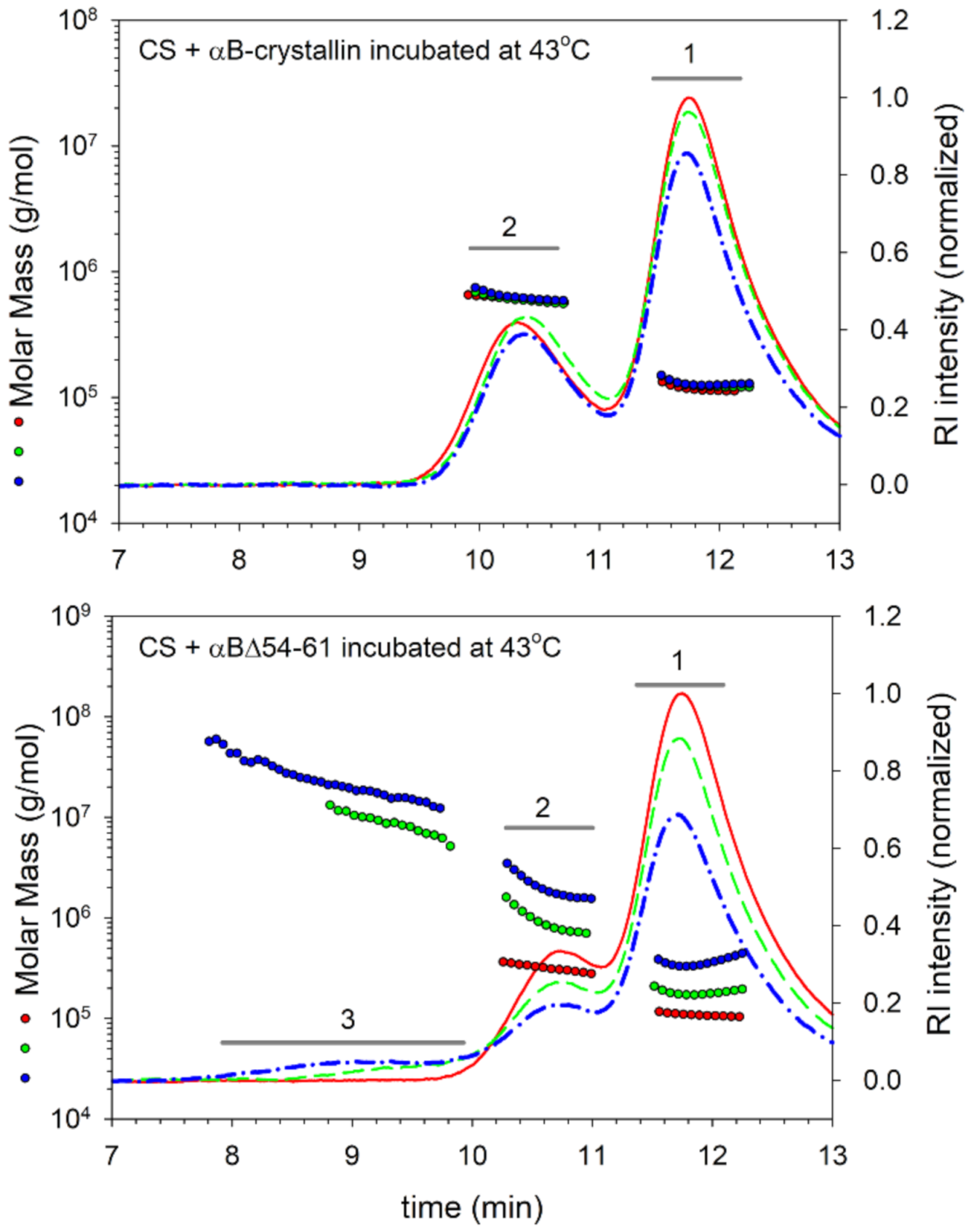
| αB-wt + CS | ||||||
| Peak 1 Citrate Synthase | Peak 2 Crystallin or Complex | Peak 3 Complex | ||||
| (11.5–12.3 min) | (9.9–10.8 min) | (7.8–9.8 min) | ||||
| Incubation time (min) | Average Mw (kDa) | Rh (nm) | Average Mw (kDa) | Rh (nm) | Average Mw (kDa) | Rh (nm) |
| 0 | 117 (± 0.5%) | 3.95 ± 0.17 | 612 (± 0.2%) | 7.23 ± 0.23 | ND | ND |
| 40 | 126 (± 0.2%) | 4.10 ± 0.17 | 588 (± 0.2%) | 7.20 ± 0.23 | ND | ND |
| 80 | 127 (± 0.2%) | 4.30 ± 0.18 | 621 (± 0.1%) | 7.39 ± 0.24 | ND | ND |
| αBΔ54–61 + CS | ||||||
| Peak 1 Citrate Synthase | Peak 2 Crystallin or Complex | Peak 3 Complex | ||||
| (11.5–12.3 min) | (10.3–11.0 min) | (7.8–9.8 min) | ||||
| Incubation time (min) | Average Mw (kDa) | Rh (nm) | Average Mw (kDa) | Rh (nm) | Average Mw (kDa) | Rh (nm) |
| 0 | 110 (± 0.5%) | 3.92 ± 0.18 | 306 (± 0.4%) | 5.3 ± 0.2 | ND | ND |
| 40 | 174 (± 0.6%) | 6.41 ± 0.25 | 784 (± 0.5%) | 12.0 ± 0.4 | 8105 (± 0.4%) | 16.9 ± 0.5 |
| 80 | 333 (± 0.5%) | 11.9 ± 0.40 | 1725 (± 0.4%) | 18.9 ± 0.5 | 21,000 (± 0.4%) | 21.9 ± 0.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahalingam, S.; Karmakar, S.; Santhoshkumar, P.; Sharma, K.K. Effect of Structural Changes Induced by Deletion of 54FLRAPSWF61 Sequence in αB-crystallin on Chaperone Function and Anti-Apoptotic Activity. Int. J. Mol. Sci. 2021, 22, 10771. https://doi.org/10.3390/ijms221910771
Mahalingam S, Karmakar S, Santhoshkumar P, Sharma KK. Effect of Structural Changes Induced by Deletion of 54FLRAPSWF61 Sequence in αB-crystallin on Chaperone Function and Anti-Apoptotic Activity. International Journal of Molecular Sciences. 2021; 22(19):10771. https://doi.org/10.3390/ijms221910771
Chicago/Turabian StyleMahalingam, Sundararajan, Srabani Karmakar, Puttur Santhoshkumar, and Krishna K. Sharma. 2021. "Effect of Structural Changes Induced by Deletion of 54FLRAPSWF61 Sequence in αB-crystallin on Chaperone Function and Anti-Apoptotic Activity" International Journal of Molecular Sciences 22, no. 19: 10771. https://doi.org/10.3390/ijms221910771
APA StyleMahalingam, S., Karmakar, S., Santhoshkumar, P., & Sharma, K. K. (2021). Effect of Structural Changes Induced by Deletion of 54FLRAPSWF61 Sequence in αB-crystallin on Chaperone Function and Anti-Apoptotic Activity. International Journal of Molecular Sciences, 22(19), 10771. https://doi.org/10.3390/ijms221910771







